Glioblastoma CUSA Fluid Protein Profiling: A Comparative Investigation of the Core and Peripheral Tumor Zones
Abstract
:Simple Summary
Abstract
1. Introduction
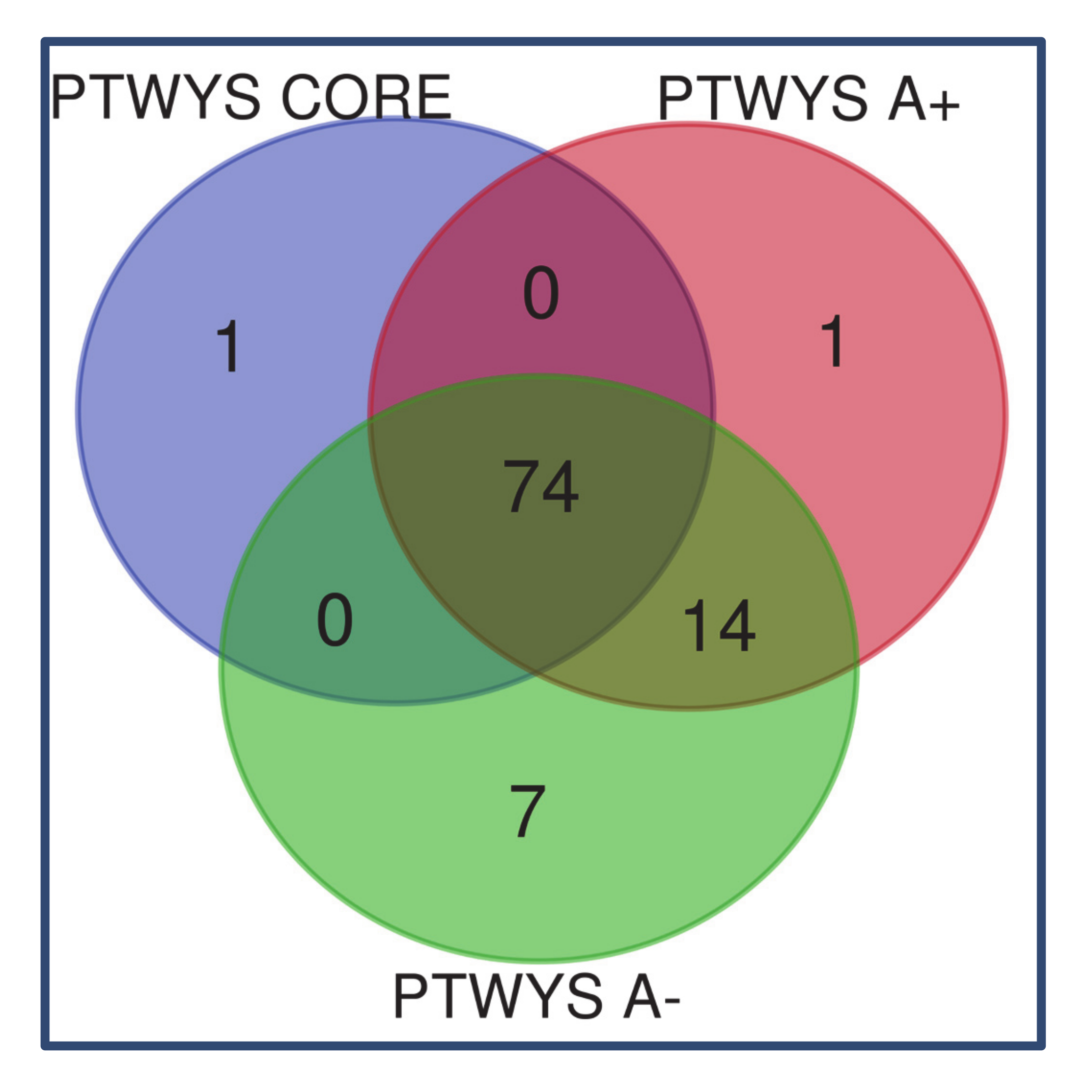
2. Results
2.1. Newly Diagnosed (ND) GBM
2.2. Recurrent (R) GBM
3. Discussion
3.1. Newly Diagnosed GBM
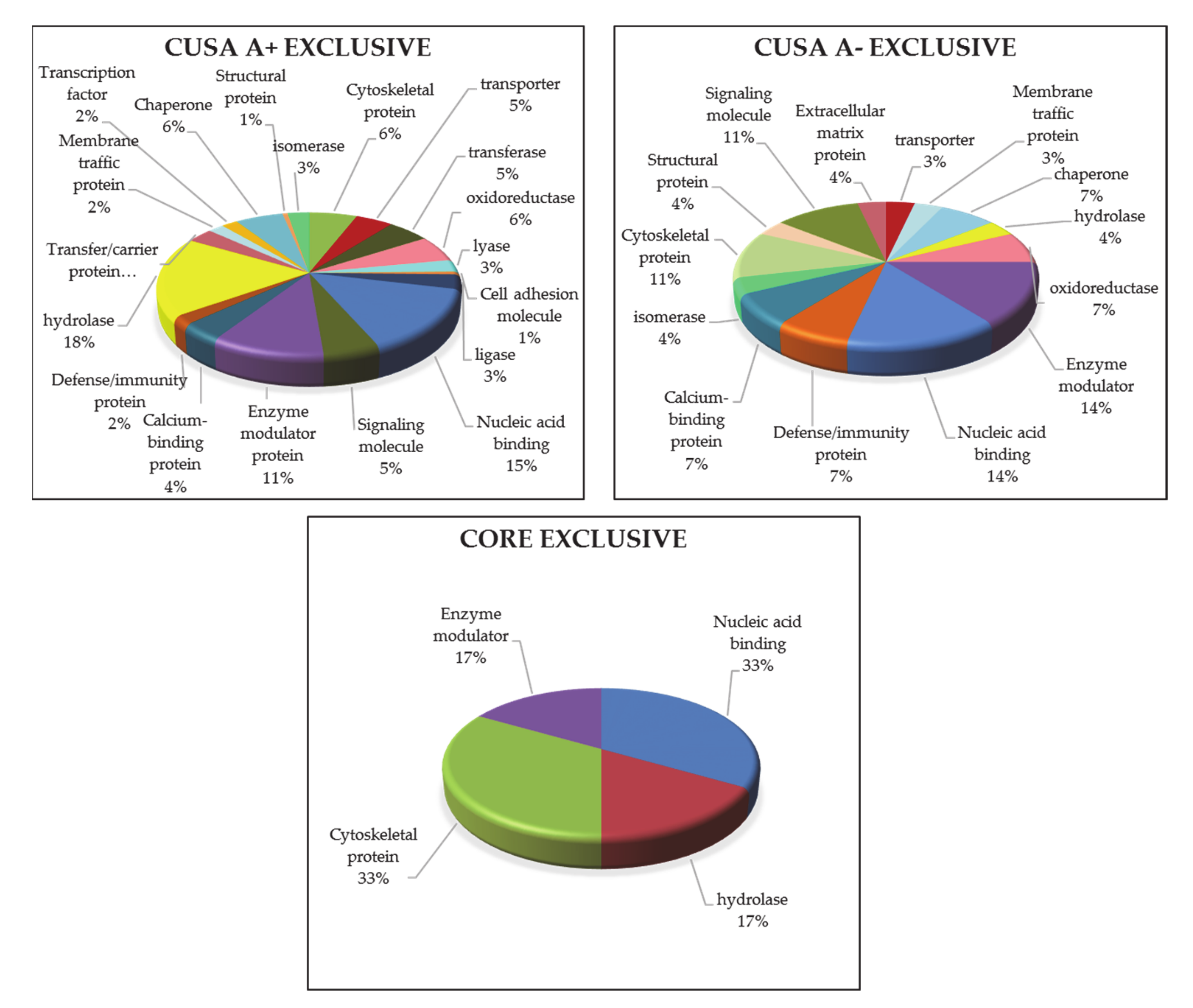
3.1.1. “Tumor Zone”
3.1.2. “Healthy Zone”
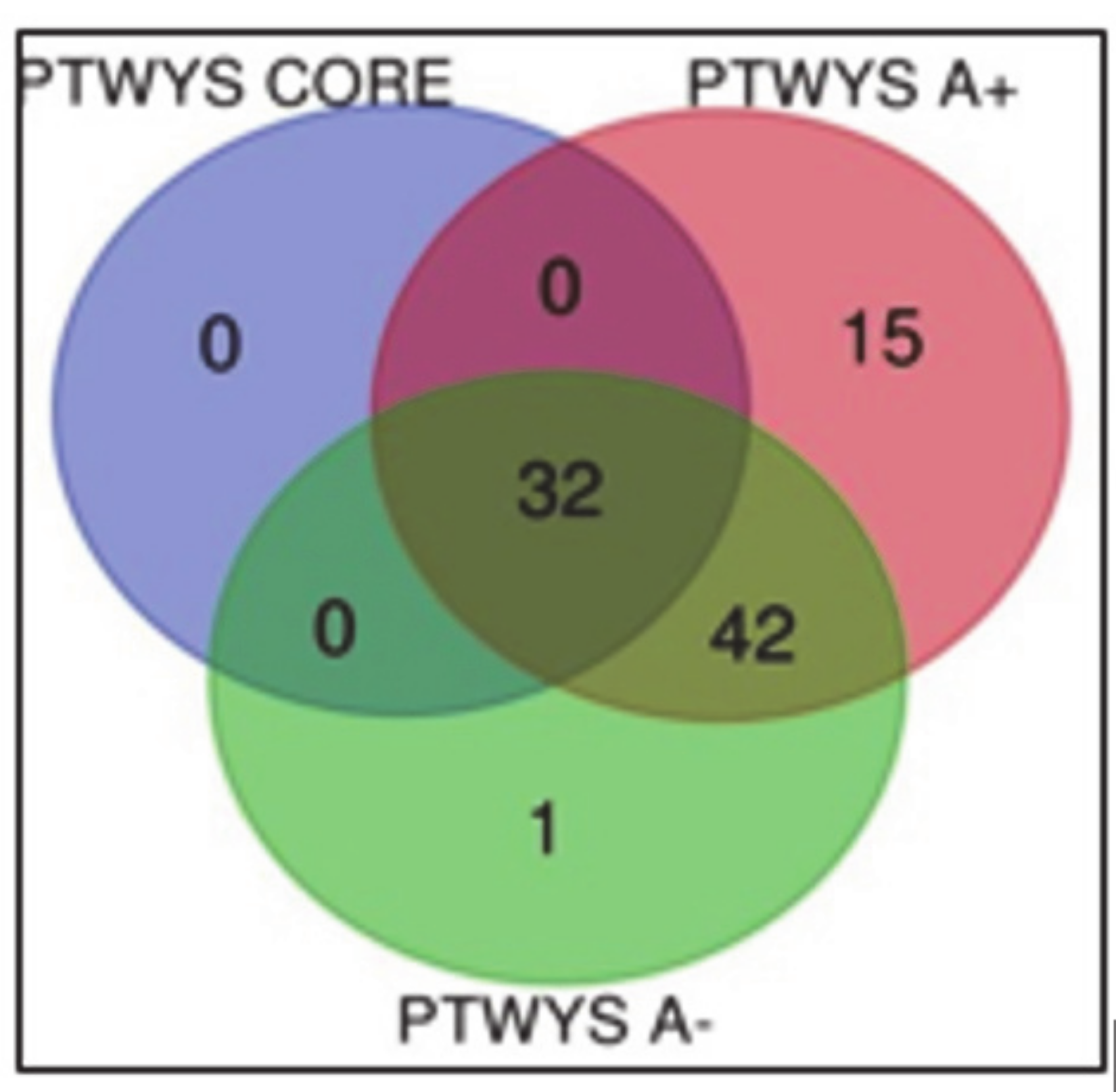
3.1.3. Overlapping A- and CUSA CORE Zones
3.1.4. Overlapping A- and A+ Zones
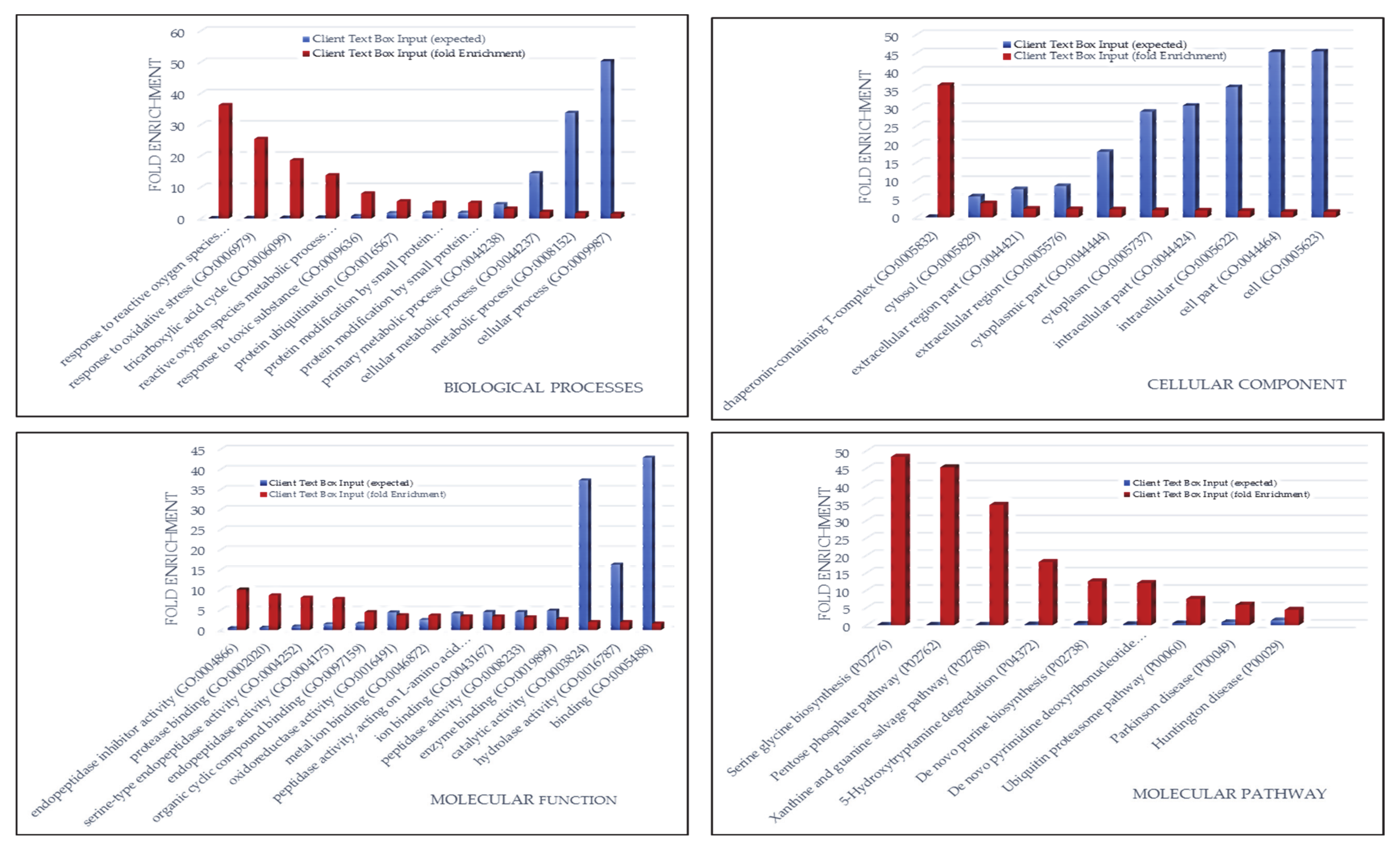
3.1.5. ND GBM Gene Ontology Classification
3.2. Recurrent GBM
3.2.1. “Tumor Zone”
3.2.2. “Healthy Zone”
3.2.3. R GBM Gene Ontology Classification
3.3. Newly Diagnosed versus Recurrent GBM
4. Materials and Methods
4.1. Patients Enrolment
4.2. Chemicals
4.3. Surgical Procedure and CUSA Aspirate Fluid Sampling
4.4. CUSA Fluid Sample Treatment
4.5. LC-MS Proteomic Analysis
4.6. Data Elaboration
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Suk, K. Proteomic Analysis of Glioma Chemoresistance. Curr. Neuropharmacol. 2012, 10, 72–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayaram, S.; Gupta, M.K.; Polisetty, R.V.; Cho, W.C.; Sirdeshmukh, R. Towards developing biomarkers for glioblastoma multiforme: A proteomics view. Expert Rev. Proteom. 2014, 11, 621–639. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.-C.; Lu, G.-X.; Zhang, H.-W.; Zhong, X.-M.; Cong, X.-L.; Xue, S.-B.; Kong, R.; Li, D.; Chang, Z.-Y.; Wang, X.-F.; et al. Proteogenomic characterization and integrative analysis of glioblastoma multiforme. Oncotarget 2017, 8, 97304–97312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silantyev, A.S.; Falzone, L.; Libra, M.; Gurina, O.I.; Kardashova, K.S.; Nikolouzakis, T.K.; Nosyrev, A.E.; Sutton, C.W.; Mitsias, P.D.; Tsatsakis, A. Current and Future Trends on Diagnosis and Prognosis of Glioblastoma: From Molecular Biology to Proteomics. Cells 2019, 8, 863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pirlog, R.; Susman, S.; Iuga, C.; Florian, I.S. Proteomic Advances in Glial Tumors through Mass Spectrometry Approaches. Medicina 2019, 55, 412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, T.; Jackson, E.; Giamas, G. Breaking through the glioblastoma micro-environment via extracellular vesicles. Oncogene 2020, 39, 4477–4490. [Google Scholar] [CrossRef]
- Yekula, A.; Yekula, A.; Muralidharan, K.; Kang, K.; Carter, B.S.; Balaj, L. Extracellular Vesicles in Glioblastoma Tumor Microenvironment. Front. Immunol. 2020, 10, 3137. [Google Scholar] [CrossRef]
- Hallal, S.; Russell, B.P.; Wei, H.; Lee, M.Y.T.; Toon, C.W.; Sy, J.; Shivalingam, B.; Buckland, M.E.; Kaufman, K.L. Extracellular Vesicles from Neurosurgical Aspirates Identifies Chaperonin Containing TCP1 Subunit 6A as a Potential Glioblastoma Biomarker with Prognostic Significance. Proteomics 2019, 19, e1800157. [Google Scholar] [CrossRef] [Green Version]
- Mallawaaratchy, D.M.; Hallal, S.; Russell, B.; Ly, L.; Ebrahimkhani, S.; Wei, H.; Christopherson, R.I.; Buckland, M.E.; Kaufman, K.L. Comprehensive proteome profiling of glioblastoma-derived extracellular vesicles identifies markers for more aggressive disease. J. Neuro Oncol. 2017, 131, 233–244. [Google Scholar] [CrossRef] [Green Version]
