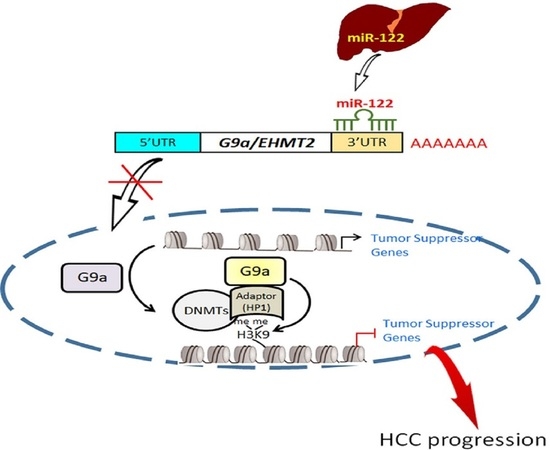
Histone Methyltransferase G9a-Promoted Progression of Hepatocellular Carcinoma Is Targeted by Liver-Specific Hsa-miR-122
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. HCC Tissue Microarray (TMA) and Immunohistochemical (IHC) Staining
2.2. Analysis of Online Available Database
2.3. Cell Lines and Cell Culture
2.4. Western Blot Analysis
2.5. Reverse-Transcriptase Quantitative Polymerase Chain Reaction (RT-qPCR)
2.6. Cell Viability and Clonogenic Assays
2.7. Migration and Invasion Assays
2.8. miRNA Mimic/Inhibitor Introduction
2.9. Lentiviral Infection
2.10. 3′-Untranslated Region (UTR) Luciferase Reporter Assay
2.11. In Vivo HCC Orthotopic Model
2.12. Statistical Analysis
3. Results
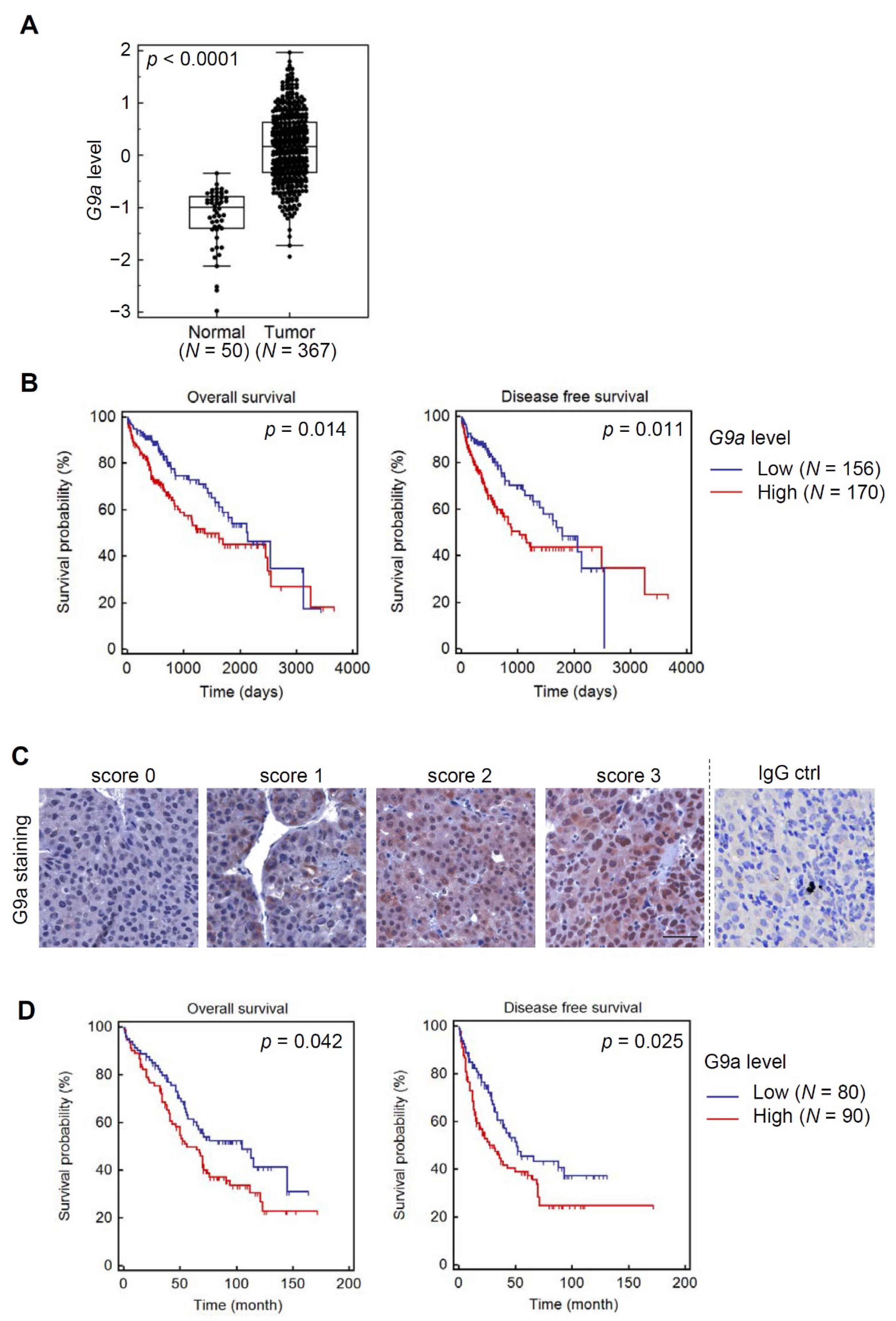
3.1. Both G9a mRNA and Protein Levels Predict Survival Outcomes of HCC Patients
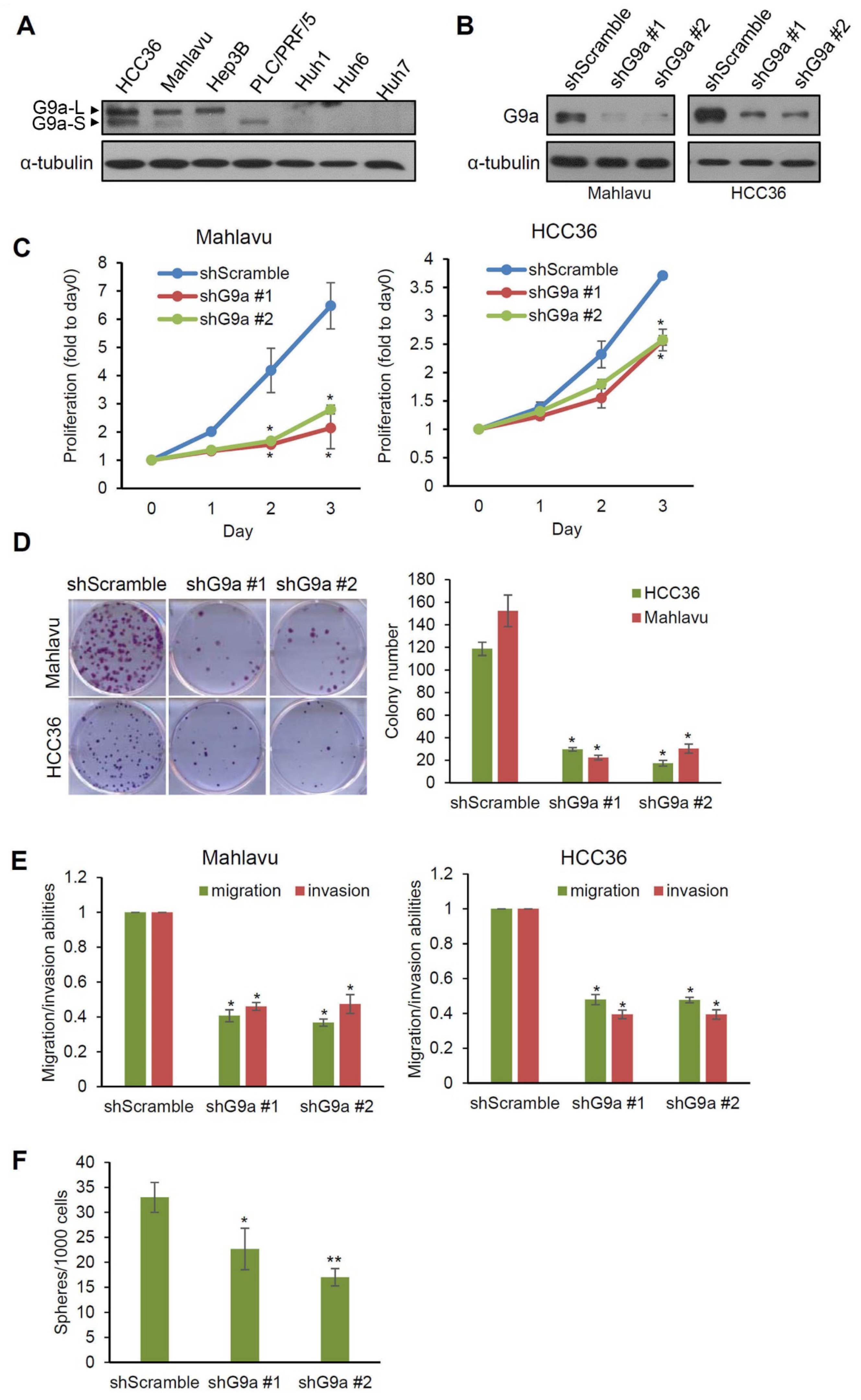
3.2. G9a Modulates Aggressive Phenotypes of HCC Cells
3.3. G9a Depletion Attenuates HCC Tumorigenicity in an Orthotopic Xenograft Model
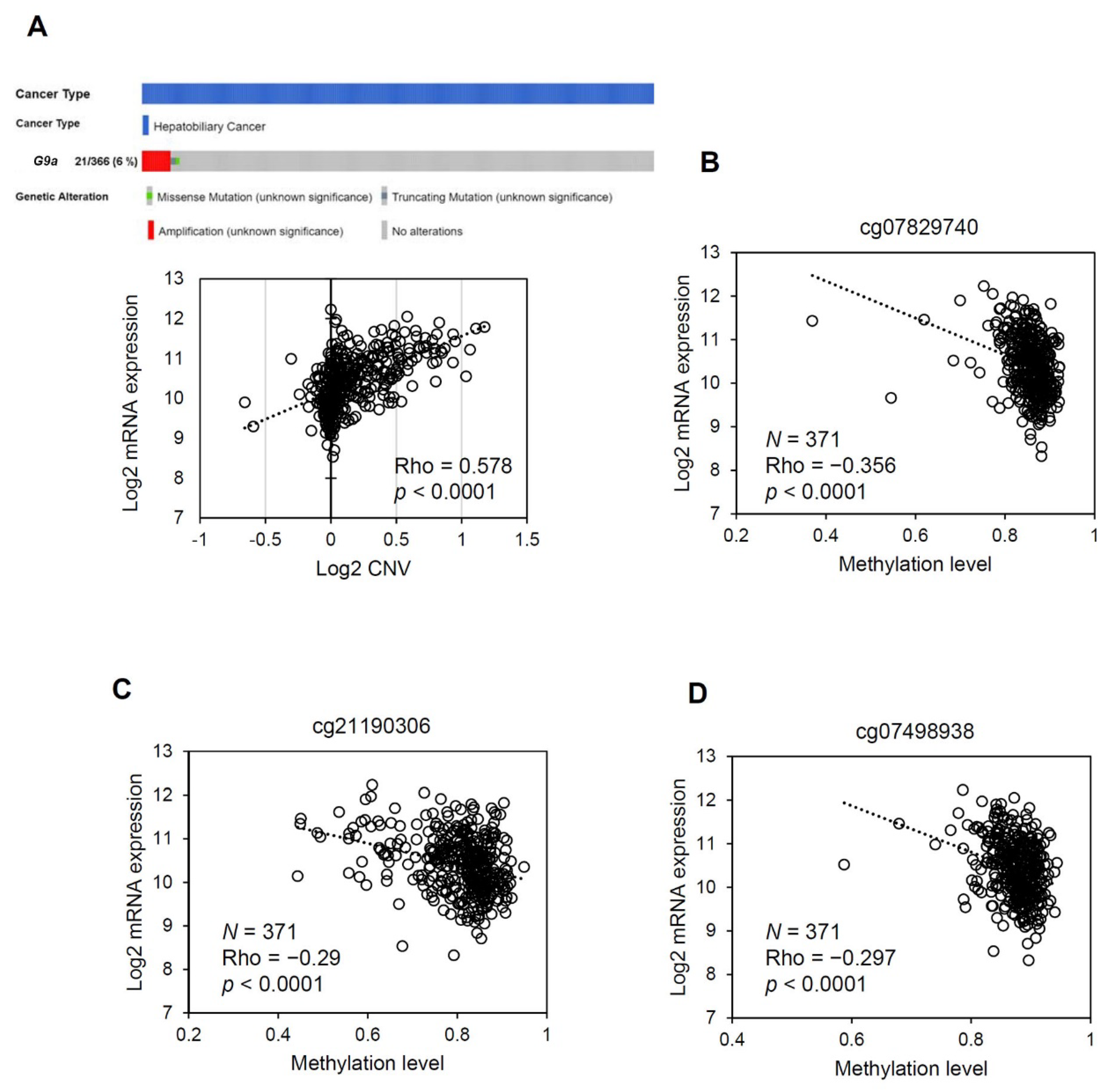
3.4. The G9a Expression Level Correlates with Its Copy Number and DNA Methylation Status in HCC
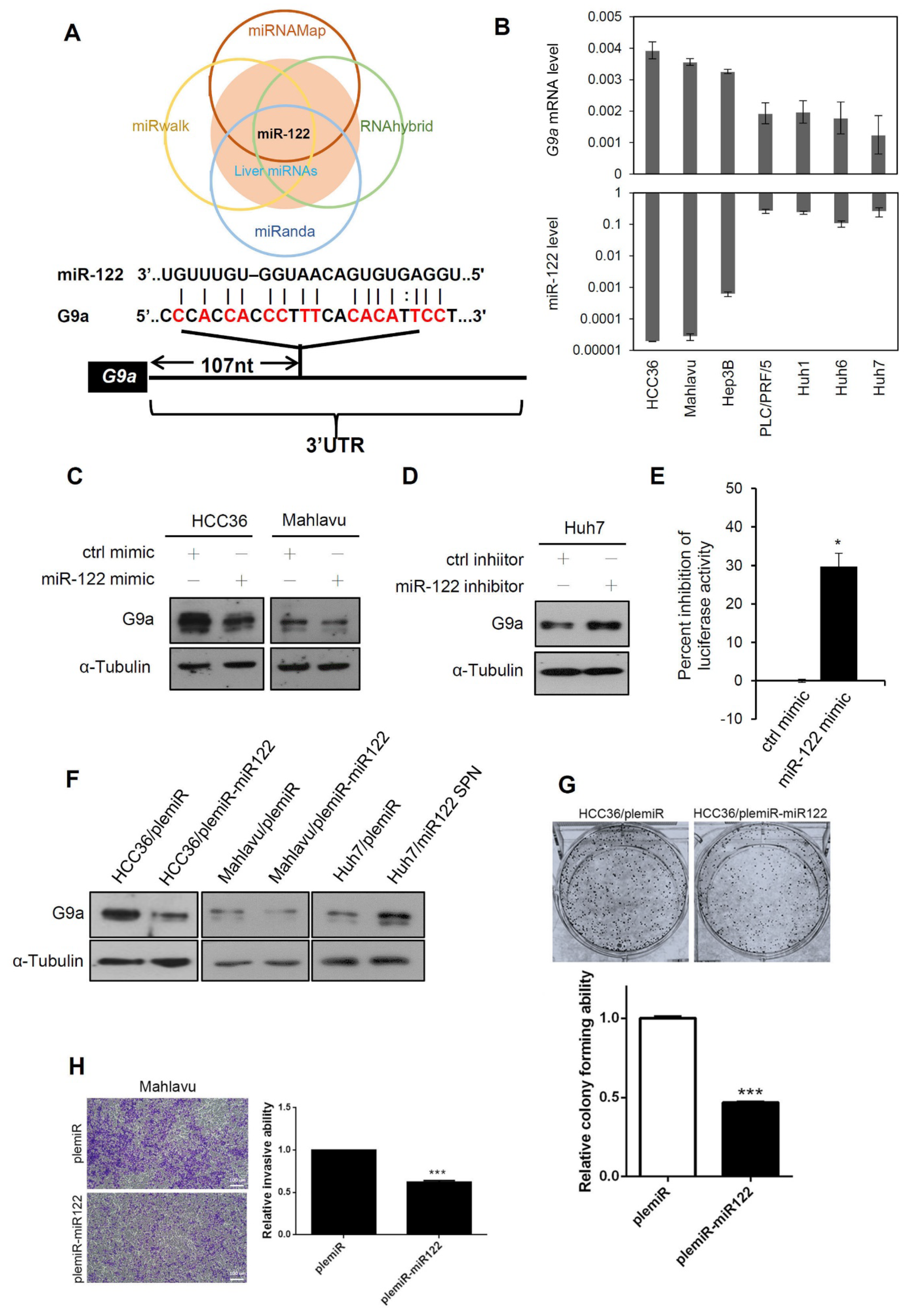
3.5. Liver-Specific miR-122 Regulates G9a Expression, and the Growth and Motility of HCC Cells
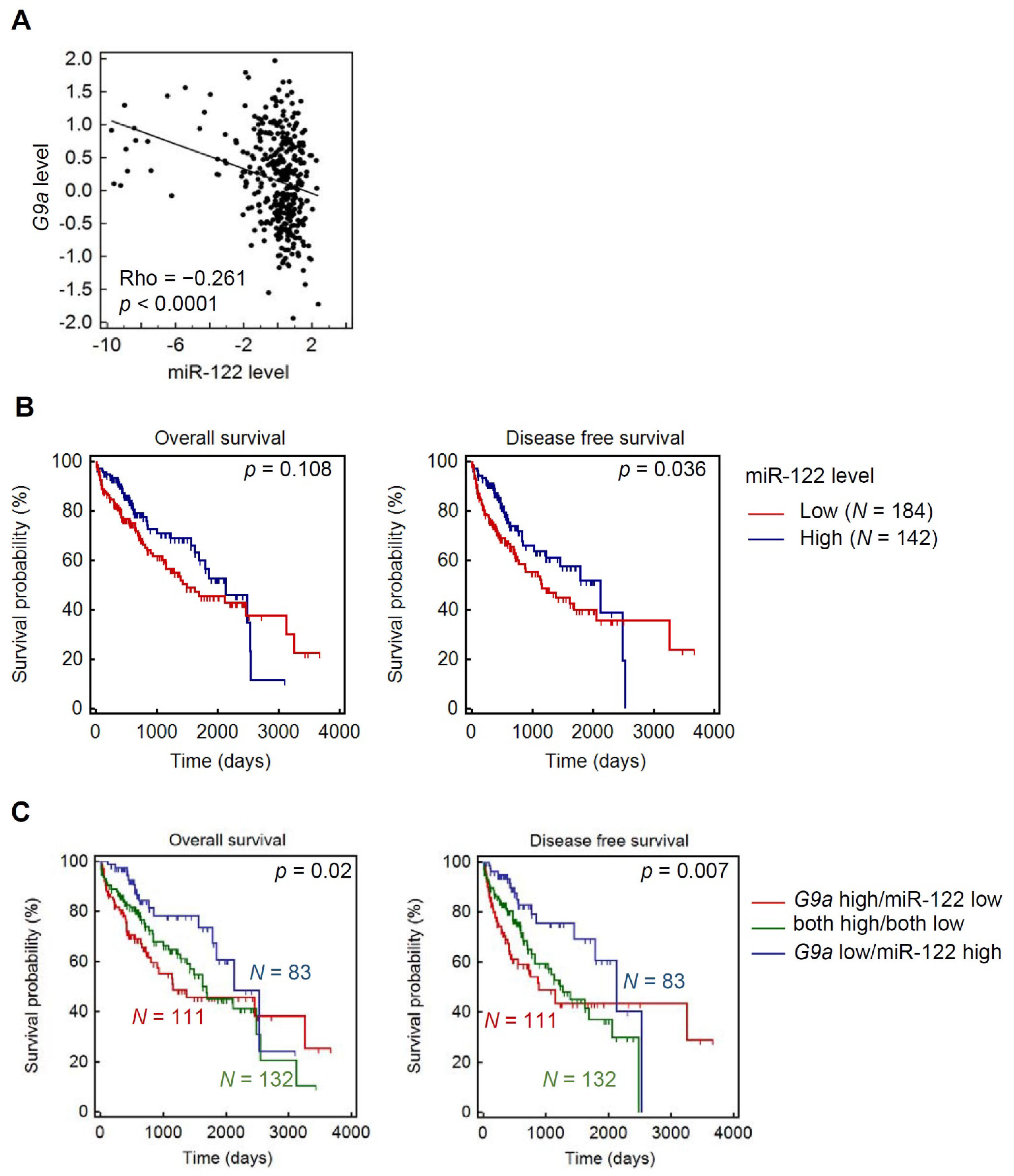
3.6. Clinical Correlations and Prognostic Significance of miR-122 and G9a in HCC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Simard, E.P.; Ward, E.M.; Siegel, R.; Jemal, A. Cancers with increasing incidence trends in the United States: 1999 through 2008. CA A Cancer J. Clin. 2012, 62, 118–128. [Google Scholar] [CrossRef]
- Kulik, L.; El-Serag, H.B. Epidemiology and Management of Hepatocellular Carcinoma. Gastroenterolgy 2019, 156, 477–491.e1. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Nishida, N.; Goel, A. Genetic and epigenetic signatures in human hepatocellular carcinoma: A systematic review. Curr. Genom. 2011, 12, 130–137. [Google Scholar] [CrossRef] [Green Version]
