Gene Expression Levels of the Prolyl Hydroxylase Domain Proteins PHD1 and PHD2 but Not PHD3 Are Decreased in Primary Tumours and Correlate with Poor Prognosis of Patients with Surgically Resected Non-Small-Cell Lung Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Collection of Samples
2.2. RNA Isolation and cDNA Synthesis
2.3. RT-qPCR
2.4. A549 Cell Culture
2.5. Statistics
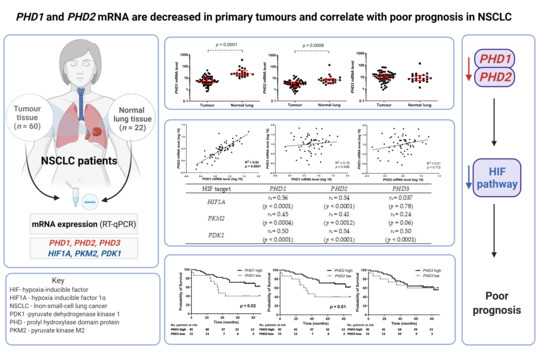
3. Results
3.1. Patient Characteristics
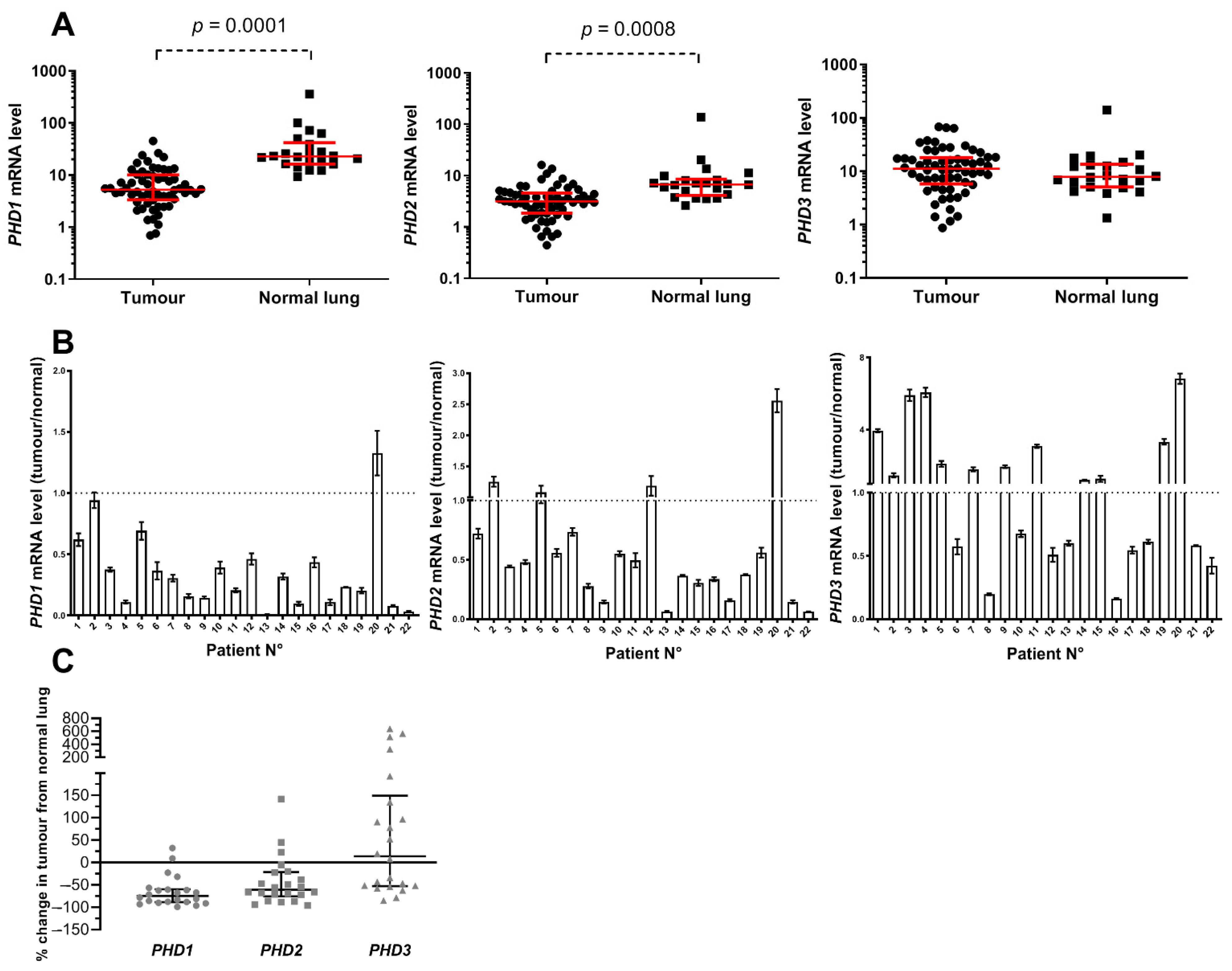
3.2. PHD mRNA Levels in NSCLC Primary Tumours and Normal Lungs
3.3. Associations between PHD mRNA Levels and HIF Pathway Downstream Targets
3.4. Association between PHD mRNA Levels and Patient Characteristics
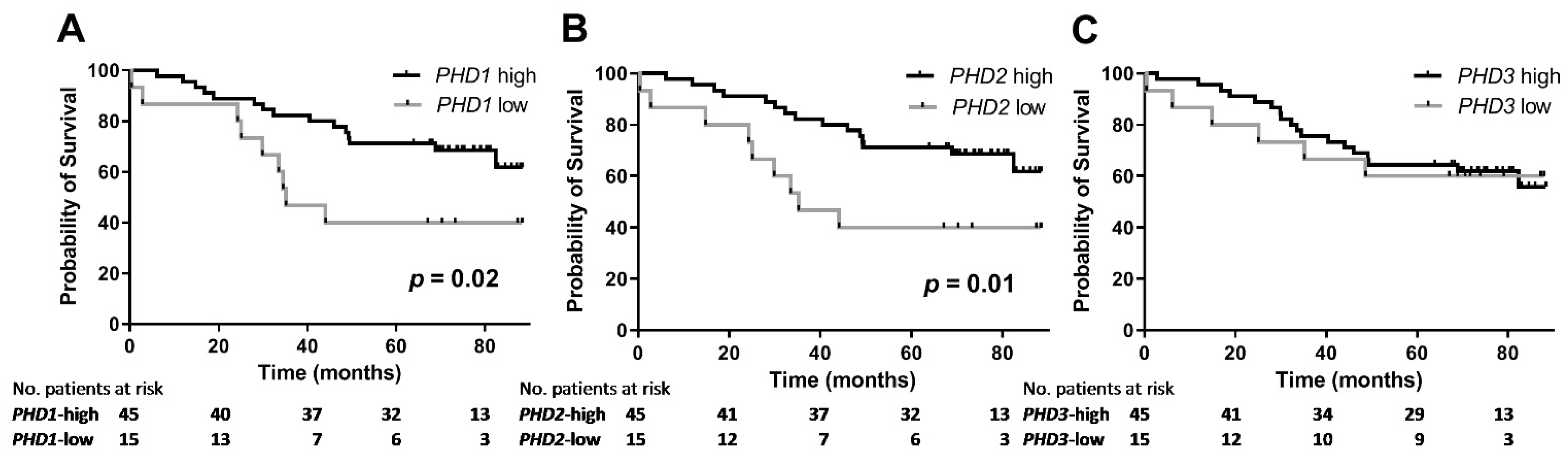
3.5. Association between PHD mRNA Levels and Survival
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brown, J.M.; Wilson, W.R. Exploiting tumour hypoxia in cancer treatment. Nat. Rev. Cancer 2004, 4, 437–447. [Google Scholar] [CrossRef]
- Rofstad, E.K.; Mathiesen, B.; Henriksen, K.; Kindem, K.; Galappathi, K. The Tumor Bed Effect: Increased Metastatic Dissemination from Hypoxia-Induced Up-regulation of Metastasis-Promoting Gene Products. Cancer Res. 2005, 65, 2387–2396. [Google Scholar] [CrossRef]
- Salem, A.; Asselin, M.C.; Reymen, B.; Jackson, A.; Lambin, P.; West, C.M.L.; O’Connor, J.P.B.; Faivre-Finn, C. Targeting hypoxia to improve non–small cell lung cancer outcome. J. Natl. Cancer Inst. 2018, 110, 14–30. [Google Scholar] [CrossRef]
- Wilson, W.R.; Hay, M.P. Targeting hypoxia in cancer therapy. Nat. Rev. Cancer 2011, 11, 393–410. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Riaz, S.P.; Lüchtenborg, M.; Coupland, V.H.; Spicer, J.; Peake, M.D.; Møller, H. Trends in incidence of small cell lung cancer and all lung cancer. Lung Cancer 2012, 75, 280–284. [Google Scholar] [CrossRef]
- Li, T.; Kung, H.-J.; Mack, P.C.; Gandara, D.R. Genotyping and Genomic Profiling of Non–Small-Cell Lung Cancer: Implications for Current and Future Therapies. J. Clin. Oncol. 2013, 31, 1039–1049. [Google Scholar] [CrossRef] [PubMed]
- Cufer, T.; Knez, L. Update on systemic therapy of advanced non-small-cell lung cancer. Expert Rev. Anticancer Ther. 2014, 14, 1189–1203. [Google Scholar] [CrossRef]