- Whitehead, C.A.; Kaye, A.H.; Drummond, K.J.; Widodo, S.S.; Mantamadiotis, T.; Vella, L.J.; Stylli, S.S. Extracellular vesicles and their role in glioblastoma. Crit. Rev. Clin. Lab. Sci. 2020, 57, 227–252. [Google Scholar] [CrossRef]
- Feldman, L.; Fuchshuber, P.; Jones, D.B. (Eds.) The SAGES Manual on the Fundamental Use of Surgical Energy (FUSE); Springer: New York, NY, USA, 2012. [Google Scholar]
- Day, B.W.; Stringer, B.W.; Wilson, J.; Jeffree, R.L.; Jamieson, P.R.; Ensbey, K.S.; Bruce, Z.C.; Inglis, P.; Allan, S.; Winter, C.; et al. Glioma Surgical Aspirate: A Viable Source of Tumor Tissue for Experimental Research. Cancers 2013, 5, 357–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louis, D.N.; Perry, A.; Reifenberger, G.; Von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monticelli, M.; Zeppa, P.; Zenga, F.; Altieri, R.; Mammi, M.; Bertero, L.; Castellano, I.; Cassoni, P.; Melcarne, A.; La Rocca, G.; et al. The post-surgical era of GBM: How molecular biology has impacted on our clinical management. A review. Clin. Neurol. Neurosurg. 2018, 170, 120–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ius, T.; Pignotti, F.; Della Pepa, G.M.; Bagatto, D.; Isola, M.; Battistella, C.; Gaudino, S.; Pegolo, E.; Chiesa, S.; Arcicasa, M.; et al. Glioblastoma: From volumetric analysis to molecular predictors. J. Neurosurg. Sci. 2020, 4, 153. [Google Scholar] [CrossRef]
- Kim, H.; Park, Y.J. Links between Serine Biosynthesis Pathway and Epigenetics in Cancer Metabolism. Clin. Nutr. Res. 2018, 7, 153. [Google Scholar] [CrossRef] [Green Version]
- Kathagen-Buhmann, A.; Schulte, A.; Weller, J.; Holz, M.; Herold-Mende, C.; Glass, R.; Lamszus, K. Glycolysis and the pentose phosphate pathway are differentially associated with the dichotomous regulation of glioblastoma cell migration versus proliferation. Neuro Oncol. 2016, 18, 1219–1229. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Yang, K.; Xie, Q.; Wu, Q.; Mack, S.C.; Shi, Y.; Kim, L.J.Y.; Prager, B.C.; Flavahan, W.A.; Liu, X.; et al. Purine synthesis promotes maintenance of brain tumor initiating cells in glioma. Nat. Neurosci. 2017, 20, 661–673. [Google Scholar] [CrossRef] [Green Version]
- Merzak, A.; Koochekpour, S.; Fillion, M.-P.; Fillion, G.; Pilkington, G.J. Expression of serotonin receptors in human fetal astrocytes and glioma cell lines: A possible role in glioma cell proliferation and migration. Mol. Brain Res. 1996, 41, 1–7. [Google Scholar] [CrossRef]
- Vasilev, A.; Sofi, R.; Tong, L.; Teschemacher, A.G.; Kasparov, S. In Search of a Breakthrough Therapy for Glioblastoma Multiforme. Neuroglia 2018, 1, 292–310. [Google Scholar] [CrossRef] [Green Version]
- Plun-Favreau, H.; Lewis, P.A.; Hardy, J.; Martins, L.M.; Wood, N. Cancer and Neurodegeneration: Between the Devil and the Deep Blue Sea. PLoS Genet. 2010, 6, e1001257. [Google Scholar] [CrossRef] [Green Version]
