Altered Expression of Secreted Mediator Genes That Mediate Aggressive Breast Cancer Metastasis to Distant Organs
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
2.1. Identifying Potential Gene Signatures in TNBC Metastatic Variants of Breast Cancer Cells
2.2. ANGPTL7 Secreted Angiogenesis-Related Protein-Encoding Gene Alteration in Breast Cancer Distant Metastasis
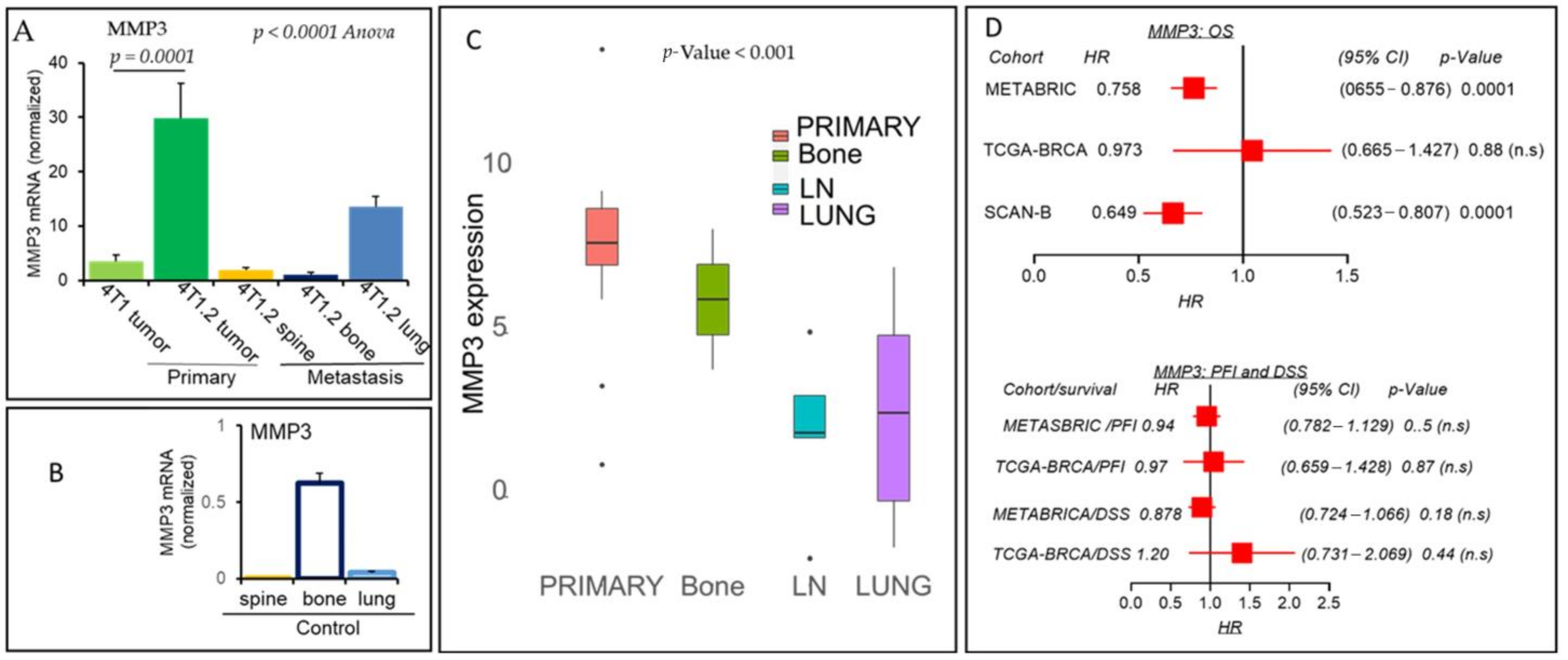
2.3. MMP3 Matrix Metalloproteinase Secreted Endopeptidase Protein-Encoding Gene Inversely Linked in Breast Cancer Distant Metastasis
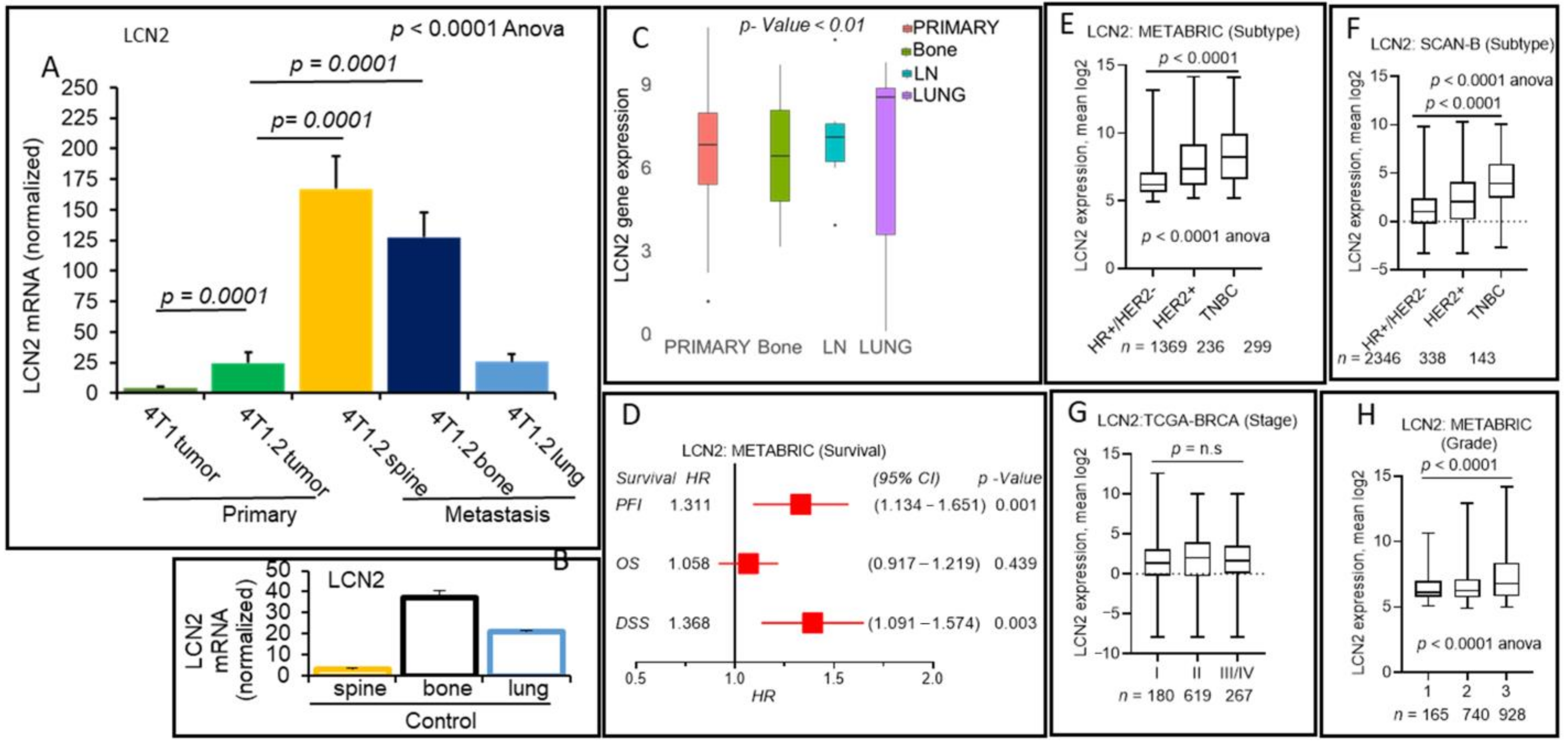
2.4. Positive Association of Lipocalin-2 Secreted Glycoprotein-Encoding Gene and Breast Cancer Distant Metastasis
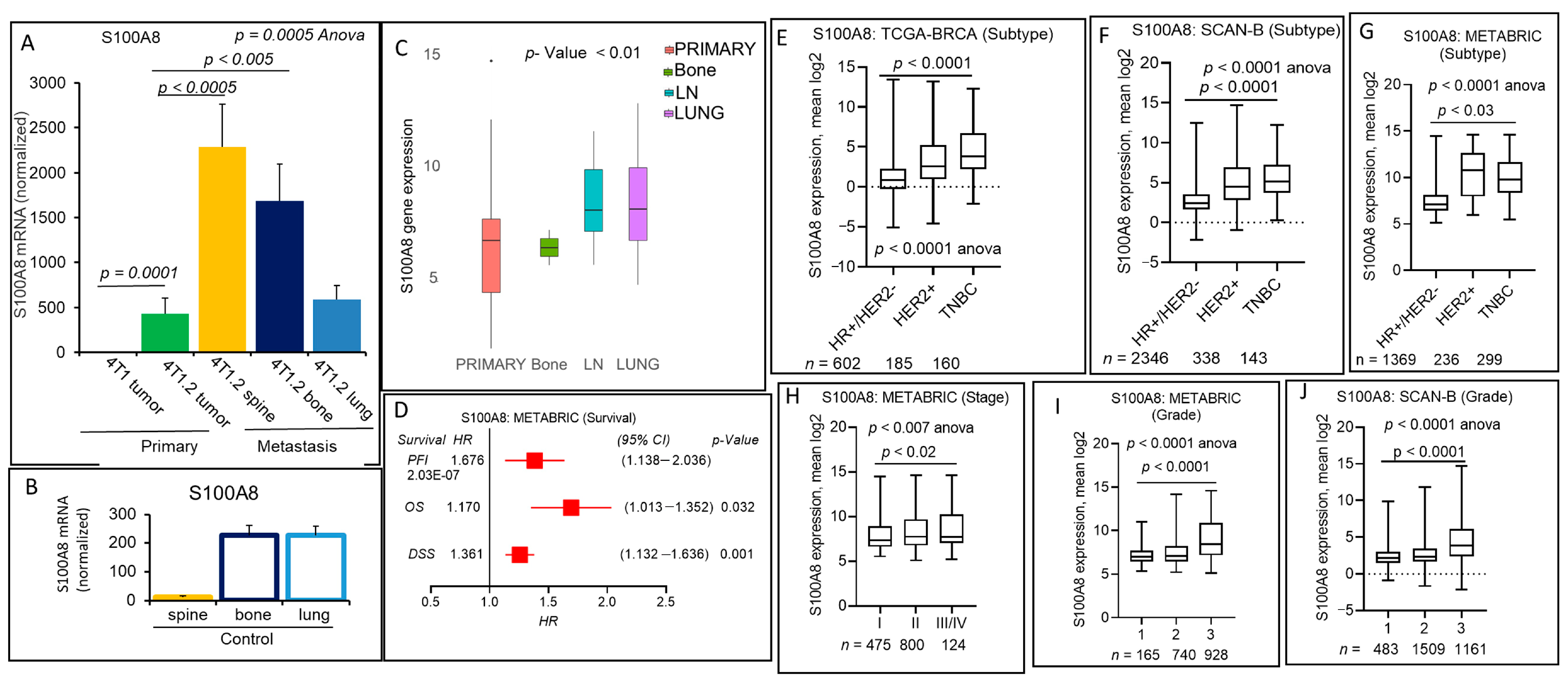
2.5. S100A8 Secreted Pro-Inflammatory Mediator Encoding Gene Expression Correlates with Breast Cancer Progression and Metastasis
2.6. ESM-1 Secreted Endothelial Proteoglycan Encoding Gene Expression Is Associated with Poor Prognosis of Aggressive Subtypes of Breast Cancers
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Cell Culture
4.3. RNA Preparation and RNA-Seq
4.4. Gene Expression Analyses
4.5. Quantitative-Real-Time PCR (qRT-PCR)
4.6. Ethical Statement
4.7. Animals and Tumor Cell Implantations
4.8. Clinical and Gene Expression Data Analyses of Breast Cancer Patient Cohorts
4.9. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chambers, A.F.; Groom, A.C.; MacDonald, I.C. Dissemination and growth of cancer cells in metastatic sites. Nat. Rev. Cancer 2002, 2, 563–572. [Google Scholar] [CrossRef]
- Klein, C.A. Parallel progression of primary tumours and metastases. Nat. Rev. Cancer 2009, 9, 302–312. [Google Scholar] [CrossRef]
- Husemann, Y.; Geigl, J.B.; Schubert, F.; Musiani, P.; Meyer, M.; Burghart, E.; Forni, G.; Eils, R.; Fehm, T.; Riethmuller, G.; et al. Systemic spread is an early step in breast cancer. Cancer Cell 2008, 13, 58–68. [Google Scholar] [CrossRef]
- Fisher, B.; Redmond, C. Systemic therapy in node-negative patients: Updated findings from NSABP clinical trials. National Surgical Adjuvant Breast and Bowel Project. J. Natl. Cancer Inst. Monogr. 1992, 105–116. [Google Scholar]
- Nagrath, S.; Sequist, L.V.; Maheswaran, S.; Bell, D.W.; Irimia, D.; Ulkus, L.; Smith, M.R.; Kwak, E.L.; Digumarthy, S.; Muzikansky, A.; et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007, 450, 1235–1239. [Google Scholar] [CrossRef]
- Matrisian, L.M.; Bowden, G.T.; Krieg, P.; Furstenberger, G.; Briand, J.P.; Leroy, P.; Breathnach, R. The mRNA coding for the secreted protease transin is expressed more abundantly in malignant than in benign tumors. Proc. Natl. Acad. Sci. USA 1986, 83, 9413–9417. [Google Scholar] [CrossRef] [PubMed]
- Gunthert, U.; Hofmann, M.; Rudy, W.; Reber, S.; Zoller, M.; Haussmann, I.; Matzku, S.; Wenzel, A.; Ponta, H.; Herrlich, P. A new variant of glycoprotein CD44 confers metastatic potential to rat carcinoma cells. Cell 1991, 65, 13–24. [Google Scholar] [CrossRef]