- Nault, J.-C.; Zucman-Rossi, J. Genetics of Hepatobiliary Carcinogenesis. Semin. Liver Dis. 2011, 31, 173–187. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.S.; Ali, I.; Afridi, U.K.; Ishtiaq, M.; Mehmood, R. Epigenetic mechanisms regulating the development of hepatocellular carcinoma and their promise for therapeutics. Hepatol. Int. 2017, 11, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Herceg, Z.; Paliwal, A. Epigenetic mechanisms in hepatocellular carcinoma: How environmental factors influence the epigenome. Mutat. Res./Rev. Mutat. Res. 2011, 727, 55–61. [Google Scholar] [CrossRef]
- Farazi, P.A.; Depinho, R.A. Hepatocellular carcinoma pathogenesis: From genes to environment. Nat. Rev. Cancer 2006, 6, 674–687. [Google Scholar] [CrossRef]
- Nakamura, M.; Chiba, T.; Kanayama, K.; Kanzaki, H.; Saito, T.; Kusakabe, Y.; Kato, N. Epigenetic dysregulation in hepatocellular carcinoma: An up-to-date review. Hepatol. Res. 2019, 49, 3–13. [Google Scholar] [CrossRef] [Green Version]
- Toh, T.B.; Lim, J.J.; Chow, E.K. Epigenetics of hepatocellular carcinoma. Clin. Transl. Med. 2019, 8, 13. [Google Scholar] [CrossRef]
- Tachibana, M.; Sugimoto, K.; Fukushima, T.; Shinkai, Y. SET Domain-containing Protein, G9a, Is a Novel Lysine-preferring Mammalian Histone Methyltransferase with Hyperactivity and Specific Selectivity to Lysines 9 and 27 of Histone H3. J. Biol. Chem. 2001, 276, 25309–25317. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.-W.; Hua, K.-T.; Kao, H.-J.; Chi, C.-C.; Wei, L.-H.; Johansson, G.; Shiah, S.-G.; Chen, P.-S.; Jeng, Y.-M.; Cheng, T.-Y.; et al. H3K9 Histone Methyltransferase G9a Promotes Lung Cancer Invasion and Metastasis by Silencing the Cell Adhesion Molecule Ep-CAM. Cancer Res. 2010, 70, 7830–7840. [Google Scholar] [CrossRef] [Green Version]
- Hua, K.-T.; Wang, M.-Y.; Chen, M.-W.; Wei, L.-H.; Chen, C.-K.; Ko, C.-H.; Jeng, Y.-M.; Sung, P.-L.; Jan, Y.-H.; Hsiao, M.; et al. The H3K9 methyltransferase G9a is a marker of aggressive ovarian cancer that promotes peritoneal metastasis. Mol. Cancer 2014, 13, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsiao, S.-M.; Chen, M.-W.; Chen, C.-A.; Chien, M.-H.; Hua, K.-T.; Hsiao, M.; Kuo, M.-L.; Wei, L.-H. The H3K9 Methyltransferase G9a Represses E-cadherin and is Associated with Myometrial Invasion in Endometrial Cancer. Ann. Surg. Oncol. 2015, 22, 1556–1565. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-W.; Hua, K.-T.; Li, K.-C.; Kao, H.-F.; Hong, R.-L.; Ko, J.-Y.; Hsiao, M.; Kuo, M.-L.; Tan, C.