- Allemani, C.; Matsuda, T.; Di Carlo, V.; Harewood, R.; Matz, M.; Nikšić, M.; Bonaventure, A.; Valkov, M.; Johnson, C.J.; Estève, J.; et al. Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): Analysis of individual records for 37513025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 2018, 391, 1023–1075. [Google Scholar] [CrossRef]
- Puderecki, M.; Szumiło, J.; Marzec-Kotarska, B. Novel prognostic molecular markers in lung cancer (Review). Oncol. Lett. 2020, 20, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Sodja, E.; Knez, L.; Kern, I.; Ovčariček, T.; Sadikov, A.; Cufer, T. Impact of ERCC1 expression on treatment outcome in small-cell lung cancer patients treated with platinum-based chemotherapy. Eur. J. Cancer 2012, 48, 3378–3385. [Google Scholar] [CrossRef] [PubMed]
- Bruick, R.K. A Conserved Family of Prolyl-4-Hydroxylases That Modify HIF. Science 2001, 294, 1337–1340. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.S. Characterization and comparative analysis of the EGLN gene family. Gene 2001, 275, 125–132. [Google Scholar] [CrossRef]
- Del Peso, L.; Castellanos, M.C.; Temes, E.; Martín-Puig, S.; Cuevas, Y.; Olmos, G.; Landázuri, M.O. The von Hippel Lindau/hypoxia-inducible factor (HIF) pathway regulates the transcription of the HIF-proline hydroxylase genes in response to low oxygen. J. Biol. Chem. 2003, 278, 48690–48695. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Su, H.; Soga, T.; Kranc, K.R.; Pollard, P.J. Prolyl hydroxylase domain enzymes: Important regulators of cancer metabolism. Hypoxia 2014, 2, 127–142. [Google Scholar]
- Fong, G.-H.; Takeda, K. Role and regulation of prolyl hydroxylase domain proteins. Cell Death Differ. 2008, 15, 635–641. [Google Scholar] [CrossRef]
- Jokilehto, T.; Jaakkola, P.M. The role of HIF prolyl hydroxylases in tumour growth. J. Cell Mol. Med. 2010, 14, 758–770. [Google Scholar] [CrossRef]
- Chan, D.A.; Kawahara, T.L.A.; Sutphin, P.D.; Chang, H.Y.; Chi, J.-T.; Giaccia, A.J. Tumor vasculature is regulated by PHD2-mediated angiogenesis and bone marrow-derived cell recruitment. Cancer Cell 2009, 15, 527–538. [Google Scholar] [CrossRef]
