Automatized Hepatic Tumor Volume Analysis of Neuroendocrine Liver Metastases by Gd-EOB MRI—A Deep-Learning Model to Support Multidisciplinary Cancer Conference Decision-Making
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Patient Cohorts
2.1.1. AI Development (AI dev) Cohort
2.1.2. MCC Cohort
2.2. Magnetic Resonance Imaging
2.3. Manual Segmentation
2.4. Model Training and Validation
2.5. Clinical Correlation
2.6. Statistics
3. Results
3.1. Patient Cohorts
3.1.1. AI dev Cohort
3.1.2. MCC Cohort
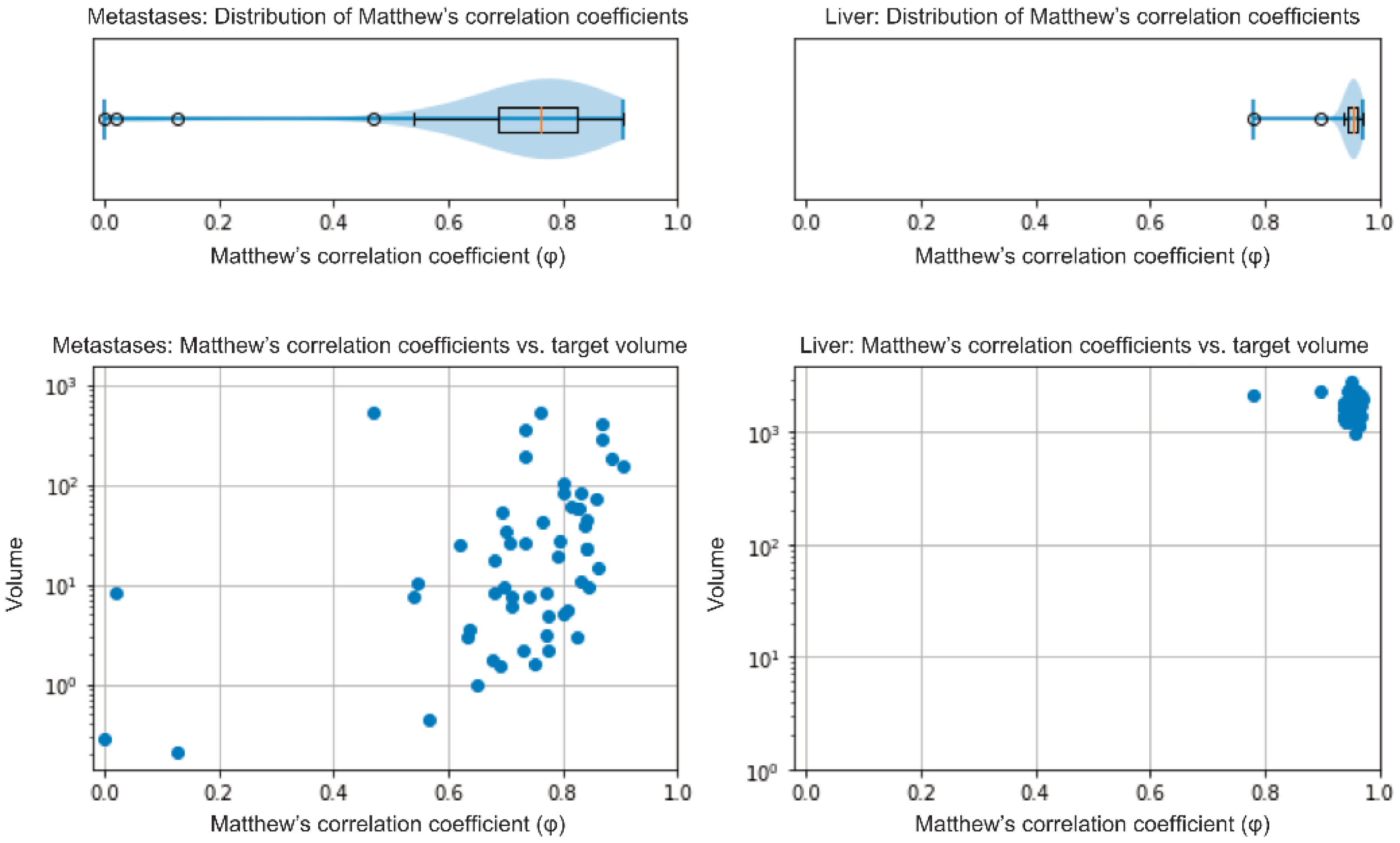
3.2. Validation of the Model
3.3. Automatized NELM Volume Analysis and Clinical Correlation (MCC Cohort)
3.4. Comparison of 3D Quantification between HBP and DWI Sequences
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Maggio, I.; Manuzzi, L.; Lamberti, G.; Ricci, A.D.; Tober, N.; Campana, D. Landscape and Future Perspectives of Immunotherapy in Neuroendocrine Neoplasia. Cancers 2020, 12, 832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, J.-H.; Zhang, Y.-Q.; Shi, S.-S.; Chen, Y.-J.; Yuan, X.-H.; Jiang, L.-M.; Wang, S.-M.; Ma, L.; Su-Sheng, S.; Feng, C.-Y.; et al. A nation-wide retrospective epidemiological study of gastroenteropancreatic neuroendocrine neoplasms in china. Oncotarget 2017, 8, 71699–71708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, T.; Igarashi, H.; Nakamura, K.; Sasano, H.; Okusaka, T.; Takano, K.; Komoto, I.; Tanaka, M.; Imamura, M.; Jensen, R.T.; et al. Epidemiological trends of pancreatic and gastrointestinal neuroendocrine tumors in Japan: A nationwide survey analysis. J. Gastroenterol. 2015, 50, 58–64. [Google Scholar] [CrossRef]
- Cives, M.; Strosberg, J.R. Gastroenteropancreatic Neuroendocrine Tumors. Cancer J. Clin. 2018, 68, 471–487. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.C.; Hassan, M.M.; Phan, A.T.; Dagohoy, C.G.; Leary, C.C.; Mares, J.E.; Abdalla, E.K.; Fleming, J.B.; Vauthey, J.-N.; Rashid, A.; et al. One Hundred Years after “Carcinoid”: Epidemiology of and Prognostic Factors for Neuroendocrine Tumors in 35,825 Cases in the United States. J. Clin. Oncol. 2008, 26, 3063–3072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Man, D.; Wu, J.; Shen, Z.; Zhu, X. Prognosis of patients with neuroendocrine tumor: A SEER database analysis. Cancer Manag. Res. 2018, 10, 5629–5638. [Google Scholar] [CrossRef] [Green Version]
- Cetinkaya, R.B.; Aagnes, B.; Thiis-Evensen, E.; Tretli, S.; Bergestuen, D.S.; Hansen, S.M. Trends in Incidence of Neuroendocrine Neoplasms in Norway: A Report of 16,075 Cases from 1993 through 2010. Neuroendocrinology 2015, 104, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Dasari, A.; Shen, C.; Halperin, D.; Zhao, B.; Zhou, S.; Xu, Y.; Shih, T.; Yao, J.C. Trends in the Incidence, Prevalence, and Survival Outcomes in Patients with Neuroendocrine Tumors in the United States. JAMA Oncol. 2017, 3, 1335–1342. [Google Scholar] [CrossRef]
- Pavel, M.; Baudin, E.; Couvelard, A.; Krenning, E.; Öberg, K.; Steinmüller, T.; Anlauf, M.; Wiedenmann, B.; Salazar, R. ENETS Consensus Guidelines for the Management of Patients with Liver and Other Distant Metastases from Neuroendocrine Neoplasms of Foregut, Midgut, Hindgut, and Unknown Primary. Neuroendocrinology 2012, 95, 157–176. [Google Scholar] [CrossRef]