-T. Histone Methyltransferase G9a Drives Chemotherapy Resistance by Regulating the Glutamate–Cysteine Ligase Catalytic Subunit in Head and Neck Squamous Cell Carcinoma. Mol. Cancer Ther. 2017, 16, 1421–1434. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Zhang, H.; Wang, P.; Mao, Z.; Feng, L.; Wang, Y.; Liu, C.; Xia, Q.; Li, B.; Zhao, H.; et al. Short-Form CDYLb but not long-form CDYLa functions cooperatively with histone methyltransferase G9a in hepatocellular carcinomas. Genes Chromosomes Cancer 2013, 52, 644–655. [Google Scholar] [CrossRef]
- Kondo, Y.; Shen, L.; Suzuki, S.; Kurokawa, T.; Masuko, K.; Tanaka, Y.; Kato, H.; Mizuno, Y.; Yokoe, M.; Sugauchi, F.; et al. Alterations of DNA methylation and histone modifications contribute to gene silencing in hepatocellular carcinomas. Hepatol. Res. 2007, 37, 974–983. [Google Scholar] [CrossRef]
- Hung, S.-Y.; Lin, H.-H.; Yeh, K.-T.; Chang, J.-G. Histone-modifying genes as biomarkers in hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 2496–2507. [Google Scholar] [PubMed]
- Yokoyama, M.; Chiba, T.; Zen, Y.; Oshima, M.; Kusakabe, Y.; Noguchi, Y.; Yuki, K.; Koide, S.; Tara, S.; Saraya, A.; et al. Histone lysine methyltransferase G9a is a novel epigenetic target for the treatment of hepatocellular carcinoma. Oncotarget 2017, 8, 21315–21326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, K.; Cao, Y.; Huang, C.; Chen, J.; Zhang, X.; Jiang, Y. Association of Histone Methyltransferase G9a and Overall Survival After Liver Resection of Patients With Hepatocellular Carcinoma With a Median Observation of 40 Months. Medicine 2016, 95, e2493. [Google Scholar] [CrossRef]
- Wei, L.; Chiu, D.K.-C.; Tsang, F.H.-C.; Law, C.-T.; Cheng, C.L.-H.; Au, S.L.-K.; Lee, J.M.-F.; Wong, C.C.-L.; Ng, I.O.-L.; Wong, C.-M. Histone methyltransferase G9a promotes liver cancer development by epigenetic silencing of tumor suppressor gene RARRES3. J. Hepatol. 2017, 67, 758–769. [Google Scholar] [CrossRef]
- Hu, Y.; Zheng, Y.; Dai, M.; Wang, X.; Wu, J.; Yu, B.; Zhang, H.; Cui, Y.; Kong, W.; Wu, H.; et al. G9a and histone deacetylases are crucial for Snail2-mediated E-cadherin repression and metastasis in hepatocellular carcinoma. Cancer Sci. 2019, 110, 3442–3452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, M.; Toh, T.B.; Hooi, L.; Lim, J.J.; Zhang, X.; Chow, E.K.-H. Nanodiamond-Mediated Delivery of a G9a Inhibitor for Hepatocellular Carcinoma Therapy. ACS Appl. Mater. Interfaces 2019, 11, 45427–45441. [Google Scholar] [CrossRef] [PubMed]