- Kim, S.H.; Choi, S.J.; Park, J.S.; Lee, J.; Cho, Y.B.; Kang, M.W.; Lee, W.Y.; Choi, Y.S.; Kim, H.K.; Han, J.; et al. Tropism between hepatic and pulmonary metastases in colorectal cancers. Oncol. Rep. 2012, 28, 459–464. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Choi, S.J.; Cho, Y.B.; Kang, M.W.; Lee, J.; Lee, W.Y.; Chun, H.K.; Choi, Y.S.; Kim, H.K.; Han, J.; et al. Differential gene expression during colon-to-lung metastasis. Oncol. Rep. 2011, 25, 629–636. [Google Scholar] [CrossRef][Green Version]
- Nishimori, H.; Yasoshima, T.; Hata, F.; Denno, R.; Yanai, Y.; Nomura, H.; Tanaka, H.; Kamiguchi, K.; Sato, N.; Hirata, K. A novel nude mouse model of liver metastasis and peritoneal dissemination from the same human pancreatic cancer line. Pancreas 2002, 24, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Minn, A.J.; Kang, Y.; Serganova, I.; Gupta, G.P.; Giri, D.D.; Doubrovin, M.; Ponomarev, V.; Gerald, W.L.; Blasberg, R.; Massague, J. Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J. Clin. Investig. 2005, 115, 44–55. [Google Scholar] [CrossRef]
- Bolin, C.; Sutherland, C.; Tawara, K.; Moselhy, J.; Jorcyk, C.L. Novel mouse mammary cell lines for in vivo bioluminescence imaging (BLI) of bone metastasis. Biol. Proced. Online 2012, 14. [Google Scholar] [CrossRef] [PubMed]
- Tawara, K.; Bolin, C.; Koncinsky, J.; Kadaba, S.; Covert, H.; Sutherland, C.; Bond, L.; Kronz, J.; Garbow, J.R.; Jorcyk, C.L. OSM potentiates preintravasation events, increases CTC counts, and promotes breast cancer metastasis to the lung. Breast Cancer Res. 2018, 20, 53. [Google Scholar] [CrossRef] [PubMed]
- Santulli, G. Angiopoietin-like proteins: A comprehensive look. Front. Endocrinol. 2014, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Morisada, T.; Kubota, Y.; Urano, T.; Suda, T.; Oike, Y. Angiopoietins and angiopoietin-like proteins in angiogenesis. Endothelium 2006, 13, 71–79. [Google Scholar] [CrossRef]
- Oike, Y.; Ito, Y.; Maekawa, H.; Morisada, T.; Kubota, Y.; Akao, M.; Urano, T.; Yasunaga, K.; Suda, T. Angiopoietin-related growth factor (AGF) promotes angiogenesis. Blood 2004, 103, 3760–3765. [Google Scholar] [CrossRef]
- Oike, Y.; Yasunaga, K.; Suda, T. Angiopoietin-related/angiopoietin-like proteins regulate angiogenesis. Int. J. Hematol. 2004, 80, 21–28. [Google Scholar] [CrossRef]
- Hato, T.; Tabata, M.; Oike, Y. The role of angiopoietin-like proteins in angiogenesis and metabolism. Trends Cardiovasc. Med. 2008, 18, 6–14. [Google Scholar] [CrossRef]
- Peek, R.; Verbraak, F.; Coevoet, H.M.; Kijlstra, A. Muller cell-specific autoantibodies in a patient with progressive loss of vision. Invest. Ophthalmol. Vis. Sci. 1998, 39, 1976–1979. [Google Scholar]
- Peek, R.; van Gelderen, B.E.; Bruinenberg, M.; Kijlstra, A. Molecular cloning of a new angiopoietinlike factor from the human cornea. Invest. Ophthalmol. Vis. Sci. 1998, 39, 1782–1788. [Google Scholar]
- Kuchtey, J.; Kallberg, M.E.; Gelatt, K.N.; Rinkoski, T.; Komaromy, A.M.; Kuchtey, R.W. Angiopoietin-like 7 secretion is induced by glaucoma stimuli and its concentration is elevated in glaucomatous aqueous humor. Invest. Ophthalmol. Vis. Sci. 2008, 49, 3438–3448. [Google Scholar] [CrossRef]
- Parri, M.; Pietrovito, L.; Grandi, A.; Campagnoli, S.; De Camilli, E.; Bianchini, F.; Marchio, S.; Bussolino, F.; Jin, B.; Sarmientos, P.; et al. Angiopoietin-like 7, a novel pro-angiogenetic factor over-expressed in cancer. Angiogenesis 2014, 17, 881–896. [Google Scholar] [CrossRef] [PubMed]