- Rindi, G.; Wiedenmann, B. Neuroendocrine neoplasia of the gastrointestinal tract revisited: Towards precision medicine. Nat. Rev. Endocrinol. 2020, 16, 590–607. [Google Scholar] [CrossRef] [PubMed]
- Saeed, A.; Buell, J.F.; Kandil, E. Surgical treatment of liver metastases in patients with neuroendocrine tumors. Ann. Transl. Med. 2013, 1, 6. [Google Scholar]
- Schwartz, L.H.; Litière, S.; De Vries, E.; Ford, R.; Gwyther, S.; Mandrekar, S.; Shankar, L.; Bogaerts, J.; Chen, A.; Dancey, J.; et al. RECIST 1.1—Update and clarification: From the RECIST committee. Eur. J. Cancer 2016, 62, 132–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnold, C.N.; Nagasaka, T.; Goel, A.; Scharf, I.; Grabowski, P.; Sosnowski, A.; Schmitt-Gräff, A.; Boland, C.R.; Arnold, R.; Blum, H.E. Molecular characteristics and predictors of survival in patients with malignant neuroendocrine tumors. Int. J. Cancer 2008, 123, 1556–1564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dromain, C.; De Baere, T.; Lumbroso, J.; Caillet, H.; Laplanche, A.; Boige, V.; Ducreux, M.; Duvillard, P.; Elias, D.; Schlumberger, M.; et al. Detection of Liver Metastases from Endocrine Tumors: A Prospective Comparison of Somatostatin Receptor Scintigraphy, Computed Tomography, and Magnetic Resonance Imaging. J. Clin. Oncol. 2005, 23, 70–78. [Google Scholar] [CrossRef] [Green Version]
- Sundin, A.; Arnold, R.; Baudin, E.; Cwikla, J.B.; Eriksson, B.; Fanti, S.; Fazio, N.; Giammarile, F.; Hicks, R.J.; Kjaer, A.; et al. ENETS Consensus Guidelines for the Standards of Care in Neuroendocrine Tumors: Radiological, Nuclear Medicine and Hybrid Imaging. Neuroendocrinology 2017, 105, 212–244. [Google Scholar] [CrossRef]
- Ronot, M.; Clift, A.K.; Baum, R.P.; Singh, A.; Kulkarni, H.R.; Frilling, A.; Vilgrain, V. Morphological and Functional Imaging for Detecting and Assessing the Resectability of Neuroendocrine Liver Metastases. Neuroendocrinology 2018, 106, 74–88. [Google Scholar] [CrossRef] [PubMed]
- Vreugdenburg, T.D.; Ma, N.; Duncan, J.K.; Riitano, D.; Cameron, A.L.; Maddern, G.J. Comparative diagnostic accuracy of hepatocyte-specific gadoxetic acid (Gd-EOB-DTPA) enhanced MR imaging and contrast enhanced CT for the detection of liver metastases: A systematic review and meta-analysis. Int. J. Colorectal Dis. 2016, 31, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Giesel, F.; Kratochwil, C.; Mehndiratta, A.; Wulfert, S.; Moltz, J.; Zechmann, C.; Kauczor, H.; Haberkorn, U.; Ley, S. Comparison of neuroendocrine tumor detection and characterization using DOTATOC-PET in correlation with contrast enhanced CT and delayed contrast enhanced MRI. Eur. J. Radiol. 2012, 81, 2820–2825. [Google Scholar] [CrossRef]
- Karaosmanoglu, A.D.; Onur, M.R.; Ozmen, M.N.; Akata, D.; Karcaaltincaba, M. Magnetic Resonance Imaging of Liver Metastasis. Semin. Ultrasound CT MRI 2016, 37, 533–548. [Google Scholar] [CrossRef]
- Feuerlein, S.; Gupta, R.T.; Boll, D.T.; Merkle, E.M. Hepatocellular MR contrast agents: Enhancement characteristics of liver parenchyma and portal vein after administration of gadoxetic acid in comparison to gadobenate dimeglumine. Eur. J. Radiol. 2012, 81, 2037–2041. [Google Scholar] [CrossRef]