- Erez, N.; Milyavsky, M.; Eilam, R.; Shats, I.; Goldfinger, N.; Rotter, V. Expression of Prolyl-Hydroxylase-1 (PHD1/EGLN2) Suppresses Hypoxia Inducible Factor-1alpha Activation and Inhibits Tumor Growth. Cancer Res. 2003, 63, 8777–8783. [Google Scholar]
- Chu, X.; Xiang, M.; Feng, L.; Liu, H.; Zhou, C. Prolyl hydroxylase 3 involvement in lung cancer progression under hypoxic conditions: Association with hypoxia-inducible factor-1α and pyruvate kinase M2. J. Thorac. Dis. 2019, 11, 3941–3950. [Google Scholar] [CrossRef]
- Giatromanolaki, A.; Koukourakis, M.I.; Pezzella, F.; Turley, H.; Sivridis, E.; Bouros, D.; Bougioukas, G.; Harris, A.L.; Gatter, K.C. Expression of prolyl-hydroxylases PHD-1, 2 and 3 and of the asparagine hydroxylase FIH in non-small cell lung cancer relates to an activated HIF pathway. Cancer Lett. 2008, 262, 87–93. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, J.; Li, X.; Luo, X.; Fang, J.; Chen, H. The expression of prolyl hydroxylase domain enzymes are upregulated and negatively correlated with Bcl-2 in non-small cell lung cancer. Mol. Cell Biochem. 2011, 358, 257–263. [Google Scholar] [CrossRef]
- Crinò, L.; Weder, W.; van Meerbeeck, J.; Felip, E. ESMO Guidelines Working Group Early stage and locally advanced (non-metastatic) non-small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2010, 21 (Suppl. 5), v103–v115. [Google Scholar] [CrossRef]
- Koren, A.; Rijavec, M.; Kern, I.; Sodja, E.; Korosec, P.; Cufer, T. BMI1, ALDH1A1, and CD133 transcripts connect epithelial-mesenchymal transition to cancer stem cells in lung carcinoma. Stem Cells Int. 2016, 2016, 9714315. [Google Scholar] [CrossRef] [PubMed]
- Koren, A.; Sodja, E.; Rijavec, M.; Jez, M.; Kovac, V.; Korosec, P.; Cufer, T. Prognostic value of cytokeratin-7 mRNA expression in peripheral whole blood of advanced lung adenocarcinoma patients. Cell Oncol. 2015, 38, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Edge, S.B.; Compton, C.C. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann. Surg. Oncol. 2010, 17, 1471–1474. [Google Scholar] [CrossRef]
- Muz, B.; de la Puente, P.; Azab, F.; Azab, A.K. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia 2015, 3, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Dehdashti, F.; Mintun, M.A.; Lewis, J.S.; Bradley, J.; Govindan, R.; Laforest, R.; Welch, M.J.; Siegel, B.A. In vivo assessment of tumor hypoxia in lung cancer with 60Cu-ATSM. Eur. J. Nucl. Med. Mol. Imaging 2003, 30, 844–850. [Google Scholar] [CrossRef]