- Bárcena-Varela, M.; Caruso, S.; Llerena, S.; Álvarez-Sola, G.; Uriarte, I.; Latasa, M.U.; Urtasun, R.; Rebouissou, S.; Alvarez, L.; Jimenez, M.; et al. Dual Targeting of Histone Methyltransferase G9a and DNA-Methyltransferase 1 for the Treatment of Experimental Hepatocellular Carcinoma. Hepatology 2019, 69, 587–603. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Piovan, C.; Croce, C.M. Interplay between microRNAs and the epigenetic machinery: An intricate network. Biochim. Biophys. Acta (BBA)-Bioenerg. 2010, 1799, 694–701. [Google Scholar] [CrossRef]
- Arif, K.M.T.; Elliott, E.K.; Haupt, L.M.; Griffiths, L.R. Regulatory Mechanisms of Epigenetic miRNA Relationships in Human Cancer and Potential as Therapeutic Targets. Cancers 2020, 12, 2922. [Google Scholar] [CrossRef] [PubMed]
- Rebouissou, S.; Zucman-Rossi, J.; Moreau, R.; Qiu, Z.; Hui, L. Note of caution: Contaminations of hepatocellular cell lines. J. Hepatol. 2017, 67, 896–897. [Google Scholar] [CrossRef] [Green Version]
- Heffelfinger, S.C.; Hawkins, H.H.; Barrish, J.; Taylor, L.; Darlington, G.J. SK HEP-1: A human cell line of endothelial origin. Vitr. Cell. Dev. Biol. Anim. 1992, 28, 136–142. [Google Scholar] [CrossRef]
- López-Terrada, D.; Cheung, S.W.; Finegold, M.J.; Knowles, B.B. Hep G2 is a hepatoblastoma-derived cell line. Hum. Pathol. 2009, 40, 1512–1515. [Google Scholar] [CrossRef]
- Oh, S.Y.; Seok, J.Y.; Choi, Y.S.; Lee, S.H.; Bae, J.-S.; Lee, Y.M. The Histone Methyltransferase Inhibitor BIX01294 Inhibits HIF-1α Stability and Angiogenesis. Mol. Cells 2015, 38, 528–534. [Google Scholar] [CrossRef] [Green Version]
- Chien, M.-H.; Lee, W.-J.; Yang, Y.-C.; Tan, P.; Pan, K.-F.; Liu, Y.-C.; Tsai, H.-C.; Hsu, C.-H.; Wen, Y.-C.; Hsiao, M.; et al. N-α-acetyltransferase 10 protein promotes metastasis by stabilizing matrix metalloproteinase-2 protein in human osteosarcomas. Cancer Lett. 2018, 433, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Tay, N.; Chan, S.-H.; Ren, E.-C. Detection of integrated hepatitis B virus DNA in hepatocellular carcinoma cell lines by nonradioactive in situ hybridization. J. Med. Virol. 1990, 30, 266–271. [Google Scholar] [CrossRef]