- D’Assignies, G.; Fina, P.; Bruno, O.; Vullierme, M.-P.; Tubach, F.; Paradis, V.; Sauvanet, A.; Ruszniewski, P.; Vilgrain, V. High Sensitivity of Diffusion-weighted MR Imaging for the Detection of Liver Metastases from Neuroendocrine Tumors: Comparison with T2-weighted and Dynamic Gadolinium-enhanced MR Imaging. Radiology 2013, 268, 390–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sankowski, A.J.; Ćwikla, J.B.; Nowicki, M.L.; Chaberek, S.; Pech, M.; Lewczuk, A.; Walecki, J. The clinical value of MRI using single-shot echoplanar DWI to identify liver involvement in patients with advanced gastroenteropancreatic-neuroendocrine tumors (GEP-NETs), compared to FSE T2 and FFE T1 weighted image after i.v. Gd-EOB-DTPA contrast enhancement. Med. Sci. Monit. 2012, 18, MT33–MT40. [Google Scholar] [CrossRef]
- Minon, M.; Soriano, C.; Morland, D.; Walter, T.; Lepage, C.; Tabarin, A.; DeBlock, M.; Rousset, P.; Barbe, C.; Hoeffel, C.; et al. Prospective comparison of whole-body MRI with diffusion-weighted and conventional imaging for the follow-up of neuroendocrine tumors. Endocrine 2019, 67, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Hayoz, R.; Vietti-Violi, N.; Duran, R.; Knebel, J.-F.; LeDoux, J.-B.; Dromain, C. The combination of hepatobiliary phase with Gd-EOB-DTPA and DWI is highly accurate for the detection and characterization of liver metastases from neuroendocrine tumor. Eur. Radiol. 2020, 30, 6593–6602. [Google Scholar] [CrossRef]
- Danet, I.-M.; Semelka, R.C.; Leonardou, P.; Braga, L.; Vaidean, G.; Woosley, J.T.; Kanematsu, M. Spectrum of MRI Appearances of Untreated Metastases of the Liver. Am. J. Roentgenol. 2003, 181, 809–817. [Google Scholar] [CrossRef]
- Khosa, F.; Khan, A.N.; Eisenberg, R.L. Hypervascular Liver Lesions on MRI. Am. J. Roentgenol. 2011, 197, W204–W220. [Google Scholar] [CrossRef]
- Luersen, G.F.; Wei, W.; Tamm, E.P.; Bhosale, P.R.; Szklaruk, J. Evaluation of Magnetic Resonance (MR) Biomarkers for Assessment of Response with Response Evaluation Criteria in Solid Tumors: Comparison of the Measurements of Neuroendocrine Tumor Liver Metastases (NETLM) with Various MR Sequences and at Multiple Phases of Contrast Administration. J. Comput. Assist. Tomogr. 2016, 40, 717–722. [Google Scholar] [CrossRef] [Green Version]
- Liang, H.; Tsui, B.Y.; Ni, H.; Valentim, C.C.S.; Baxter, S.L.; Liu, G.; Cai, W.; Kermany, D.S.; Sun, X.; Chen, J.; et al. Evaluation and accurate diagnoses of pediatric diseases using artificial intelligence. Nat. Med. 2019, 25, 433–438. [Google Scholar] [CrossRef]
- Rauschecker, A.M.; Rudie, J.D.; Xie, L.; Wang, J.; Duong, M.T.; Botzolakis, E.J.; Kovalovich, A.M.; Egan, J.; Cook, T.C.; Bryan, R.N.; et al. Artificial Intelligence System Approaching Neuroradiologist-level Differential Diagnosis Accuracy at Brain MRI. Radiology 2020, 295, 626–637. [Google Scholar] [CrossRef]
- Tschandl, P.; Codella, N.; Akay, B.N.; Argenziano, G.; Braun, R.P.; Cabo, H.; Gutman, D.; Halpern, A.; Helba, B.; Hofmann-Wellenhof, R.; et al. Comparison of the accuracy of human readers versus machine-learning algorithms for pigmented skin lesion classification: An open, web-based, international, diagnostic study. Lancet Oncol. 2019, 20, 938–947. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, C.J.P.; Jiang, B.; Chen, J.; Song, J.; Liu, Z.; He, Z.; Krittanawong, C.; Fang, P.-H.; Ming, W.-K. Artificial Intelligence Versus Clinicians in Disease Diagnosis: Systematic Review. JMIR Med. Inform. 2019, 7, e10010. [Google Scholar] [CrossRef] [Green Version]