- Carbone, C.; Piro, G.; Merz, V.; Simionato, F.; Santoro, R.; Zecchetto, C.; Tortora, G.; Melisi, D. Angiopoietin-Like Proteins in Angiogenesis, Inflammation and Cancer. Int. J. Mol. Sci. 2018, 19, 431. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- Siegel, M.B.; He, X.; Hoadley, K.A.; Hoyle, A.; Pearce, J.B.; Garrett, A.L.; Kumar, S.; Moylan, V.J.; Brady, C.M.; Van Swearingen, A.E.; et al. Integrated RNA and DNA sequencing reveals early drivers of metastatic breast cancer. J. Clin. Investig. 2018, 128, 1371–1383. [Google Scholar] [CrossRef] [PubMed]
- Brueffer, C.; Vallon-Christersson, J.; Grabau, D.; Ehinger, A.; Hakkinen, J.; Hegardt, C.; Malina, J.; Chen, Y.; Bendahl, P.O.; Manjer, J.; et al. Clinical Value of RNA Sequencing-Based Classifiers for Prediction of the Five Conventional Breast Cancer Biomarkers: A Report From the Population-Based Multicenter Sweden Cancerome Analysis Network-Breast Initiative. JCO Precis. Oncol. 2018, 2. [Google Scholar] [CrossRef]
- Lochter, A.; Galosy, S.; Muschler, J.; Freedman, N.; Werb, Z.; Bissell, M.J. Matrix metalloproteinase stromelysin-1 triggers a cascade of molecular alterations that leads to stable epithelial-to-mesenchymal conversion and a premalignant phenotype in mammary epithelial cells. J. Cell Biol. 1997, 139, 1861–1872. [Google Scholar] [CrossRef]
- Duffy, M.J.; Maguire, T.M.; Hill, A.; McDermott, E.; O’Higgins, N. Metalloproteinases: Role in breast carcinogenesis, invasion and metastasis. Breast Cancer Res. 2000, 2, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef]
- Lindy, O.; Konttinen, Y.T.; Sorsa, T.; Ding, Y.; Santavirta, S.; Ceponis, A.; Lopez-Otin, C. Matrix metalloproteinase 13 (collagenase 3) in human rheumatoid synovium. Arthritis Rheum. 1997, 40, 1391–1399. [Google Scholar] [CrossRef] [PubMed]
- Neuhold, L.A.; Killar, L.; Zhao, W.; Sung, M.L.; Warner, L.; Kulik, J.; Turner, J.; Wu, W.; Billinghurst, C.; Meijers, T.; et al. Postnatal expression in hyaline cartilage of constitutively active human collagenase-3 (MMP-13) induces osteoarthritis in mice. J. Clin. Investig. 2001, 107, 35–44. [Google Scholar] [CrossRef]
- Wang, X.; Cao, X. Regulation of metastasis of pediatric multiple myeloma by MMP13. Tumor Biol. 2014, 35, 8715–8720. [Google Scholar] [CrossRef]
- Nickolas, T.L.; Barasch, J.; Devarajan, P. Biomarkers in acute and chronic kidney disease. Curr. Opin. Nephrol. Hypertens. 2008, 17, 127–132. [Google Scholar] [CrossRef]
- Yan, Q.W.; Yang, Q.; Mody, N.; Graham, T.E.; Hsu, C.H.; Xu, Z.; Houstis, N.E.; Kahn, B.B.; Rosen, E.D. The adipokine lipocalin 2 is regulated by obesity and promotes insulin resistance. Diabetes 2007, 56, 2533–2540. [Google Scholar] [CrossRef]
- Devarajan, P. Neutrophil gelatinase-associated lipocalin: New paths for an old shuttle. Cancer Ther. 2007, 5, 463–470. [Google Scholar] [PubMed]
- Yang, J.; Moses, M.A. Lipocalin 2: A multifaceted modulator of human cancer. Cell Cycle 2009, 8, 2347–2352. [Google Scholar] [CrossRef]