- Chen, J.-Y.; Harrison, T.J.; Tsuei, D.-J.; Hsu, T.-Y.; Zuckerman, A.J.; Chan, T.-S.; Yang, C.-S. Analysis of Integrated Hepatitis B Virus DNA and Flanking Cellular Sequences in the Hepatocellular Carcinoma Cell Line HCC36. Intervirology 1994, 37, 41–46. [Google Scholar] [CrossRef]
- Sobolewski, C.; Calo, N.V.; Portius, D.; Foti, M. MicroRNAs in Fatty Liver Disease. Semin. Liver Dis. 2015, 35, 012–025. [Google Scholar] [CrossRef]
- Satishchandran, A.; Szabo, G. MicroRNAs in Alcoholic Liver Disease. Semin. Liver Dis. 2015, 35, 036–042. [Google Scholar] [CrossRef] [Green Version]
- Otsuka, M.; Kishikawa, T.; Yoshikawa, T.; Yamagami, M.; Ohno, M.; Takata, A.; Shibata, C.; Ishibashi, R.; Koike, K. MicroRNAs and liver disease. J. Hum. Genet. 2016, 62, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Xie, H.; Hu, M.; Huang, T.; Hu, Y.; Sang, N.; Zhao, Y. Recent progress in treatment of hepatocellular carcinoma. Am. J. Cancer Res. 2020, 10, 2993–3036. [Google Scholar] [PubMed]
- Bayo, J.; Fiore, E.J.; Dominguez, L.M.; Real, A.; Malvicini, M.; Rizzo, M.; Atorrasagasti, C.; García, M.G.; Argemi, J.; Martinez, E.D.; et al. A comprehensive study of epigenetic alterations in hepatocellular carcinoma identifies potential therapeutic targets. J. Hepatol. 2019, 71, 78–90. [Google Scholar] [CrossRef]
- Schulze, K.; Imbeaud, S.; Letouzé, E.; Alexandrov, L.B.; Calderaro, J.; Rebouissou, S.; Couchy, G.; Meiller, C.; Shinde, J.; Soysouvanh, F.; et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet. 2015, 47, 505–511. [Google Scholar] [CrossRef]
- Wen, B.; Wu, H.; Shinkai, Y.A.; Irizarry, R.; Feinberg, A.P. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat. Genet. 2009, 41, 246–250. [Google Scholar] [CrossRef] [Green Version]
- Lu, H.; Lei, X.; Zhang, Q. Liver-specific knockout of histone methyltransferase G9a impairs liver maturation and dysregulates inflammatory, cytoprotective, and drug-processing genes. Xenobiotica 2019, 49, 740–752. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xue, W.; Zhang, W.; Yuan, Y.; Zhu, X.; Wang, Q.; Wei, Y.; Yang, D.; Yang, C.; Chen, Y.; et al. Histone methyltransferase G9a protects against acute liver injury through GSTP1. Cell Death Differ. 2020, 27, 1243–1258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Yang, D.; Yuan, Y.; Liu, C.; Chen, H.; Zhang, Y.; Wang, Q.; Petersen, R.B.; Huang, K.; Zheng, L. Muscular G9a Regulates Muscle-Liver-Fat Axis by Musclin Under Overnutrition in Female Mice. Diabetes 2020, 69, 2642–2654. [Google Scholar] [CrossRef]