- Ardila, D.; Kiraly, A.P.; Bharadwaj, S.; Choi, B.; Reicher, J.J.; Peng, L.; Tse, D.; Etemadi, M.; Ye, W.; Corrado, G.; et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med. 2019, 25, 954–961. [Google Scholar] [CrossRef] [PubMed]
- Sumkin, J.H.; Berg, W.A.; Carter, G.J.; Bandos, A.I.; Chough, D.M.; Ganott, M.A.; Hakim, C.M.; Kelly, A.E.; Zuley, M.L.; Houshmand, G.; et al. Diagnostic Performance of MRI, Molecular Breast Imaging, and Contrast-enhanced Mammography in Women with Newly Diagnosed Breast Cancer. Radiology 2019, 293, 531–540. [Google Scholar] [CrossRef]
- Azer, S.A. Deep learning with convolutional neural networks for identification of liver masses and hepatocellular carcinoma: A systematic review. World J. Gastrointest. Oncol. 2019, 11, 1218–1230. [Google Scholar] [CrossRef]
- Zhou, L.-Q.; Wang, J.-Y.; Yu, S.-Y.; Wu, G.-G.; Wei, Q.; Deng, Y.-B.; Wu, X.-L.; Cui, X.-W.; Dietrich, C.F. Artificial intelligence in medical imaging of the liver. World J. Gastroenterol. 2019, 25, 672–682. [Google Scholar] [CrossRef] [PubMed]
- Kaya, B.; Koc, Z. Diffusion-weighted MRI and optimal b-value for characterization of liver lesions. Acta Radiol. 2014, 55, 532–542. [Google Scholar] [CrossRef]
- Nolden, M.; Zelzer, S.; Seitel, A.; Wald, D.; Müller, M.; Franz, A.M.; Maleike, D.; Fangerau, M.; Baumhauer, M.; Maier-Hein, L.; et al. The Medical Imaging Interaction Toolkit: Challenges and advances. Int. J. Comput. Assist. Radiol. Surg. 2013, 8, 607–620. [Google Scholar] [CrossRef]
- MIC-DKFZ nnUNet. Available online: https://github.com/MIC-DKFZ/nnUNet (accessed on 1 October 2020).
- Isensee, F.; Jäger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. Automated Design of Deep Learning Methods for Biomedical Image Segmentation. arXiv 2019, arXiv:1904.08128v2. [Google Scholar]
- Chicco, D.; Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020, 21, 6. [Google Scholar] [CrossRef] [Green Version]
- Goehler, A.; Hsu, T.-M.H.; Lacson, R.; Gujrathi, I.; Hashemi, R.; Chlebus, G.; Szolovits, P.; Khorasani, R. Three-Dimensional Neural Network to Automatically Assess Liver Tumor Burden Change on Consecutive Liver MRIs. J. Am. Coll. Radiol. 2020, 17, 1475–1484. [Google Scholar] [CrossRef] [PubMed]
- Larouche, V.; Akirov, A.; AlShehri, S.; Ezzat, S. Management of Small Bowel Neuroendocrine Tumors. Cancers 2019, 11, 1395. [Google Scholar] [CrossRef] [Green Version]
- Akirov, A.; Larouche, V.; AlShehri, S.; Asa, S.L.; Ezzat, S. Treatment Options for Pancreatic Neuroendocrine Tumors. Cancers 2019, 11, 828. [Google Scholar] [CrossRef] [Green Version]
- Tsurusaki, M.; Sofue, K.; Murakami, T. Current evidence for the diagnostic value of gadoxetic acid-enhanced magnetic resonance imaging for liver metastasis. Hepatol. Res. 2016, 46, 853–861. [Google Scholar] [CrossRef]
- Morse, B.; Jeong, D.; Thomas, K.; Diallo, D.; Strosberg, J.R. Magnetic Resonance Imaging of Neuroendocrine Tumor Hepatic Metastases. Pancreas 2017, 46, 1219–1224. [Google Scholar] [CrossRef]
- Tirumani, S.H.; Jagannathan, J.P.; Braschi-Amirfarzan, M.; Qin, L.; Balthazar, P.; Ramaiya, N.H.; Shinagare, A.B. Value of hepatocellular phase imaging after intravenous gadoxetate disodium for assessing hepatic metastases from gastroenteropancreatic neuroendocrine tumors: Comparison with other MRI pulse sequences and with extracellular agent. Abdom. Radiol. 2018, 43, 2329–2339. [Google Scholar] [CrossRef]