- Shi, H.; Gu, Y.; Yang, J.; Xu, L.; Mi, W.; Yu, W. Lipocalin 2 promotes lung metastasis of murine breast cancer cells. J. Exp. Clin. Cancer Res. 2008, 27. [Google Scholar] [CrossRef]
- Yang, J.; Bielenberg, D.R.; Rodig, S.J.; Doiron, R.; Clifton, M.C.; Kung, A.L.; Strong, R.K.; Zurakowski, D.; Moses, M.A. Lipocalin 2 promotes breast cancer progression. Proc. Natl. Acad. Sci. USA 2009, 106, 3913–3918. [Google Scholar] [CrossRef]
- Leclerc, E.; Fritz, G.; Vetter, S.W.; Heizmann, C.W. Binding of S100 proteins to RAGE: An update. Biochim. Biophys. Acta 2009, 1793, 993–1007. [Google Scholar] [CrossRef]
- Donato, R.; Cannon, B.R.; Sorci, G.; Riuzzi, F.; Hsu, K.; Weber, D.J.; Geczy, C.L. Functions of S100 proteins. Curr. Mol. Med. 2013, 13, 24–57. [Google Scholar] [CrossRef]
- Gross, S.R.; Sin, C.G.; Barraclough, R.; Rudland, P.S. Joining S100 proteins and migration: For better or for worse, in sickness and in health. Cell Mol. Life Sci. 2014, 71, 1551–1579. [Google Scholar] [CrossRef]
- Funk, S.; Mark, R.; Bayo, P.; Flechtenmacher, C.; Grabe, N.; Angel, P.; Plinkert, P.K.; Hess, J. High S100A8 and S100A12 protein expression is a favorable prognostic factor for survival of oropharyngeal squamous cell carcinoma. Int. J. Cancer 2015, 136, 2037–2046. [Google Scholar] [CrossRef]
- Fujita, Y.; Khateb, A.; Li, Y.; Tinoco, R.; Zhang, T.; Bar-Yoseph, H.; Tam, M.A.; Chowers, Y.; Sabo, E.; Gerassy-Vainberg, S.; et al. Regulation of S100A8 Stability by RNF5 in Intestinal Epithelial Cells Determines Intestinal Inflammation and Severity of Colitis. Cell Rep. 2018, 24, 3296–3311. [Google Scholar] [CrossRef]
- Tajmul, M.; Parween, F.; Singh, L.; Mathur, S.R.; Sharma, J.B.; Kumar, S.; Sharma, D.N.; Yadav, S. Identification and validation of salivary proteomic signatures for non-invasive detection of ovarian cancer. Int. J. Biol. Macromol. 2018, 108, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Miller, P.; Kidwell, K.M.; Thomas, D.; Sabel, M.; Rae, J.M.; Hayes, D.F.; Hudson, B.I.; El-Ashry, D.; Lippman, M.E. Elevated S100A8 protein expression in breast cancer cells and breast tumor stroma is prognostic of poor disease outcome. Breast Cancer Res. Treat. 2017, 166, 85–94. [Google Scholar] [CrossRef]
- Donato, R. Intracellular and extracellular roles of S100 proteins. Microsc. Res. Tech. 2003, 60, 540–551. [Google Scholar] [CrossRef]
- Bechard, D.; Gentina, T.; Delehedde, M.; Scherpereel, A.; Lyon, M.; Aumercier, M.; Vazeux, R.; Richet, C.; Degand, P.; Jude, B.; et al. Endocan is a novel chondroitin sulfate/dermatan sulfate proteoglycan that promotes hepatocyte growth factor/scatter factor mitogenic activity. J. Biol. Chem. 2001, 276, 48341–48349. [Google Scholar] [CrossRef] [PubMed]
- Bechard, D.; Meignin, V.; Scherpereel, A.; Oudin, S.; Kervoaze, G.; Bertheau, P.; Janin, A.; Tonnel, A.; Lassalle, P. Characterization of the secreted form of endothelial-cell-specific molecule 1 by specific monoclonal antibodies. J. Vasc. Res. 2000, 37, 417–425. [Google Scholar] [CrossRef]
- Rennel, E.; Mellberg, S.; Dimberg, A.; Petersson, L.; Botling, J.; Ameur, A.; Westholm, J.O.; Komorowski, J.; Lassalle, P.; Cross, M.J.; et al. Endocan is a VEGF-A and PI3K regulated gene with increased expression in human renal cancer. Exp. Cell Res. 2007, 313, 1285–1294. [Google Scholar] [CrossRef]