- Mooney, K.L.; Choy, W.; Sidhu, S.; Pelargos, P.; Bui, T.T.; Voth, B.; Barnette, N.; Yang, I. The role of CD44 in glioblastoma multiforme. J. Clin. Neurosci. 2016, 34, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Hegi, M.E.; Mason, W.P.; Bent, M.J.V.D.; Taphoorn, M.J.B.; Janzer, R.C.; Ludwin, S.K.; Allgeier, A.; Fisher, B.; Belanger, K.; et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009, 10, 459–466. [Google Scholar] [CrossRef]
- Pietras, A.; Katz, A.M.; Ekström, E.J.; Wee, B.; Halliday, J.J.; Pitter, K.L.; Werbeck, J.L.; Amankulor, N.M.; Huse, J.T.; Holland, E.C. Osteopontin-CD44 Signaling in the Glioma Perivascular Niche Enhances Cancer Stem Cell Phenotypes and Promotes Aggressive Tumor Growth. Cell Stem Cell 2014, 14, 357–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaillant, B.D.; Bhat, K.; Sulman, E.P.; Balasubramaniyan, V.; Wang, S.; Aldape, K.D.; Colman, H. CD44 as a prognostic and predictive marker for GBM. J. Clin. Oncol. 2011, 29, 2049. [Google Scholar] [CrossRef]
- Lim, S.; Kim, D.; Ju, S.; Shin, S.; Cho, I.-J.; Park, S.; Grailhe, R.; Lee, C.; Kim, Y.K. Glioblastoma-secreted soluble CD44 activates tau pathology in the brain. Exp. Mol. Med. 2018, 50, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, J.; Marisetty, A.; Schrand, B.; Gabrusiewicz, K.; Hashimoto, Y.; Ott, M.; Grami, Z.; Kong, L.-Y.; Ling, X.; Caruso, H.G.; et al. Osteopontin mediates glioblastoma-associated macrophage infiltration and is a potential therapeutic target. J. Clin. Investig. 2018, 129, 137–149. [Google Scholar] [CrossRef] [PubMed]
- D’Alessio, A.; Proietti, G.; Sica, G.; Scicchitano, B.M. Pathological and Molecular Features of Glioblastoma and Its Peritumoral Tissue. Cancers 2019, 11, 469. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Liu, Q.; Liao, Q.; Zhao, Y. CD59: A promising target for tumor immunotherapy. Futur. Oncol. 2018, 14, 781–791. [Google Scholar] [CrossRef]
- Zheng, P.-P.; Sieuwerts, A.M.; Luider, T.M.; Van Der Weiden, M.; Sillevis-Smitt, P.A.; Kros, J.M. Differential Expression of Splicing Variants of the Human Caldesmon Gene (CALD1) in Glioma Neovascularization versus Normal Brain Microvasculature. Am. J. Pathol. 2004, 164, 2217–2228. [Google Scholar] [CrossRef] [Green Version]
- Uhlén, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef] [Green Version]
- Pontén, F.; Jirström, K.; Uhlen, M. The Human Protein Atlas—A tool for pathology. J. Pathol. 2008, 216, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Data Available from v19.proteinatlas.org. Available online: https://www.proteinatlas.org/ENSG00000137285-TUBB2B/pathology (accessed on 13 July 2019).
- Masiulionytė, B.; Valiulyte, I.; Tamašauskas, A.; Skiriutė, D. Metallothionein Genes are Highly Expressed in Malignant Astrocytomas and Associated with Patient Survival. Sci. Rep. 2019, 9, 5406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Si, M.; Lang, J. The roles of metallothioneins in carcinogenesis. J. Hematol. Oncol. 2018, 11, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Rupp, T.; Langlois, B.; Koczorowska, M.M.; Radwanska, A.; Sun, Z.; Hussenet, T.; Lefebvre, O.; Murdamoothoo, D.; Arnold, C.; Klein, A.; et al. Tenascin-C Orchestrates Glioblastoma Angiogenesis by Modulation of Pro- and Anti-angiogenic Signaling. Cell Rep. 2016, 17, 2607–2619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morales, F.C.; Molina, J.R.; Hayashi, Y.; Georgescu, M.-M. Overexpression of ezrin inactivates NF2 tumor suppressor in glioblastoma. Neuro Oncol. 2010, 12, 528–539. [Google Scholar] [CrossRef] [Green Version]