- Barcena-Varela, M.; Paish, H.; Alvarez, L.; Uriarte, I.; Latasa, M.U.; Santamaria, E.; Recalde, M.; Garate, M.; Claveria, A.; Colyn, L.; et al. Epigenetic mechanisms and metabolic reprogramming in fibrogenesis: Dual targeting of G9a and DNMT1 for the inhibition of liver fibrosis. Gut 2020. [Google Scholar] [CrossRef]
- Zeisel, M.B.; Pfeffer, S.; Baumert, T.F. miR-122 acts as a tumor suppressor in hepatocarcinogenesis in vivo. J. Hepatol. 2013, 58, 821–823. [Google Scholar] [CrossRef] [PubMed]
- Bandiera, S.; Pfeffer, S.; Baumert, T.F.; Zeisel, M.B. miR-122–A key factor and therapeutic target in liver disease. J. Hepatol. 2015, 62, 448–457. [Google Scholar] [CrossRef] [Green Version]
- Girard, M.; Jacquemin, E.; Munnich, A.; Lyonnet, S.; Henrion-Caude, A. miR-122, a paradigm for the role of microRNAs in the liver. J. Hepatol. 2008, 48, 648–656. [Google Scholar] [CrossRef] [Green Version]
- Liu, A.M.; Xu, Z.; Shek, F.H.; Wong, K.-F.; Lee, N.P.; Poon, R.T.; Chen, J.; Luk, J.M. miR-122 Targets Pyruvate Kinase M2 and Affects Metabolism of Hepatocellular Carcinoma. PLoS ONE 2014, 9, e86872. [Google Scholar] [CrossRef] [Green Version]
- Lei, Y.; Chen, L.; Zhang, G.; Shan, A.; Ye, C.; Liang, B.; Deng, L. MicroRNAs target the Wnt/β-catenin signaling pathway to regulate epithelial-mesenchymal transition in cancer (Review). Oncol. Rep. 2020, 44, 1299–1313. [Google Scholar] [CrossRef]
- Bai, S.; Nasser, M.W.; Wang, B.; Hsu, S.-H.; Datta, J.; Kutay, H.; Yadav, A.; Nuovo, G.; Kumar, P.; Ghoshal, K. MicroRNA-122 Inhibits Tumorigenic Properties of Hepatocellular Carcinoma Cells and Sensitizes These Cells to Sorafenib. J. Biol. Chem. 2009, 284, 32015–32027. [Google Scholar] [CrossRef] [Green Version]
- Nakatsuka, T.; Tateishi, K.; Kato, H.; Fujiwara, H.; Yamamoto, K.; Kudo, Y.; Nakagawa, H.; Tanaka, Y.; Ijichi, H.; Ikenoue, T.; et al. Inhibition of histone methyltransferase G9a attenuates liver cancer initiation by sensitizing DNA-damaged hepatocytes to p53-induced apoptosis. Cell Death Dis. 2021, 12, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.-H.; Wang, B.; Kutay, H.; Bid, H.; Shreve, J.; Zhang, X.; Costinean, S.; Bratasz, A.; Houghton, P.; Ghoshal, K. Hepatic Loss of miR-122 Predisposes Mice to Hepatobiliary Cyst and Hepatocellular Carcinoma upon Diethylnitrosamine Exposure. Am. J. Pathol. 2013, 183, 1719–1730. [Google Scholar] [CrossRef] [Green Version]