- Grieser, C.; Denecke, T.; Rothe, J.-H.; Geisel, D.; Stelter, L.; Walter, T.C.; Seehofer, D.; Steffen, I.G. Gd-EOB enhanced MRI T1-weighted 3D-GRE with and without elevated flip angle modulation for threshold-based liver volume segmentation. Acta Radiol. 2014, 56, 1419–1427. [Google Scholar] [CrossRef]
- Kahn, J.; Posch, H.; Steffen, I.G.; Geisel, D.; Bauknecht, C.; Liebig, T.; Denecke, T. Is There Long-term Signal Intensity Increase in the Central Nervous System on T1-weighted Images after MR Imaging with the Hepatospecific Contrast Agent Gadoxetic Acid? A Cross-sectional Study in 91 Patients. Radiology 2017, 282, 708–716. [Google Scholar] [CrossRef] [PubMed]
- Lestra, T.; Kanagaratnam, L.; Mulé, S.; Janvier, A.; Brixi, H.; Cadiot, G.; Dohan, A.; Hoeffel, C. Measurement variability of liver metastases from neuroendocrine tumors on different magnetic resonance imaging sequences. Diagn. Interv. Imaging 2018, 99, 73–81. [Google Scholar] [CrossRef]
- Lavelle, L.; O’Neill, A.; McMahon, C.; Cantwell, C.; Heffernan, E.; Malone, D.; Daly, L.; Skehan, S. Is diffusion-weighted MRI sufficient for follow-up of neuroendocrine tumour liver metastases? Clin. Radiol. 2016, 71, 863–868. [Google Scholar] [CrossRef]
- Kaye, E.A.; Cornelis, F.H.; Petre, E.N.; Tyagi, N.; Shady, W.; Shi, W.; Zhang, Z.; Solomon, S.B.; Sofocleous, C.T.; Durack, J.C. Volumetric 3D assessment of ablation zones after thermal ablation of colorectal liver metastases to improve prediction of local tumor progression. Eur. Radiol. 2019, 29, 2698–2705. [Google Scholar] [CrossRef]
- Sakakibara, M.; Ohkawa, K.; Katayama, K.; Imanaka, K.; Ishihara, A.; Hasegawa, N.; Kimura, H. Three-Dimensional Registration of Images Obtained before and after Radiofrequency Ablation of Hepatocellular Carcinoma to Assess Treatment Adequacy. Am. J. Roentgenol. 2014, 202, W487–W495. [Google Scholar] [CrossRef]
- Wang, L.; Tan, J.; Ge, Y.; Tao, X.; Cui, Z.; Fei, Z.; Lu, J.; Zhang, H.; Pan, Z. Assessment of liver metastases radiomic feature reproducibility with deep-learning-based semi-automatic segmentation software. Acta Radiol. 2021, 62, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Solis-Hernandez, M.P.; Del Valle, A.F.; Carmona-Bayonas, A.; Garcia-Carbonero, R.; Custodio, A.; Benavent, M.; Gordoa, T.A.; Nuñez-Valdovino, B.; Canovas, M.S.; Matos, I.; et al. Evaluating radiological response in pancreatic neuroendocrine tumours treated with sunitinib: Comparison of Choi versus RECIST criteria (CRIPNET_ GETNE1504 study). Br. J. Cancer 2019, 121, 537–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamm, C.A.; Wang, C.J.; Savic, L.J.; Ferrante, M.; Schobert, I.; Schlachter, T.; Lin, M.; Duncan, J.S.; Weinreb, J.C.; Chapiro, J.; et al. Deep learning for liver tumor diagnosis part I: Development of a convolutional neural network classifier for multi-phasic MRI. Eur. Radiol. 2019, 29, 3338–3347. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.J.; Hamm, C.A.; Savic, L.J.; Ferrante, M.; Schobert, I.; Schlachter, T.; Lin, M.; Weinreb, J.C.; Duncan, J.S.; Chapiro, J.; et al. Deep learning for liver tumor diagnosis part II: Convolutional neural network interpretation using radiologic imaging features. Eur. Radiol. 2019, 29, 3348–3357. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wang, J.; Ng, C.W.; Ma, Y.; Mo, S.; Fong, E.L.S.; Xing, J.; Song, Z.; Xie, Y.; Si, K.; et al. Deep learning enables automated scoring of liver fibrosis stages. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef]