- Shin, J.W.; Huggenberger, R.; Detmar, M. Transcriptional profiling of VEGF-A and VEGF-C target genes in lymphatic endothelium reveals endothelial-specific molecule-1 as a novel mediator of lymphangiogenesis. Blood 2008, 112, 2318–2326. [Google Scholar] [CrossRef]
- Gamarra, F.; Noel, J.L.; Brunelli, A.; Dingemans, A.C.; Felip, E.; Gaga, M.; Grigoriu, B.D.; Hardavella, G.; Huber, R.M.; Janes, S.; et al. Thoracic oncology HERMES: European curriculum recommendations for training in thoracic oncology. Breathe 2016, 12, 249–255. [Google Scholar] [CrossRef]
- Leroy, X.; Aubert, S.; Zini, L.; Franquet, H.; Kervoaze, G.; Villers, A.; Delehedde, M.; Copin, M.C.; Lassalle, P. Vascular endocan (ESM-1) is markedly overexpressed in clear cell renal cell carcinoma. Histopathology 2010, 56, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.H.; Ji, N.Y.; Han, S.R.; Lee, C.I.; Kim, J.W.; Yeom, Y.I.; Kim, Y.H.; Chun, H.K.; Kim, J.W.; Chung, J.W.; et al. ESM-1 regulates cell growth and metastatic process through activation of NF-kappaB in colorectal cancer. Cell. Signal. 2012, 24, 1940–1949. [Google Scholar] [CrossRef]
- Maurage, C.A.; Adam, E.; Mineo, J.F.; Sarrazin, S.; Debunne, M.; Siminski, R.M.; Baroncini, M.; Lassalle, P.; Blond, S.; Delehedde, M. Endocan expression and localization in human glioblastomas. J. Neuropathol. Exp. Neurol. 2009, 68, 633–641. [Google Scholar] [CrossRef]
- Sagara, A.; Igarashi, K.; Otsuka, M.; Kodama, A.; Yamashita, M.; Sugiura, R.; Karasawa, T.; Arakawa, K.; Narita, M.; Kuzumaki, N.; et al. Endocan as a prognostic biomarker of triple-negative breast cancer. Breast Cancer Res. Treat. 2017, 161, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Zhang, L.H.; Du, H.; Hu, Y.; Zhang, G.G.; Wang, X.H.; Li, J.Y.; Ji, J.F. Overexpression of endothelial cell specific molecule-1 (ESM-1) in gastric cancer. Ann. Surg. Oncol. 2010, 17, 2628–2639. [Google Scholar] [CrossRef]
- Kang, Y.H.; Ji, N.Y.; Lee, C.I.; Lee, H.G.; Kim, J.W.; Yeom, Y.I.; Kim, D.G.; Yoon, S.K.; Kim, J.W.; Park, P.J.; et al. ESM-1 silencing decreased cell survival, migration, and invasion and modulated cell cycle progression in hepatocellular carcinoma. Amino Acids 2011, 40, 1003–1013. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Mu, P. Targeting Breast Cancer Metastasis. Breast Cancer 2015, 9, 23–34. [Google Scholar] [CrossRef]
- Klemm, F.; Joyce, J.A. Microenvironmental regulation of therapeutic response in cancer. Trends Cell Biol. 2015, 25, 198–213. [Google Scholar] [CrossRef]
- Nguyen, D.X.; Bos, P.D.; Massague, J. Metastasis: From dissemination to organ-specific colonization. Nat. Rev. Cancer 2009, 9, 274–284. [Google Scholar] [CrossRef]
- Bouis, D.R.; Dam, W.A.; Meijer, C.; Mulder, N.H.; Hospers, G.A. Effect of CDT6 on factors of angiogenic balance in tumour cell lines. Anticancer Res. 2007, 27, 2325–2329. [Google Scholar]
- Peek, R.; Kammerer, R.A.; Frank, S.; Otte-Holler, I.; Westphal, J.R. The angiopoietin-like factor cornea-derived transcript 6 is a putative morphogen for human cornea. J. Biol. Chem. 2002, 277, 686–693. [Google Scholar] [CrossRef]