- Kotb, A.; Hyndman, M.E.; Patel, T.R. The role of zyxin in regulation of malignancies. Heliyon 2018, 4, e00695. [Google Scholar] [CrossRef] [Green Version]
- Wen, X.-M.; Luo, T.; Jiang, Y.; Wang, L.-H.; Luo, Y.; Chen, Q.; Yang, K.; Yuan, Y.; Luo, C.; Zhang, X.; et al. Zyxin (ZYX) promotes invasion and acts as a biomarker for aggressive phenotypes of human glioblastoma multiforme. Lab. Investig. 2020, 100, 812–823. [Google Scholar] [CrossRef]
- Data Available from v19.proteinatlas.org. Available online: https://www.proteinatlas.org/ENSG00000152795-HNRNPDL/pathology#gene_information (accessed on 13 July 2019).
- Data Available from v19.proteinatlas.org. Available online: https://www.proteinatlas.org/search/protein_class:Cancer-related+genes (accessed on 13 July 2019).
- Data Available from v19.proteinatlas.org. Available online: https://www.proteinatlas.org/ENSG00000117632-STMN1/pathology (accessed on 13 July 2019).
- Data Available from v19.proteinatlas.org. Available online: https://www.proteinatlas.org/ENSG00000148672-GLUD1/pathology (accessed on 13 July 2019).
- Kutwin, M.; Sawosz, E.; Jaworski, S.; Wierzbicki, M.; Strojny, B.; Grodzik, M.; Chwalibog, A. Assessment of the proliferation status of glioblastoma cell and tumour tissue after nanoplatinum treatment. PLoS ONE 2017, 12, e0178277. [Google Scholar] [CrossRef] [Green Version]
- Data Available from v19.proteinatlas.org. Available online: https://www.proteinatlas.org/ENSG00000008056-SYN1/pathology#gene_information (accessed on 13 July 2019).
- Cesca, F.; Baldelli, P.; Valtorta, F.; Benfenati, F. The synapsins: Key actors of synapse function and plasticity. Prog. Neurobiol. 2010, 91, 313–348. [Google Scholar] [CrossRef]
- Conti, F. Fisiologia Medica 1, 2nd ed.; Edi. Ermes: Milano, Italy, 2010; Volume 1–2. [Google Scholar]
- Tanji, K.; Mori, F.; Nakajo, S.; Imaizumi, T.; Yoshida, H.; Hirabayashi, T.; Yoshimoto, M.; Satoh, K.; Takahashi, H.; Wakabayashi, K. Expression of β-synuclein in normal human astrocytes. NeuroReport 2001, 12, 2845–2848. [Google Scholar] [CrossRef]
- Data Available from v19.proteinatlas.org. Available online: https://www.proteinatlas.org/ENSG00000132639-SNAP25/pathology (accessed on 13 July 2019).
- Strzalka, W.; Ziemienowicz, A. Proliferating cell nuclear antigen (PCNA): A key factor in DNA replication and cell cycle regulation. Ann. Bot. 2010, 107, 1127–1140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maga, G. Proliferating cell nuclear antigen (PCNA): A dancer with many partners. J. Cell Sci. 2003, 116, 3051–3060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Data Available from v19.proteinatlas.org. Available online: https://www.proteinatlas.org/ENSG00000178802-MPI/pathology (accessed on 13 July 2019).