- Gehan, E.A.; Tefft, M.C. Will There Be Resistance to the RECIST (Response Evaluation Criteria in Solid Tumors)? J. Natl. Cancer Inst. 2000, 92, 179–181. [Google Scholar] [CrossRef]
- Lamarca, A.; Barriuso, J.; Kulke, M.; Borbath, I.; Lenz, H.-J.; Raoul, J.L.; Meropol, N.J.; Lombard-Bohas, C.; Posey, J.; Faivre, S.; et al. Determination of an optimal response cut-off able to predict progression-free survival in patients with well-differentiated advanced pancreatic neuroendocrine tumours treated with sunitinib: An alternative to the current RECIST-defined response. Br. J. Cancer 2017, 118, 181–188. [Google Scholar] [CrossRef] [Green Version]
- Lamarca, A.; Crona, J.; Ronot, M.; Opalinska, M.; Lopez, C.L.; Pezzutti, D.; Najran, P.; Carvhalo, L.; Bezerra, R.O.F.; Borg, P.; et al. Value of Tumor Growth Rate (TGR) as an Early Biomarker Predictor of Patients’ Outcome in Neuroendocrine Tumors (NET)—The GREPONET Study. Oncology 2019, 24, e1082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, M.; Kuhl, C.K.; Engelke, H.; Bettermann, G.; Keil, S. Factors That Drive Heterogeneity of Response-to-Treatment of Different Metastatic Deposits Within the Same Patients as Measured by RECIST 1.1 Analyses. Acad. Radiol. 2020. [Google Scholar] [CrossRef]
- Rothe, J.H.; Steffen, I.G.; Lehmkuhl, L.; Grieser, C.; Mußler, A.; Schnapauff, D.; Stelter, L.; Denecke, T. Volume Measurement of Liver Metastases Using Multidetector Computed Tomography: Comparison of Lesion Diameter and Volume segmentation—A Phantom Study. In RöFo—Fortschritte auf dem Gebiet der Röntgenstrahlen und der bildgebenden Verfahren; Thieme: Stuttgart, Germany, 2010; Volume 182, pp. 1082–1090. [Google Scholar] [CrossRef]
- Palazzo, M.; Lombard-Bohas, C.; Cadiot, G.; Matysiak-Budnik, T.; Rebours, V.; Vullierme, M.-P.; Couvelard, A.; Hentic, O.; Ruszniewski, P. Ki67 proliferation index, hepatic tumor load, and pretreatment tumor growth predict the antitumoral efficacy of lanreotide in patients with malignant digestive neuroendocrine tumors. Eur. J. Gastroenterol. Hepatol. 2013, 25, 232–238. [Google Scholar] [CrossRef]
- Beleù, A.; Rizzo, G.; De Robertis, R.; Drudi, A.; Aluffi, G.; Longo, C.; Sarno, A.; Cingarlini, S.; Capelli, P.; Landoni, L.; et al. Liver Tumor Burden in Pancreatic Neuroendocrine Tumors: CT Features and Texture Analysis in the Prediction of Tumor Grade and 18F-FDG Uptake. Cancers 2020, 12, 1486. [Google Scholar] [CrossRef] [PubMed]
- Cieciera, M.; Kratochwil, C.; Moltz, J.; Kauczor, H.-U.; Holland-Letz, T.; Choyke, P.; Mier, W.; Haberkorn, U.; Giesel, F.L. Semi-automatic 3D-volumetry of liver metastases from neuroendocrine tumors to improve combination therapy with 177Lu-DOTATOC and 90Y-DOTATOC. Diagn. Interv. Radiol. 2016, 22, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Kuhl, C.K. RECIST Needs Revision: A Wake-up Call for Radiologists. Radiology 2019, 292, 110–111. [Google Scholar] [CrossRef]
- Keating, N.L.; Landrum, M.B.; Lamont, E.B.; Bozeman, S.R.; Shulman, L.N.; McNeil, B.J. Tumor Boards and the Quality of Cancer Care. J. Natl. Cancer Inst. 2012, 105, 113–121. [Google Scholar] [CrossRef] [Green Version]