- Lim, S.Y.; Gordon-Weeks, A.; Allen, D.; Kersemans, V.; Beech, J.; Smart, S.; Muschel, R.J. Cd11b(+) myeloid cells support hepatic metastasis through down-regulation of angiopoietin-like 7 in cancer cells. Hepatology 2015, 62, 521–533. [Google Scholar] [CrossRef]
- Fukushima, R.; Kasamatsu, A.; Nakashima, D.; Higo, M.; Fushimi, K.; Kasama, H.; Endo-Sakamoto, Y.; Shiiba, M.; Tanzawa, H.; Uzawa, K. Overexpression of Translocation Associated Membrane Protein 2 Leading to Cancer-Associated Matrix Metalloproteinase Activation as a Putative Metastatic Factor for Human Oral Cancer. J. Cancer 2018, 9, 3326–3333. [Google Scholar] [CrossRef]
- Miyake, M.; Goodison, S.; Lawton, A.; Gomes-Giacoia, E.; Rosser, C.J. Angiogenin promotes tumoral growth and angiogenesis by regulating matrix metallopeptidase-2 expression via the ERK1/2 pathway. Oncogene 2015, 34, 890–901. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Kasamatsu, A.; Ogawara, K.; Miyamoto, I.; Saito, K.; Iyoda, M.; Suzuki, T.; Endo-Sakamoto, Y.; Shiiba, M.; Tanzawa, H.; et al. Semaphorin7A Promotion of Tumoral Growth and Metastasis in Human Oral Cancer by Regulation of G1 Cell Cycle and Matrix Metalloproteases: Possible Contribution to Tumoral Angiogenesis. PLoS ONE 2015, 10, e0137923. [Google Scholar] [CrossRef] [PubMed]
- Gyorffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 2010, 123, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Mehner, C.; Miller, E.; Nassar, A.; Bamlet, W.R.; Radisky, E.S.; Radisky, D.C. Tumor cell expression of MMP3 as a prognostic factor for poor survival in pancreatic, pulmonary, and mammary carcinoma. Genes Cancer 2015, 6, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Stoesz, S.P.; Friedl, A.; Haag, J.D.; Lindstrom, M.J.; Clark, G.M.; Gould, M.N. Heterogeneous expression of the lipocalin NGAL in primary breast cancers. Int. J. Cancer 1998, 79, 565–572. [Google Scholar] [CrossRef]
- Wenners, A.S.; Mehta, K.; Loibl, S.; Park, H.; Mueller, B.; Arnold, N.; Hamann, S.; Weimer, J.; Ataseven, B.; Darb-Esfahani, S.; et al. Neutrophil gelatinase-associated lipocalin (NGAL) predicts response to neoadjuvant chemotherapy and clinical outcome in primary human breast cancer. PLoS ONE 2012, 7, e45826. [Google Scholar] [CrossRef]
- Cheng, G.; Sun, X.; Wang, J.; Xiao, G.; Wang, X.; Fan, X.; Zu, L.; Hao, M.; Qu, Q.; Mao, Y.; et al. HIC1 silencing in triple-negative breast cancer drives progression through misregulation of LCN2. Cancer Res. 2014, 74, 862–872. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; McNeish, B.; Butterfield, C.; Moses, M.A. Lipocalin 2 is a novel regulator of angiogenesis in human breast cancer. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2013, 27, 45–50. [Google Scholar] [CrossRef]
- Kurozumi, S.; Alsaeed, S.; Orah, N.; Miligy, I.M.; Joseph, C.; Aljohani, A.; Toss, M.S.; Fujii, T.; Shirabe, K.; Green, A.R.; et al. Clinicopathological significance of lipocalin 2 nuclear expression in invasive breast cancer. Breast Cancer Res. Treat. 2020, 179, 557–564. [Google Scholar] [CrossRef]
- Rodvold, J.J.; Mahadevan, N.R.; Zanetti, M. Lipocalin 2 in cancer: When good immunity goes bad. Cancer Lett. 2012, 316, 132–138. [Google Scholar] [CrossRef]