- Mi, H.; Huang, X.; Muruganujan, A.; Tang, H.; Mills, C.; Kang, D.; Thomas, P.D. PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017, 45, D183–D189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Accession P00019. Available online: http://www.pantherdb.org/pathway/pathDetail.do?clsAccession=P00019 (accessed on 9 July 2019).
- Accession P00047. Available online: http://www.pantherdb.org/pathway/pathDetail.do?clsAccession=P00047 (accessed on 9 July 2019).
- Popescu, A.M.; Alexandru, O.; Brindusa, C.; Purcaru, S.O.; Tache, D.E.; Tataranu, L.G.; Taisescu, C.; Dricu, A. Targeting the VEGF and PDGF signaling pathway in glioblastoma treatment. Int. J. Clin. Exp. Pathol. 2015, 8, 7825–7837. [Google Scholar]
- Della Pepa, G.M.; Menna, G.; Ius, T.; Di Bonaventura, R.; Altieri, R.; Marchese, E.; Olivi, A.; Sabatino, G.; La Rocca, G. Contrast enhanced ultrasound (CEUS) applications in neurosurgical and neurological settings—New scenarios for brain and spinal cord ultrasonography. A systematic review. Clin. Neurol. Neurosurg. 2020, 198, 106105. [Google Scholar] [CrossRef]
- Altieri, R.; Melcarne, A.; Di Perna, G.; Specchia, F.M.C.; Fronda, C.; La Rocca, G.; Cofano, F.; Sabatino, G.; Della Pepa, G.M.; Olivi, A.; et al. Intra-Operative Ultrasound: Tips and Tricks for Making the Most in Neurosurgery. Surg. Technol. Int. 2018, 33, 353–360. [Google Scholar]
- Della Pepa, G.M.; Ius, T.; La Rocca, G.; Gaudino, S.; Isola, M.; Pignotti, F.; Rapisarda, A.; Mazzucchi, E.; Giordano, C.; Dragonetti, V.; et al. 5-Aminolevulinic Acid and Contrast-Enhanced Ultrasound: The Combination of the Two Techniques to Optimize the Extent of Resection in Glioblastoma Surgery. Neurosurgery 2020, 86, E529–E540. [Google Scholar] [CrossRef]
- Della Pepa, G.M.; Sabatino, G.; La Rocca, G. “Enhancing Vision” in High Grade Glioma Surgery: A Feasible Integrated 5-ALA + CEUS Protocol to Improve Radicality. World Neurosurg. 2019, 129, 401–403. [Google Scholar] [CrossRef]
- La Rocca, G.; Della Pepa, G.M.; Menna, G.; Altieri, R.; Ius, T.; Rapisarda, A.; Olivi, A.; Sabatino, G. State of the art of fluorescence guided techniques in neurosurgery. J. Neurosurg. Sci. 2019, 63, 619–624. [Google Scholar] [CrossRef]
- Manini, I.; Caponnetto, F.; Dalla, E.; Ius, T.; Della Pepa, G.M.; Pegolo, E.; Bartolini, A.; La Rocca, G.; Menna, G.; Di Loreto, C.; et al. Heterogeneity Matters: Different Regions of Glioblastoma Are Characterized by Distinctive Tumor-Supporting Pathways. Cancers 2020, 12, 2960. [Google Scholar] [CrossRef]
- Deutsch, E.W.; Overall, C.M.; van Eyk, J.E.; Baker, M.S.; Paik, Y.-K.; Weintraub, S.T.; Lane, L.; Martens, L.; Vandenbrouck, Y.; Kusebauch, U.; et al. Human Proteome Project Mass Spectrometry Data Interpretation Guidelines 2.1. J. Proteome Res. 2016, 15, 3961–3970. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, A.; Sidiropoulos, K.; Viteri, G.; Forner-Martinez, O.; Marin-Garcia, P.; Arnau, V.; D’Eustachio, P.; Stein, L.; Hermjakob, H. Reactome pathway analysis: A high-performance in-memory approach. BMC Bioinform. 2017, 18, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–Protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
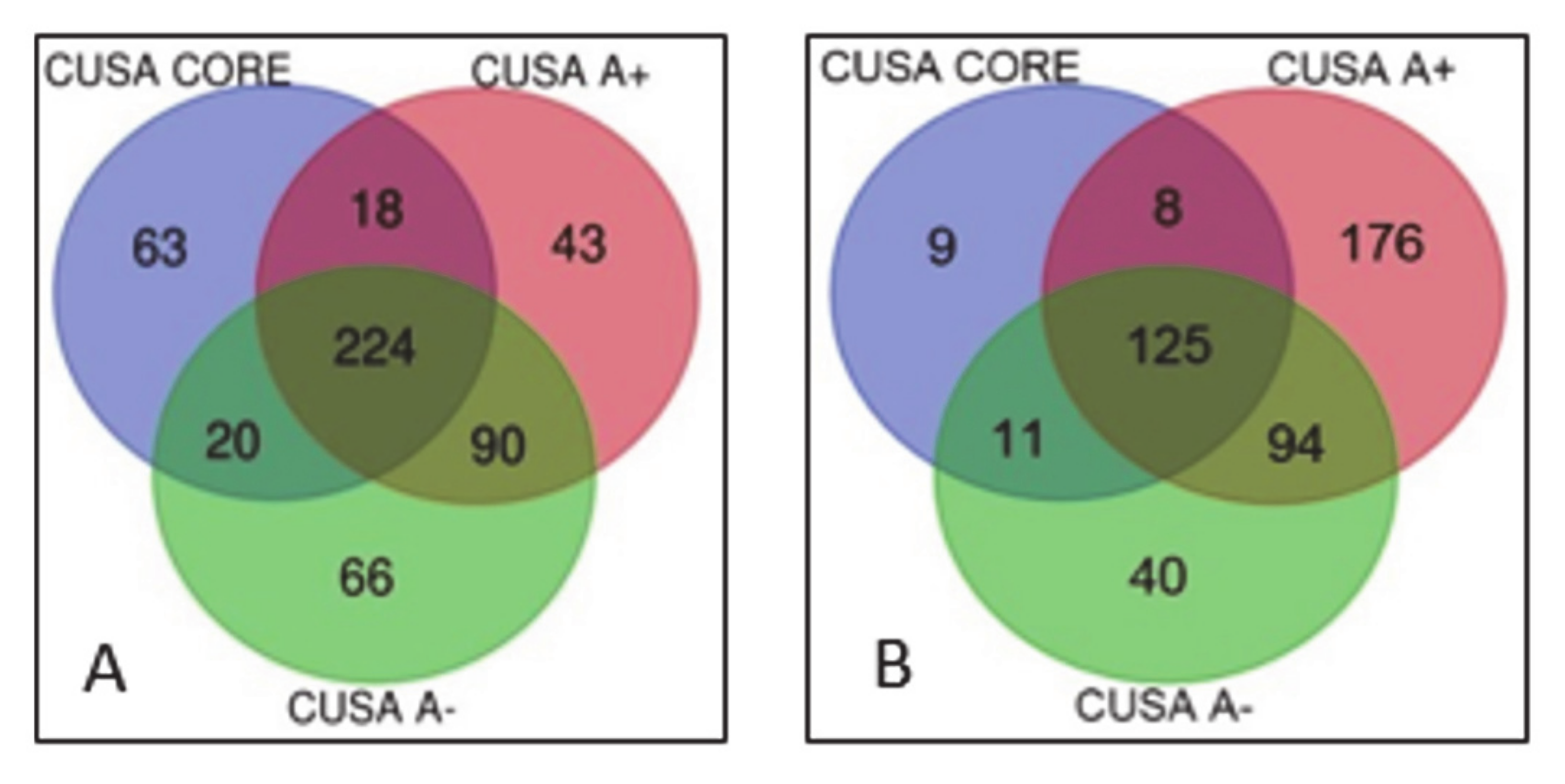
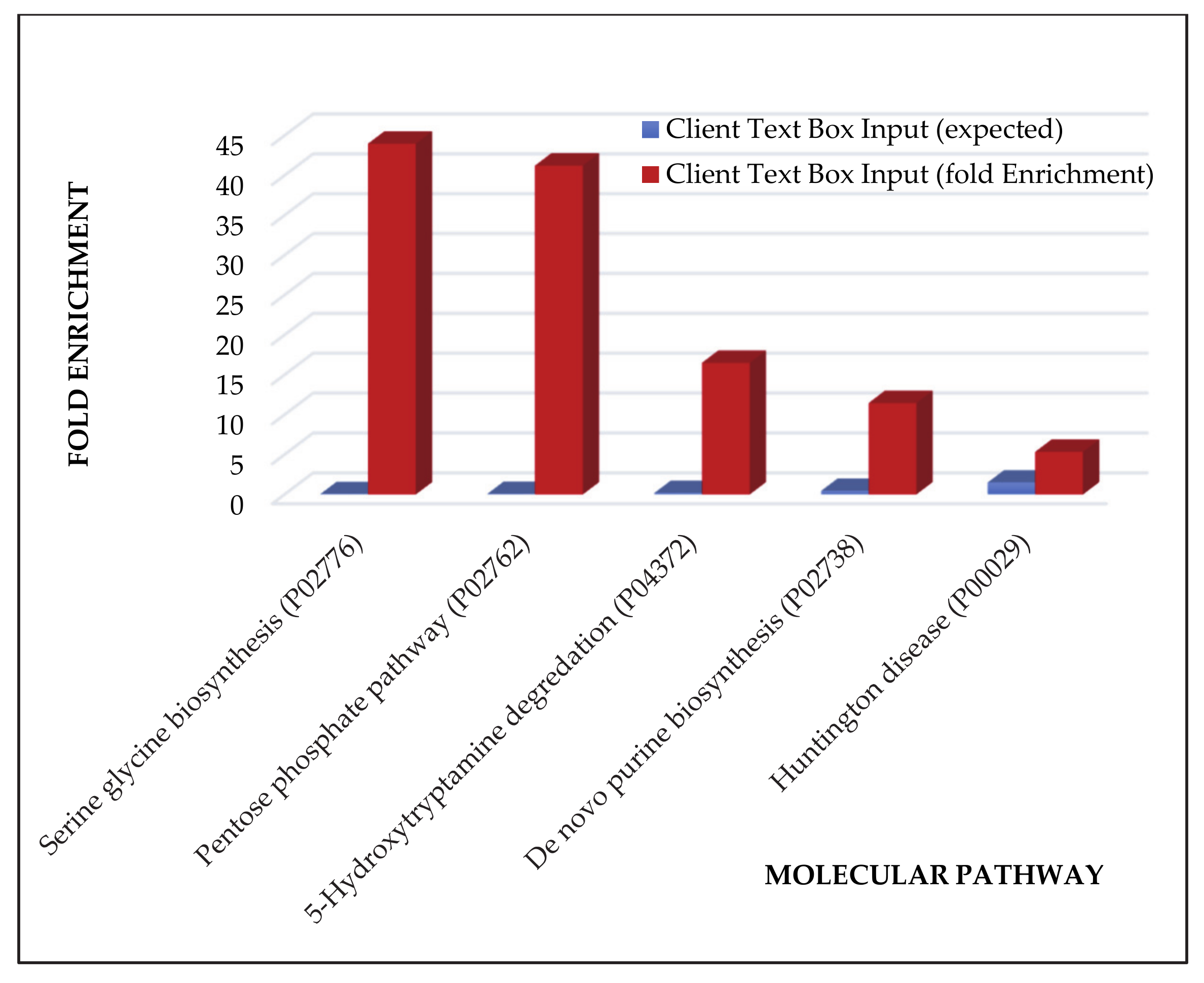
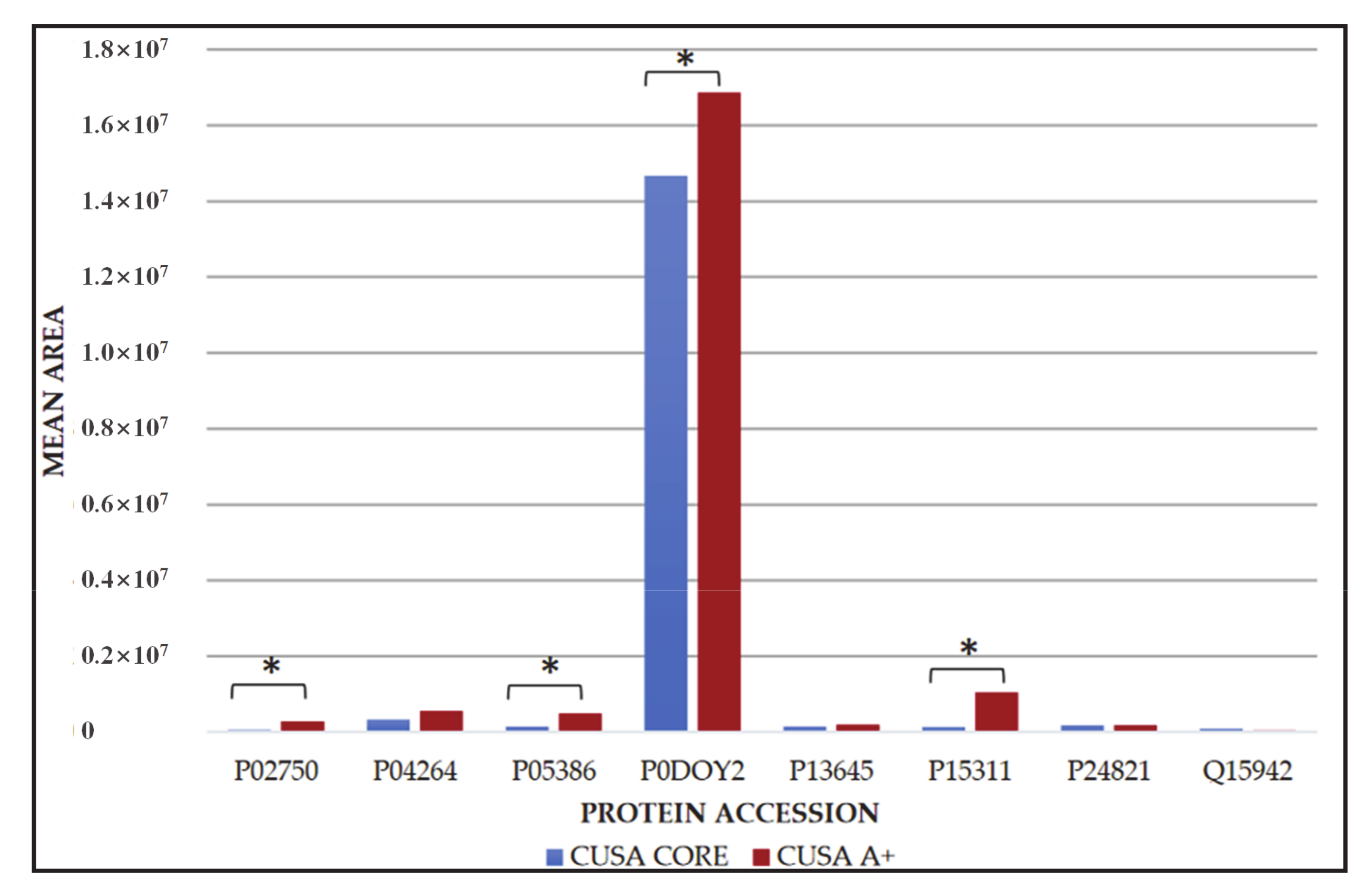



| Pathway Name | Entities Found | Entities Total | Entities Ratio | Entities p-Value | Entities FDR | Reactions Found | Reactions Total | Reactions Ratio |