- Chen, B.; Li, L.; Li, M.; Wang, X. HIF1A expression correlates with increased tumor immune and stromal signatures and aggressive phenotypes in human cancers. Cell Oncol. 2020, 43, 877–888. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.; Zhu, C.C.; Liu, H.; Wang, J.C. Expression of hypoxia-inducible factor prolyl hydroxylase 3 HIFPH3 in human non-small cell lung cancer (NSCLC) and its correlation with prognosis. Asian Pac. J. Cancer Prev. 2014, 15, 5819–5823. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, S.; Tamayo, P.; Rifkin, R.; Mukherjee, S.; Yeang, C.-H.; Angelo, M.; Ladd, C.; Reich, M.; Latulippe, E.; Mesirov, J.P.; et al. Multiclass cancer diagnosis using tumor gene expression signatures. Proc. Natl. Acad. Sci. USA 2001, 98, 15149–15154. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Xiao, H.; Ding, F.; Zhong, H.; Zhu, J.; Ma, N.; Mei, J. Over-expression of prolyl hydroxylase-1 blocks NF-κB-mediated cyclin D1 expression and proliferation in lung carcinoma cells. Cancer Genet. 2014, 207, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Kawase, A.; Yoshida, J.; Ishii, G.; Nakao, M.; Aokage, K.; Hishida, T.; Nishimura, M.; Nagai, K. Differences between squamous cell carcinoma and adenocarcinoma of the lung: Are adenocarcinoma and squamous cell carcinoma prognostically equal? Jpn. J. Clin. Oncol. 2012, 42, 189–195. [Google Scholar] [CrossRef]
- Luo, W.; Hu, H.; Chang, R.; Zhong, J.; Knabel, M.; O’Meally, R.; Cole, R.N.; Pandey, A.; Semenza, G.L. Pyruvate Kinase M2 Is a PHD3-Stimulated Coactivator for Hypoxia-Inducible Factor 1. Cell 2011, 145, 732–744. [Google Scholar] [CrossRef]
- Zahra, K.; Dey, T.; Ashish; Mishra, S.P.; Pandey, U. Pyruvate Kinase M2 and Cancer: The Role of PKM2 in Promoting Tumorigenesis. Front. Oncol. 2020, 10, 159. [Google Scholar] [CrossRef]
- Kim, J.W.; Tchernyshyov, I.; Semenza, G.L.; Dang, C.V. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006, 3, 177–185. [Google Scholar] [CrossRef]