- Croke, J.M.; El-Sayed, S. Multidisciplinary Management of Cancer Patients: Chasing a Shadow or Real Value? An Overview of the Literature. Curr. Oncol. 2012, 19, 232–238. [Google Scholar] [CrossRef] [Green Version]
- Hofland, J.; Kaltsas, G.; De Herder, W.W. Advances in the Diagnosis and Management of Well-Differentiated Neuroendocrine Neoplasms. Endocr. Rev. 2020, 41, 371–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Feature | Subgroups | AI dev Cohort | MCC Cohort | p-Value |
|---|---|---|---|---|
| Number of patients | - | 149 | 33 | - |
| Number of scans | - | 278 (of 398) | 66 | - |
| Gender (M: F) | - | 66:83 | 18:15 | 0.285 |
| Age (median) | - | 58.92 (48.86–66.38) | 56.45 (48.62–67.40) | 0.631 |
| Ki67 (%, median) | - | 5.0 (2.0–10.0) | 7.0 (2.5–13.0) | 0.139 |
| Primary site | 0.001 | |||
| Pancreas Ileum Other | 64 (43.0%) 76 (51.0%) 9 (6.0%) | 12 (36.4%) 12 (36.4%) 9 (27.2%) | ||
| Grading | 0.406 | |||
| 1 2 3 | 52 (34.9%) 85 (57.0%) 12 (8.1%) | 8 (24.2%) 23 (69.7%) 2 (6.1%) | ||
| NET: NEC | - | 144:5 | 31:2 | 0.612 |
| Functionality | yes no | 42 (28.2%) 107 (71.8%) | 12 (36.4%) 21 (63.6%) | 0.401 |
| Extrahepatic metastases | - | 92 (61.7%) | 27 (81.8%) | 0.042 |
| Somatostatin receptor (SR) | 0.004 | |||
| pos neg | 110 (73.8%) * 37 (24.9%) * | 32 (97.0%) 1 (3.0%) |
| Variable | Overall | Significance | |||
|---|---|---|---|---|---|
| BL | FU | ||||
| n | 33 | 33 | - | ||
| NELM (cm3) | 23.48 (10.45–113.17) | 86.93 (12.08–204.50) | - | ||
| Liver (cm3) | 1582.23 (1336.25–2030.03) | 1716.75 (1477.12–2092.94) | - | ||
| HTL (vol.-%) | 1.57 (0.55–7.05) | 5.93 (0.99–11.74) | - | ||
| ΔabsNELM (%) | 14.70 (0.76–96.35) | - | |||
| ΔabsHTL (%) | 0.98 (−0.03–5.41) | - | |||
| ΔrelNELM (%) | 58.51 (3.93–245.64) | - | |||
| ΔrelHTL (%) | 64.97 (−3.44–223.31) | - | |||
| Therapy Success | Therapy Failure | ||||
| BL | FU | BL | FU | ||
| n | 16 | 16 | 17 | 17 | - |
| NELM (cm3) | 75.45 (12.35–141.65) | 66.78 (11.64–167.82) | 19.15 (7.04–78.44) | 86.93 (24.40–253.32) | - |
| Liver (cm3) | 1692.26 (1475.09–2061.63) | 1725.30 (1471.78–2130.28) | 1580.35 (1290.13–1902.53) | 1716.75 (1451.10–2106.81) | - |
| HTL (vol.-%) | 4.41 (0.87–7.83) | 3.75 (0.75–8.88) | 1.46 (0.34–5.97) | 5.93 (1.47–16.78) | - |
| ΔabsNELM (%) | 0.76 (−18.07–39.32) | 59.70 (16.49–156.59) | p < 0.001 | ||
| ΔabsHTL (%) | −0.03 (−1.28–0.23) | 4.94 (1.07–9.78) | p < 0.001 | ||
| ΔrelNELM (%) | 3.93 (−15.75–10.36) | 242.68 (124.56–463.87) | p < 0.001 | ||
| ΔrelHTL (%) | −3.45 (−18.11–11.15) | 204.49 (109.39–490.19) | p < 0.001 | ||
| Case # | Baseline | Follow-up | Response Variables | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | Liver Volume (cm3) | NELM Volume (cm3) | HTL (vol.-%) | Liver Volume (cm3) | NELM Volume (cm3) | HTL (vol.-%) | MCC | ΔabsNELM (cm3) | ΔabsHTL (vol.-%) | ΔrelNELM (%) | ΔrelHTL (%) |
| 0001 | 1649.3 | 100.5 | 6.5 | 1796.0 | 232.7 | 14.9 | PD | 132.2 | 8.4 | 131.6 | 129.5 |
| 0002 | 1582.2 | 69.5 | 4.6 | 1783.7 | 242.4 | 15.7 | PD | 172.9 | 11.1 | 248.6 | 242.1 |
| 0003 | 1516.3 | 23.5 | 1.6 | 1766.8 | 160.7 | 10.0 | PD | 137.2 | 8.4 | 584.3 | 536.1 |
| 0004 | 1476.7 | 2.5 | 0.2 | 1487.8 | 2.0 | 0.1 | SD | −0.5 | −0.0 | −20.2 | −20.8 |
| 0005 | 1304.2 | 9.5 | 0.7 | 1350.2 | 10.5 | 0.8 | SD | 0.9 | 0.1 | 9.8 | 6.1 |
| 0006 | 1247.6 | 87.3 | 7.5 | 1656.0 | 308.8 | 22.9 | PD | 221.5 | 15.4 | 253.5 | 204.5 |