|---|---|---|---|---|---|---|---|---|
| Innate Immune System | 51 | 1187 | 0.103940455 | 5.27 × 10−10 | 5.64 × 10−7 | 131 | 662 | 0.052414885 |
| Neutrophil degranulation | 30 | 480 | 0.042031524 | 1.13 × 10−9 | 6.06 × 10−7 | 10 | 10 | 7.92 × 10−4 |
| Platelet degranulation | 14 | 128 | 0.011208406 | 6.47 × 10−8 | 2.30 × 10−5 | 6 | 11 | 8.71 × 10−4 |
| Response to elevated platelet cytosolic Ca2+ | 14 | 133 | 0.011646235 | 1.03 × 10−7 | 2.74 × 10−5 | 6 | 14 | 0.001108472 |
| Detoxification of Reactive Oxygen Species | 8 | 39 | 0.003415061 | 4.89 × 10−7 | 1.05 × 10−4 | 11 | 32 | 0.00253365 |
| Cellular responses to external stimuli | 27 | 579 | 0.050700525 | 2.63 × 10−6 | 4.69 × 10−4 | 50 | 255 | 0.020190024 |
| Cellular responses to stress | 26 | 565 | 0.049474606 | 5.20 × 10−6 | 7.90 × 10−4 | 47 | 224 | 0.01773555 |
| Immune System | 68 | 2398 | 0.209982487 | 6.58 × 10−6 | 8.75 × 10−4 | 254 | 1598 | 0.126524149 |
| Platelet activation, signaling and aggregation | 16 | 265 | 0.023204904 | 1.66 × 10−5 | 0.001955007 | 9 | 115 | 0.009105305 |
| Hemostasis | 28 | 726 | 0.06357268 | 5.36 × 10−5 | 0.005219396 | 27 | 332 | 0.026286619 |
| Pyruvate metabolism and Citric Acid (TCA) cycle | 7 | 55 | 0.004816112 | 5.38 × 10−5 | 0.005219396 | 9 | 36 | 0.002850356 |
| Smooth Muscle Contraction | 6 | 39 | 0.003415061 | 6.65 × 10−5 | 0.005915444 | 5 | 11 | 8.71 × 10−4 |
| Formation of tubulin folding intermediates by CCT/TriC | 5 | 26 | 0.002276708 | 9.76 × 10−5 | 0.008006187 | 2 | 2 | 1.58 × 10−4 |
| Prefoldin mediated transfer of substrate to CCT/TriC | 5 | 28 | 0.002451839 | 1.38 × 10−4 | 0.010455726 | 2 | 2 | 1.58 × 10−4 |
| Metabolism | 58 | 2142 | 0.187565674 | 1.63 × 10−4 | 0.011603817 | 89 | 2001 | 0.158432304 |
| The citric acid (TCA) cycle and respiratory electron transport | 11 | 176 | 0.015411559 | 2.66 × 10−4 | 0.017547815 | 11 | 64 | 0.0050673 |
| Cooperation of Prefoldin and TriC/CCT in actin and tubulin folding | 5 | 33 | 0.002889667 | 2.92 × 10−4 | 0.018103534 | 6 | 6 | 4.75 × 10−4 |
| Interleukin-12 family signaling | 6 | 56 | 0.004903678 | 4.58 × 10−4 | 0.027046947 | 12 | 114 | 0.009026128 |
| Protein ubiquitination | 7 | 80 | 0.007005254 | 5.18 × 10−4 | 0.029024722 | 30 | 32 | 0.00253365 |
| Selective autophagy | 7 | 82 | 0.007180385 | 5.99 × 10−4 | 0.030593862 | 23 | 48 | 0.003800475 |
| Citric acid cycle (TCA cycle) | 4 | 22 | 0.001926445 | 6.12 × 10−4 | 0.030593862 | 5 | 17 | 0.001346002 |
| Folding of actin by CCT/TriC | 3 | 10 | 8.76 × 10−4 | 7.31 × 10−4 | 0.035103663 | 2 | 2 | 1.58 × 10−4 |
| RHO GTPases activate PKNs | 6 | 63 | 0.005516637 | 8.42 × 10−4 | 0.038746044 | 12 | 20 | 0.001583531 |
| TP53 Regulates Metabolic Genes | 7 | 88 | 0.007705779 | 9.01 × 10−4 | 0.039649516 | 5 | 34 | 0.002692003 |
| Aggrephagy | 5 | 44 | 0.00385289 | 0.001057731 | 0.044424721 | 13 | 15 | 0.001187648 |
| Pathway Name | Entities Found | Entities Total | Entities Ratio | Entities p-Value | Entities FDR | Reactions Found | Reactions Total | Reactions Ratio |
|---|---|---|---|---|---|---|---|---|
| Cellular responses to external stimuli | 26 | 579 | 0.051 | 9.22 × 10−10 | 8.19 × 10−7 | 50 | 255 | 0.020190024 |
| Cellular responses to stress | 25 | 565 | 0.049 | 2.76 × 10−9 | 1.23 × 10−6 | 47 | 224 | 0.01773555 |