- Andersen, S.; Donnem, T.; Stenvold, H.; Al-Saad, S.; Al-Shibli, K.; Busund, L.T.; Bremnes, R.M. Overexpression of the HIF hydroxylases PHD1, PHD2, PHD3 and FIH are individually and collectively unfavorable prognosticators for NSCLC survival. PLoS ONE 2011, 6, 4–11. [Google Scholar] [CrossRef]
- Couvelard, A.; Deschamps, L.; Rebours, V.; Sauvanet, A.; Gatter, K.; Pezzella, F.; Ruszniewski, P.; Bedossa, P. Overexpression of the oxygen sensors PHD-1, PHD-2, PHD-3, and FIH is associated with tumor aggressiveness in pancreatic endocrine tumors. Clin. Cancer Res. 2008, 14, 6634–6639. [Google Scholar] [CrossRef]
- Kamphues, C.; Wittschieber, D.; Klauschen, F.; Kasajima, A.; Dietel, M.; Schmidt, S.-C.; Glanemann, M.; Bahra, M.; Neuhaus, P.; Weichert, W.; et al. Prolyl hydroxylase domain 2 protein is a strong prognostic marker in human gastric cancer. Pathobiology 2012, 79, 11–17. [Google Scholar] [CrossRef]
- Ma, M.; Hua, S.; Li, G.; Wang, S.; Cheng, X.; He, S.; Wu, P.; Chen, X. Prolyl hydroxylase domain protein 3 and asparaginyl hydroxylase factor inhibiting HIF-1 levels are predictive of tumoral behavior and prognosis in hepatocellular carcinoma. Oncotarget 2017, 8, 12983–13002. [Google Scholar] [CrossRef] [PubMed]
- Giatromanolaki, A.; Koukourakis, M.I.; Sivridis, E.; Turley, H.; Talks, K.; Pezzella, F.; Gatter, K.C.; Harris, A.L. Relation of hypoxia inducible factor 1 alpha and 2 alpha in operable non-small cell lung cancer to angiogenic/molecular profile of tumours and survival. Br. J. Cancer 2001, 85, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Appelhoffl, R.J.; Tian, Y.M.; Raval, R.R.; Turley, H.; Harris, A.L.; Pugh, C.W.; Ratcliffe, P.J.; Gleadle, J.M. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J. Biol. Chem. 2004, 279, 38458–38465. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]

| Characteristics | NSCLC N (%) | |
|---|---|---|
| N° of patients | 60 | |
| Age in years: median (range) | 61 (42–79) | |
| Sex | ||
| Male | 33 (55.0) | |
| Female | 27 (45.0) | |
| Smoking history | ||
| Yes | 51 (85.0) | |
| No | 9 (15.0) | |
| Histology | ||
| Adenocarcinoma | 35 (58.4) | |
| Squamous cell carcinoma | 20 (33.3) | |
| Other histotype | 5 (8.3) | |
| pTNM stage a | ||
| I | 31 (51.6) | |
| II | 16 (26.7) | |
| III | 13 (21.7) | |
| Tumour size | ||
| pT1 (<3 cm) | 24 | |
| pT2 (3–7 cm) | 28 | |
| pT3 (>7 cm) | 8 | |
| Lymph node involvement Ϯ | ||