- Cheng, D.; Deng, J.; Zhang, B.; He, X.; Meng, Z.; Li, G.; Ye, H.; Zheng, S.; Wei, L.; Deng, X.; et al. LncRNA HOTAIR epigenetically suppresses miR-122 expression in hepatocellular carcinoma via DNA methylation. EBioMedicine 2018, 36, 159–170. [Google Scholar] [CrossRef] [Green Version]
- Pangeni, R.P.; Yang, L.; Zhang, K.; Wang, J.; Li, W.; Guo, C.; Yun, X.; Sun, T.; Wang, J.; Raz, D.J. G9a regulates tumorigenicity and stemness through genome-wide DNA methylation reprogramming in non-small cell lung cancer. Clin. Epigenet. 2020, 12, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Thienpont, B.; Aronsen, J.M.; Robinson, E.L.; Okkenhaug, H.; Loche, E.; Ferrini, A.; Brien, P.; Alkass, K.; Tomasso, A.; Agrawal, A.; et al. The H3K9 dimethyltransferases EHMT1/2 protect against pathological cardiac hypertrophy. J. Clin. Investig. 2017, 127, 335–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, J.; Li, Q.; Zeng, Z.; Wu, P.; Jiang, Y.; Luo, T.; Ji, X.; Zhang, Q.; Hao, Y.; Chen, L. Increased expression of G9A contributes to carcinogenesis and indicates poor prognosis in hepatocellular carcinoma. Oncol. Lett. 2018, 15, 9757–9765. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Parameter | G9a Low | G9a High | p-Value |
|---|---|---|---|
| Chi-Squared | |||
| Age (years) | 60.3 ± 12.7 | 62.2 ± 11.6 | |
| Sex, no. of patients | |||
| Male | 66 | 80 | 0.3303 |
| Female | 14 | 10 | |
| Tumor size | |||
| <5 cm | 53 | 52 | 0.3288 |
| >5 cm | 27 | 38 | |
| Stage | |||
| I or II | 61 | 65 | 0.6723 |
| III or IV | 19 | 25 | |
| Portal vein involvement | |||
| No | 72 | 79 | 0.8296 |
| Yes | 8 | 11 | |
| α-Fetoprotein (ng/mL) | |||
| 0–500 | 67 | 66 | 0.0916 |
| >500 | 11 | 23 | |
| Microvessel invasion | |||
| No | 31 | 29 | 0.4665 |
| Yes | 49 | 61 | |
| Cirrhosis | |||
| No | 51 | 42 | 0.0507 |
| Yes | 28 | 45 | |
| Hepatitis B virus status | |||
| Negative | 25 | 30 | 0.9449 |
| Positive | 54 | 60 | |
| Hepatitis C virus status | |||
| Negative | 61 | 66 | 0.6860 |