| Eukaryotic Translation Elongation | 8 | 95 | 0.008 | 1.21 × 10−5 | 0.003568961 | 6 | 9 | 7.13 × 10−4 |
| L13a-mediated translational silencing of Ceruloplasmin expression | 8 | 112 | 0.01 | 3.85 × 10−5 | 0.006094382 | 2 | 3 | 2.38 × 10−4 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | 8 | 113 | 0.01 | 4.10 × 10−5 | 0.006094382 | 3 | 3 | 2.38 × 10−4 |
| HSP90 chaperone cycle for steroid hormone receptors (SHR) | 6 | 57 | 0.005 | 4.57 × 10−5 | 0.006094382 | 7 | 12 | 9.50 × 10−4 |
| Metabolism of RNA | 20 | 675 | 0.059 | 5.23 × 10−5 | 0.006094382 | 40 | 187 | 0.014806017 |
| Cap-dependent Translation Initiation | 8 | 120 | 0.011 | 6.22 × 10−5 | 0.006094382 | 17 | 18 | 0.001425178 |
| Eukaryotic Translation Initiation | 8 | 120 | 0.011 | 6.22 × 10−5 | 0.006094382 | 19 | 21 | 0.001662708 |
| Regulation of Complement cascade | 8 | 135 | 0.012 | 1.39 × 10−4 | 0.011833692 | 7 | 42 | 0.003325416 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | 7 | 102 | 0.009 | 1.52 × 10−4 | 0.011833692 | 6 | 16 | 0.001266825 |
| Signaling by the B Cell Receptor (BCR) | 9 | 176 | 0.015 | 1.60 × 10−4 | 0.011833692 | 18 | 43 | 0.003404592 |
| Complement cascade | 8 | 146 | 0.013 | 2.36 × 10−4 | 0.016016904 | 16 | 71 | 0.005621536 |
| COPI-independent Golgi-to-ER retrograde traffic | 5 | 53 | 0.005 | 3.30 × 10−4 | 0.019025205 | 2 | 7 | 5.54 × 10−4 |
| Nonsense-Mediated Decay (NMD) | 7 | 117 | 0.01 | 3.46 × 10−4 | 0.019025205 | 6 | 6 | 4.75 × 10−4 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | 7 | 117 | 0.01 | 3.46 × 10−4 | 0.019025205 | 5 | 5 | 3.96 × 10−4 |
| Hemostasis | 19 | 726 | 0.064 | 3.98 × 10−4 | 0.019131188 | 13 | 332 | 0.026286619 |
| Advanced glycosylation endproduct receptor signaling | 3 | 13 | 0.001 | 4.32 × 10−4 | 0.019131188 | 3 | 4 | 3.17 × 10−4 |
| Apoptosis induced DNA fragmentation | 3 | 13 | 0.001 | 4.32 × 10−4 | 0.019131188 | 5 | 12 | 9.50 × 10−4 |
| Translation | 11 | 294 | 0.026 | 4.42 × 10−4 | 0.019131188 | 34 | 99 | 0.00783848 |
| Innate Immune System | 26 | 1187 | 0.104 | 5.06 × 10−4 | 0.019131188 | 125 | 662 | 0.052414885 |
| Peptide chain elongation | 6 | 90 | 0.008 | 5.27 × 10−4 | 0.019131188 | 4 | 5 | 3.96 × 10−4 |
| Translation initiation complex formation | 5 | 59 | 0.005 | 5.35 × 10−4 | 0.019131188 | 2 | 2 | 1.58 × 10−4 |
| Ribosomal scanning and start codon recognition | 5 | 59 | 0.005 | 5.35 × 10−4 | 0.019131188 | 2 | 2 | 1.58 × 10−4 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | 5 | 60 | 0.005 | 5.77 × 10−4 | 0.019131188 | 5 | 6 | 4.75 × 10−4 |
| Uniprot Accession | Protein Name |
|---|---|
| P02656 | Apolipoprotein CIII |
| P16949 | Stathmin |
| O75347 | Tubulin-specific Chaperone A |
| P23527 | Histone H2B type 1-O |
| O14979 | Heterogeneous nuclear ribonucleoprotein Dlike |
| P25713 | Metallothionein-3 * |
| Q16799 | Reticulon-1 |
| P68363 | Tubulin -1B chain |
| P68032 | Actin, α cardiac muscle 1 * |
| P0DOY3 | Immunoglobulin Lambda Constant 3 * |
| Q13526 | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 * |
| Uniprot Accession | Protein Name |
|---|---|
| P00367 | Glutamate dehydrogenase1, mytochondrial |
| P05164 | Myeloperoxidase |
| P22177 | Proliferating cell nuclear antigen |
| P17600 | Synapsin-1 |
| P60880 | Synaptosomal-associated protein 25 |
| Q16143 | Beta-synuclein |
| Pathways CUSA A- Exclusive | Pathways CUSA A+ Exclusive |
|---|---|
| TGFβ signaling pathway | Adenine and hypoxanthine salvage pathway |