| pN0 | 39 | |
| pN1 | 7 | |
| pN2 | 13 | |
| Performance status b | ||
| 0 | 21 (35.0) | |
| 1 | 37 (61.7) | |
| ≥2 | 2 (3.3) | |
| HIF Target | PHD1 | PHD2 | PHD3 |
|---|---|---|---|
| HIF1A | rs = 0.56 (p < 0.0001) | rs = 0.54 (p < 0.0001) | rs = 0.037 (p = 0.78) |
| PKM2 | rs = 0.45 (p = 0.0004) | rs = 0.41 (p = 0.0012) | rs = 0.24 (p = 0.06) |
| PDK1 | rs = 0.50 (p < 0.0001) | rs = 0.54 (p < 0.0001) | rs = 0.50 (p < 0.0001) |
| Parameter | N° of ps | PHD1 mRNA Level | p | PHD2 mRNA Level | p | PHD3 mRNA Level | p | |
|---|---|---|---|---|---|---|---|---|
| Age | ||||||||
| <60 | 29 | 5.60 (3.40–9.88) | 0.7561 | 3.49 (2.05–4.51) | 0.4159 | 10.85 (7.40–17.79) | 0.4506 | |
| ≥60 | 31 | 5.13 (2.47–10.30) | 2.95 (1.51–4.80) | 11.47 (3.93–18.32) | ||||
| Sex | ||||||||
| Male | 33 | 5.35 (2.46–9.88) | 0.6030 | 2.95 (1.65–4.35) | 0.5132 | 9.72 (5.93–17.77) | 0.6665 | |
| Female | 27 | 5.11 (3.95–11.69) | 3.49 (1.90–4.80) | 12.53 (4.98–18.32) | ||||
| Smoking history | ||||||||
| Yes | 51 | 5.11 (3.04–8.37) | 0.1654 | 2.95 (1.79–4.33) | 0.1020 | 10.78 (4.98–18.29) | 0.7719 | |
| No | 9 | 9.53 (4.29–13.64) | 4.07 (3.34–5.49) | 11.65 (8.60–17.81) | ||||
| Histology a | ||||||||
| Adenocarcinoma | 35 | 6.70 (4.51–12.41) | 0.0065 | 3.68 (2.76–5.35) | 0.0050 | 10.78 (4.27–16.22) | 0.0624 | |
| Squamous cell carcinoma | 20 | 3.71 (1.59–6.33) | 2.23 (1.25–3.63) | 15.44 (8.69–25.22) | ||||
| pTNM stage | ||||||||
| I | 31 | 7.10 (4.20–13.29) | 0.0089 | 3.52 (2.23–5.35) | 0.0890 | 12.09 (3.19–18.48) | 0.5303 | |
| II + III | 29 | 4.43 (2.36–7.17) | 3.17 (2.21–4.66) | 10.78 (7.40–18.30) | ||||
| Tumour size | ||||||||
| pT1 | 24 | 8.36 (4.78–16.36) | 0.0002 | 3.98 (2.99–6.31) | 0.0026 | 12.87 (4.28–21.40) | 0.5845 | |
| pT2-3 | 36 | 4.27 (2.32–6.68) | 2.66 (1.53–3.77) | 9.72 (5.77–17.29) | ||||
| Nodular involvement Ϯ | ||||||||
| Yes | 20 | 4.38 (2.61–6.68) | 0.0795 | 2.66 (1.32–3.87) | 0.2166 | 10.82 (7.51–18.04) | 0.5620 | |
| No | 39 | 5.53 (3.96–11.69) | 3.17 (2.21–4.66) | 11.47 (4.48–24.00) | ||||
| Parameter | Overall Survival | |
|---|---|---|
| UV p-Value HR (95% CI) | MV p-Value HR (95% CI) | |
| PHD1 mRNA level | 0.060 0.908 (0.822–1.004) | 0.060 0.908 (0.822–1.004) |
| PHD2 mRNA level | 0.481 0.940 (0.792–1.116) | n/i |