| Positive | 18 | 24 |
| Parameter | Comparison | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|---|
| HR (95% CI) | p-Value | HR (95% CI) | p-Value | ||
| G9a score | low (≥240); high (<240) | 1.52 (1.01–2.27) | 0.0442 * | 1.45 (0.96–2.20) | 0.0805 |
| AJCC stage | I or II; III or IV | 1.71 (1.11–2.63) | 0.0148 * | 1.50 (0.83–2.72) | 0.184 |
| Sex | male; female | 0.87 (0.49–1.55) | 0.6314 | 1.10 (0.59–2.05) | 0.7714 |
| AFP level (ng/mL) | ≥500; <500 | 1.56 (0.97–2.51) | 0.0662 | 1.45 (0.85–2.49) | 0.1762 |
| Tumor size (cm) | ≥5; <5 | 1.48 (0.99–2.20) | 0.0546 | 1.12 (0.64–1.97) | 0.6843 |
| Portal vein involvement | yes; no | 1.74 (1.15–2.63) | 0.0096 * | 1.81 (1.13–2.90) | 0.0147 * |
| Microvessel invasion | yes; no | 0.88 (0.59–1.33) | 0.5579 | 0.64 (0.40–1.03) | 0.0659 |
| HBV | positive; negative | 1.29 (0.86–1.95) | 0.2237 | 1.32 (0.82–2.15) | 0.2589 |
| HCV | positive; negative | 0.89 (0.57–1.41) | 0.6348 | 1.15 (0.67–1.97) | 0.6102 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, L.-T.; Lee, W.-J.; Yang, Y.-C.; Chen, B.-R.; Yang, C.-Y.; Chen, M.-W.; Chen, J.-Q.; Hsiao, M.; Chien, M.-H.; Hua, K.-T. Histone Methyltransferase G9a-Promoted Progression of Hepatocellular Carcinoma Is Targeted by Liver-Specific Hsa-miR-122. Cancers 2021, 13, 2376. https://doi.org/10.3390/cancers13102376
Yuan L-T, Lee W-J, Yang Y-C, Chen B-R, Yang C-Y, Chen M-W, Chen J-Q, Hsiao M, Chien M-H, Hua K-T. Histone Methyltransferase G9a-Promoted Progression of Hepatocellular Carcinoma Is Targeted by Liver-Specific Hsa-miR-122. Cancers. 2021; 13(10):2376. https://doi.org/10.3390/cancers13102376
Chicago/Turabian StyleYuan, Lan-Ting, Wei-Jiunn Lee, Yi-Chieh Yang, Bo-Rong Chen, Ching-Yao Yang, Min-Wei Chen, Ji-Qing Chen, Michael Hsiao, Ming-Hsien Chien, and Kuo-Tai Hua. 2021. "Histone Methyltransferase G9a-Promoted Progression of Hepatocellular Carcinoma Is Targeted by Liver-Specific Hsa-miR-122" Cancers 13, no. 10: 2376. https://doi.org/10.3390/cancers13102376
APA StyleYuan, L.-T., Lee, W.-J., Yang, Y.-C., Chen, B.-R., Yang, C.-Y., Chen, M.-W., Chen, J.-Q., Hsiao, M., Chien, M.-H., & Hua, K.-T. (2021). Histone Methyltransferase G9a-Promoted Progression of Hepatocellular Carcinoma Is Targeted by Liver-Specific Hsa-miR-122. Cancers, 13(10), 2376. https://doi.org/10.3390/cancers13102376