| 5-Hydroxytryptamine degredation | |
| Vitamin B6 metabolism | |
| Toll receptor signaling pathway | |
| p53 pathway by glucose deprivation | |
| Heme biosynthesis | |
| Axon guidance mediated by Slit/Robo | |
| Gamma-aminobutyric acid synthesis | |
| Xanthine and guanine salvage pathway | |
| Serine glycine biosynthesis | |
| Pyridoxal-5-phosphate biosynthesis | |
| Axon guidance mediated by netrin | |
| Aminobutyrate degradation | |
| p53 pathway feedback loops 2 | |
| Ras Pathway |
| CUSA CORE Exclusive Pathway | CUSA A+ Exclusive Pathway | CUSA A- Exclusive Pathways |
|---|---|---|
| TGFβ signaling pathway | Mannose metabolism | Pyridoxal phosphate salvage pathway |
| Insulin/IGF pathway-protein kinase B signaling cascade | ||
| Interleukin signaling Pathway | ||
| Gamma-aminobutyric acid synthesis | ||
| Endothelin signaling pathway | ||
| PDGF signaling pathway | ||
| Aminobutyrate degradation |
| (A) Exclusive CORE Proteins (ND vs. R, Figure 12) | (B) Exclusive CORE Proteins (ND CORE vs ND A+ vs. ND A-) (from Table S1) |
|---|---|
| Q9BVA1: Tubulin beta 2B chain | Q9BVA1: Tubulin beta 2B chain |
| P13987: CD59 glycoprotein | P13987: CD59 glycoprotein |
| P10451: Osteopontin | P10451:Osteopontin |
| P07951:Tropomyosin beta chain | P07951:Tropomyosin beta chain |
| Q14315: Filamin-C | P10412:Histone H1.4 |
| P04264: Keratin, type II cytoskeletal 1 | Q05682:Caldesmon |
| P0DOY2: Immunoglobulin lambda constant 2 | P16070:CD44 antigen |
| P13645: Keratin, type I cytoskeletal 10 | Q92945:Far upstream element-binding protein 2 |
| P59665: Neutrophil defensin 1 | P02795:Metallothionein-2 |
| Patient | Age | Location of Tumor | State of GBM Newly Diagnosed (ND) Recurrent (R) |
|---|---|---|---|
| PP1 | 57 | Left parieto-occipital | ND |
| PP2 | 74 | Left parietal | ND |
| PP3 | 52 | Left parietal | ND |
| PP4 | 50 | Left temporal | R |
| PP5 | 56 | Right parietal | R |
| PP6 | 40 | Right frontal | R |
| PP7 | 75 | Right parieto-occipital | ND |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
La Rocca, G.; Simboli, G.A.; Vincenzoni, F.; Rossetti, D.V.; Urbani, A.; Ius, T.; Della Pepa, G.M.; Olivi, A.; Sabatino, G.; Desiderio, C. Glioblastoma CUSA Fluid Protein Profiling: A Comparative Investigation of the Core and Peripheral Tumor Zones. Cancers 2021, 13, 30. https://doi.org/10.3390/cancers13010030
La Rocca G, Simboli GA, Vincenzoni F, Rossetti DV, Urbani A, Ius T, Della Pepa GM, Olivi A, Sabatino G, Desiderio C. Glioblastoma CUSA Fluid Protein Profiling: A Comparative Investigation of the Core and Peripheral Tumor Zones. Cancers. 2021; 13(1):30. https://doi.org/10.3390/cancers13010030
Chicago/Turabian StyleLa Rocca, Giuseppe, Giorgia Antonia Simboli, Federica Vincenzoni, Diana Valeria Rossetti, Andrea Urbani, Tamara Ius, Giuseppe Maria Della Pepa, Alessandro Olivi, Giovanni Sabatino, and Claudia Desiderio. 2021. "Glioblastoma CUSA Fluid Protein Profiling: A Comparative Investigation of the Core and Peripheral Tumor Zones" Cancers 13, no. 1: 30. https://doi.org/10.3390/cancers13010030
APA StyleLa Rocca, G., Simboli, G. A., Vincenzoni, F., Rossetti, D. V., Urbani, A., Ius, T., Della Pepa, G. M., Olivi, A., Sabatino, G., & Desiderio, C. (2021). Glioblastoma CUSA Fluid Protein Profiling: A Comparative Investigation of the Core and Peripheral Tumor Zones. Cancers, 13(1), 30. https://doi.org/10.3390/cancers13010030