- Acharyya, S.; Oskarsson, T.; Vanharanta, S.; Malladi, S.; Kim, J.; Morris, P.G.; Manova-Todorova, K.; Leversha, M.; Hogg, N.; Seshan, V.E.; et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell 2012, 150, 165–178. [Google Scholar] [CrossRef]
- Pitteri, S.J.; Kelly-Spratt, K.S.; Gurley, K.E.; Kennedy, J.; Buson, T.B.; Chin, A.; Wang, H.; Zhang, Q.; Wong, C.H.; Chodosh, L.A.; et al. Tumor microenvironment-derived proteins dominate the plasma proteome response during breast cancer induction and progression. Cancer Res. 2011, 71, 5090–5100. [Google Scholar] [CrossRef]
- Ichikawa, M.; Williams, R.; Wang, L.; Vogl, T.; Srikrishna, G. S100A8/A9 activate key genes and pathways in colon tumor progression. Mol. Cancer Res. 2011, 9, 133–148. [Google Scholar] [CrossRef]
- Gebhardt, C.; Nemeth, J.; Angel, P.; Hess, J. S100A8 and S100A9 in inflammation and cancer. Biochem. Pharmacol. 2006, 72, 1622–1631. [Google Scholar] [CrossRef]
- Hait, N.C.; Allegood, J.; Maceyka, M.; Strub, G.M.; Harikumar, K.B.; Singh, S.K.; Luo, C.; Marmorstein, R.; Kordula, T.; Milstien, S.; et al. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science 2009, 325, 1254–1257. [Google Scholar] [CrossRef]
- Hait, N.C.; Maiti, A.; Xu, P.; Qi, Q.; Kawaguchi, T.; Okano, M.; Takabe, K.; Yan, L.; Luo, C. Regulation of hypoxia-inducible factor functions in the nucleus by sphingosine-1-phosphate. FASEB J. 2020, 34, 4293–4310. [Google Scholar] [CrossRef]
- Maiti, A.; Qi, Q.; Peng, X.; Yan, L.; Takabe, K.; Hait, N.C. Class I histone deacetylase inhibitor suppresses vasculogenic mimicry by enhancing the expression of tumor suppressor and anti-angiogenesis genes in aggressive human TNBC cells. Int. J. Oncol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Mudge, J.M.; Jungreis, I.; Hunt, T.; Gonzalez, J.M.; Wright, J.C.; Kay, M.; Davidson, C.; Fitzgerald, S.; Seal, R.; Tweedie, S.; et al. Discovery of high-confidence human protein-coding genes and exons by whole-genome PhyloCSF helps elucidate 118 GWAS loci. Genome Res. 2019, 29, 2073–2087. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Maiti, A.; Takabe, K.; Hait, N.C. Metastatic triple-negative breast cancer is dependent on SphKs/S1P signaling for growth and survival. Cell. Signal. 2017, 32, 85–92. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Rashid, O.M.; Nagahashi, M.; Ramachandran, S.; Graham, L.; Yamada, A.; Spiegel, S.; Bear, H.D.; Takabe, K. Resection of the primary tumor improves survival in metastatic breast cancer by reducing overall tumor burden. Surgery 2013, 153, 771–778. [Google Scholar] [CrossRef] [PubMed]
- Hait, N.C.; Avni, D.; Yamada, A.; Nagahashi, M.; Aoyagi, T.; Aoki, H.; Dumur, C.I.; Zelenko, Z.; Gallagher, E.J.; Leroith, D.; et al. The phosphorylated prodrug FTY720 is a histone deacetylase inhibitor that reactivates ERalpha expression and enhances hormonal therapy for breast cancer. Oncogenesis 2015, 4, e156. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]



| ESM1 | |||||
|---|---|---|---|---|---|
| Cohorts (No. of Patients) | Survival | p-Value | HR | Lower 95% CI of HR | Upper 95% CI of HR |