| PHD3 mRNA level | 0.458 0.988 (0.958–1.020) | n/i |
| Age (>60 vs. ≤60) | 0.866 0.933 (0.417–2.086) | n/i |
| Sex (M vs. F) | 0.399 1.428 (0.624–3.265) | n/i |
| Smoking history (Yes vs. No) | 0.654 0.758 (0.225–2.548) | n/i |
| Histology (AC vs. SCC) | 0.565 0.778 (0.332–1.826) | n/i |
| PS a (≥2 vs. <2) | 0.217 0.558 (0.221–1.408) | Eliminated b |
| pTNM stage (I vs. II vs. III) I vs. II II vs. III | 0.102 0.364 (0.140–0.944) 0.439 (0.149–1.295) | Eliminated b |
| Tumour size (pT1 vs. pT2-3) | 0.231 0.584 (0.242–1.409) | Eliminated b |
| Nodular involvement (Yes vs. No) | 0.051 0.439 (0.192–1.003) | Eliminated b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koren, A.; Rijavec, M.; Krumpestar, T.; Kern, I.; Sadikov, A.; Čufer, T.; Korošec, P. Gene Expression Levels of the Prolyl Hydroxylase Domain Proteins PHD1 and PHD2 but Not PHD3 Are Decreased in Primary Tumours and Correlate with Poor Prognosis of Patients with Surgically Resected Non-Small-Cell Lung Cancer. Cancers 2021, 13, 2309. https://doi.org/10.3390/cancers13102309
Koren A, Rijavec M, Krumpestar T, Kern I, Sadikov A, Čufer T, Korošec P. Gene Expression Levels of the Prolyl Hydroxylase Domain Proteins PHD1 and PHD2 but Not PHD3 Are Decreased in Primary Tumours and Correlate with Poor Prognosis of Patients with Surgically Resected Non-Small-Cell Lung Cancer. Cancers. 2021; 13(10):2309. https://doi.org/10.3390/cancers13102309
Chicago/Turabian StyleKoren, Ana, Matija Rijavec, Tomaž Krumpestar, Izidor Kern, Aleksander Sadikov, Tanja Čufer, and Peter Korošec. 2021. "Gene Expression Levels of the Prolyl Hydroxylase Domain Proteins PHD1 and PHD2 but Not PHD3 Are Decreased in Primary Tumours and Correlate with Poor Prognosis of Patients with Surgically Resected Non-Small-Cell Lung Cancer" Cancers 13, no. 10: 2309. https://doi.org/10.3390/cancers13102309
APA StyleKoren, A., Rijavec, M., Krumpestar, T., Kern, I., Sadikov, A., Čufer, T., & Korošec, P. (2021). Gene Expression Levels of the Prolyl Hydroxylase Domain Proteins PHD1 and PHD2 but Not PHD3 Are Decreased in Primary Tumours and Correlate with Poor Prognosis of Patients with Surgically Resected Non-Small-Cell Lung Cancer. Cancers, 13(10), 2309. https://doi.org/10.3390/cancers13102309