| TCGA-BRCA (1091) | OS | 0.2651561 | 1.241098 | 0.847944 | 1.81654 |
| DSS | 0.0260737 | 1.802839 | 1.081905 | 3.00417 | |
| PFI | 0.0404124 | 1.507879 | 1.018059 | 2.23336 | |
| SCAN-B (3273) | OS | 0.0027642 | 1.393542 | 1.122641 | 1.72981 |
| METABRIC (1904) | OS | 0.0002073 | 1.313813 | 1.135676 | 1.51989 |
| DSS | 0.0000432 | 1.488734 | 1.229534 | 1.80257 | |
| PFI | 0.0001991 | 1.421232 | 1.180513 | 1.71103 | |
| ESM1 Gene | ||||||
|---|---|---|---|---|---|---|
| Breast Cancer Cohorts | Subtypes | Survival | p-Value | HR | Lower 95% CI of HR | Upper 95% CI of HR |
| METABRIC | TNBC | OS | 0.0678599 | 1.405336 | 0.9650234 | 2.046551 |
| DSS | 0.0271319 | 1.671391 | 1.05759 | 2.64142 | ||
| RFS | 0.06816303 | 1.531018 | 0.9659142 | 2.426733 | ||
| HER2+ | OS | 0.2023138 | 1.304144 | 0.8662699 | 1.963351 | |
| DSS | 0.4882853 | 1.170697 | 0.7486584 | 1.830649 | ||
| RFS | 0.4749246 | 1.177973 | 0.7512301 | 1.847131 | ||
| HR+/HER2- | OS | 0.0084293 | 1.255485 | 1.058645 | 1.488925 | |
| DSS | 0.0060594 | 1.392947 | 1.098825 | 1.765796 | ||
| RFS | 0.0075160 | 1.357008 | 1.084659 | 1.697742 | ||
| TCGA-BRCA | TNBC | OS | 0.5326945 | 1.329982 | 0.5391484 | 3.280826 |
| DSS | 0.3297474 | 1.753202 | 0.5614073 | 5.47502 | ||
| PFI | 0.690275 | 1.205813 | 0.476718 | 3.049989 | ||
| HER2+ | OS | 0.2602478 | 1.738325 | 0.6522829 | 4.632614 | |
| DSS | 0.0343557 | 6.291787 | 1.570462 | 25.20697 | ||
| PFI | 0.0204855 | 4.075747 | 1.366649 | 12.15506 | ||
| HR+/HER2- | OS | 0.4214821 | 1.284964 | 0.6960809 | 2.37204 | |
| DSS | 0.2860177 | 1.617747 | 0.6729301 | 3.889119 | ||
| PFI | 0.304155 | 1.384029 | 0.7437917 | 2.575366 | ||
| SCAN-B | TNBC | OS | 0.4688024 | 1.321067 | 0.6211149 | 2.809815 |
| HER2+ | OS | 0.0171431 | 2.245766 | 1.188923 | 4.242043 | |
| HR+/HER2- | OS | 0.2057267 | 1.190957 | 0.9086451 | 1.560983 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maiti, A.; Okano, I.; Oshi, M.; Okano, M.; Tian, W.; Kawaguchi, T.; Katsuta, E.; Takabe, K.; Yan, L.; Patnaik, S.K.; et al. Altered Expression of Secreted Mediator Genes That Mediate Aggressive Breast Cancer Metastasis to Distant Organs. Cancers 2021, 13, 2641. https://doi.org/10.3390/cancers13112641
Maiti A, Okano I, Oshi M, Okano M, Tian W, Kawaguchi T, Katsuta E, Takabe K, Yan L, Patnaik SK, et al. Altered Expression of Secreted Mediator Genes That Mediate Aggressive Breast Cancer Metastasis to Distant Organs. Cancers. 2021; 13(11):2641. https://doi.org/10.3390/cancers13112641
Chicago/Turabian StyleMaiti, Aparna, Ichiro Okano, Masanori Oshi, Maiko Okano, Wanqing Tian, Tsutomu Kawaguchi, Eriko Katsuta, Kazuaki Takabe, Li Yan, Santosh K. Patnaik, and et al. 2021. "Altered Expression of Secreted Mediator Genes That Mediate Aggressive Breast Cancer Metastasis to Distant Organs" Cancers 13, no. 11: 2641. https://doi.org/10.3390/cancers13112641
APA StyleMaiti, A., Okano, I., Oshi, M., Okano, M., Tian, W., Kawaguchi, T., Katsuta, E., Takabe, K., Yan, L., Patnaik, S. K., & Hait, N. C. (2021). Altered Expression of Secreted Mediator Genes That Mediate Aggressive Breast Cancer Metastasis to Distant Organs. Cancers, 13(11), 2641. https://doi.org/10.3390/cancers13112641