| 0007 | 1092.4 | 0.6 | 0.1 | 1111.6 | 7.0 | 0.6 | PD | 6.4 | 0.6 | 1080.0 | 1066.3 |
| 0008 | 1597.5 | 137.5 | 9.4 | 1977.1 | 299.1 | 17.8 | PD | 161.7 | 8.4 | 117.6 | 89.3 |
| 0009 | 1474.5 | 21.2 | 1.5 | 1567.0 | 22.1 | 1.4 | SD | 0.9 | −0.0 | 4.2 | −2.0 |
| 0010 | 1579.1 | 3.3 | 0.2 | 1610.6 | 6.3 | 0.4 | PD | 3.0 | 0.2 | 91.9 | 88.5 |
| 0011 | 2067.7 | 148.0 | 7.7 | 2167.6 | 124.1 | 6.1 | SD | −23.9 | −1.6 | −16.1 | −21.2 |
| 0012 | 1959.7 | 46.5 | 2.4 | 1689.6 | 11.8 | 0.7 | PR | −34.8 | −1.7 | −74.7 | −71.1 |
| 0013 | 1695.2 | 111.4 | 7.0 | 1817.3 | 173.0 | 10.5 | SD | 61.6 | 3.5 | 55.3 | 49.5 |
| 0014 | 1332.6 | 19.2 | 1.5 | 3216.9 | 883.6 | 37.9 | PD | 864.5 | 36.4 | 4513.6 | 2497.2 |
| 0015 | 2209.1 | 9.4 | 0.4 | 2236.5 | 47.0 | 2.2 | PD | 37.6 | 1.7 | 398.2 | 400.5 |
| 0016 | 2326.4 | 13.0 | 0.6 | 2421.6 | 12.4 | 0.5 | SD | −0.6 | −0.1 | −4.5 | −8.3 |
| 0017 | 969.3 | 12.1 | 1.3 | 972.4 | 11.6 | 1.2 | SD | −0.6 | −0.1 | −4.5 | −4.9 |
| 0018 | 2523.2 | 17.7 | 0.7 | 2364.0 | 43.3 | 1.9 | PD | 25.5 | 1.2 | 144.2 | 163.6 |
| 0019 | 1580.4 | 54.8 | 3.6 | 1552.8 | 86.9 | 5.9 | PD | 32.1 | 2.3 | 58.5 | 65.0 |
| 0020 | 1703.4 | 244.9 | 16.8 | 1572.5 | 209.2 | 15.3 | SD | −35.7 | −1.5 | −14.9 | −8.6 |
| 0021 | 1575.0 | 114.9 | 7.9 | 1801.2 | 119.2 | 7.1 | SD | 4.2 | −0.8 | 3.7 | −10.0 |
| 0022 | 1082.3 | 14.0 | 1.3 | 1071.6 | 14.6 | 1.4 | SD | 0.6 | 0.1 | 4.5 | 5.7 |
| 0023 | 2016.6 | 133.1 | 7.1 | 2303.4 | 264.3 | 13.0 | PD | 131.1 | 5.9 | 98.5 | 83.4 |
| 0024 | 1788.5 | 4.7 | 0.3 | 1639.5 | 22.9 | 1.4 | PD | 18.3 | 1.2 | 393.3 | 444.3 |
| 0025 | 2043.5 | 122.5 | 6.4 | 2018.2 | 152.3 | 8.2 | SD | 29.8 | 1.8 | 24.3 | 28.0 |
| 0026 | 2416.2 | 180.8 | 8.1 | 2389.9 | 199.9 | 9.1 | SD | 19.1 | 1.0 | 10.5 | 12.8 |
| 0027 | 1339.9 | 11.3 | 0.9 | 1297.2 | 71.0 | 5.8 | PD | 59.7 | 4.9 | 529.6 | 582.1 |
| 0028 | 2653.8 | 549.0 | 26.1 | 2386.0 | 199.4 | 9.1 | PR | −349.6 | −17.0 | −63.7 | −65.0 |
| 0029 | 1477.6 | 7.2 | 0.5 | 1466.5 | 10.8 | 0.7 | SD | 3.5 | 0.3 | 49.0 | 50.5 |
| 0030 | 2054.2 | 11.2 | 0.6 | 1716.8 | 25.9 | 1.5 | PD | 14.7 | 1.0 | 131.5 | 179.8 |
| 0031 | 1689.3 | 104.4 | 6.6 | 1761.0 | 111.5 | 6.8 | SD | 7.1 | 0.2 | 6.8 | 2.6 |
| 0032 | 1207.2 | 62.4 | 5.5 | 1322.4 | 214.0 | 19.3 | PD | 151.5 | 13.9 | 242.7 | 253.9 |
| 0033 | 1146.0 | 2.1 | 0.2 | 1349.4 | 6.7 | 0.5 | PD | 4.6 | 0.3 | 222.3 | 174.6 |
| Variable | HBP | DWI | Significance |
|---|---|---|---|
| NELM volume (cm3) | 63.24 (12.12–174.23) | 76.28 (12.61–182.48) | p = 0.002 |
| Liver volume (cm3) | 1659.28 (1387.73–2052.00) | 1595.00 (1324.17–1977.54) | p < 0.001 |
| HTL (vol %) | 4.05 (0.76–9.23) | 5.45 (0.88–11.49) | p < 0.001 |
| ΔabsNELM (cm3) | 19.57 (17.27–132.52) | 30.06 (18.91–142.13) | p = 0.072 |
| ΔrelNELM (%) | 107.76 (5.28–245.04) | 78.35 (11.22–221.21) | p = 0.719 |
| ΔabsHTL (vol %) | 1.20 (−0.01–8.87) | 1.25 (0.10–10.47) | p = 0.151 |
| ΔrelHTL (%) | 111.36 (−0.36–254.49) | 67.76 (4.20–198.88) | p = 0.151 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fehrenbach, U.; Xin, S.; Hartenstein, A.; Auer, T.A.; Dräger, F.; Froböse, K.; Jann, H.; Mogl, M.; Amthauer, H.; Geisel, D.; et al. Automatized Hepatic Tumor Volume Analysis of Neuroendocrine Liver Metastases by Gd-EOB MRI—A Deep-Learning Model to Support Multidisciplinary Cancer Conference Decision-Making. Cancers 2021, 13, 2726. https://doi.org/10.3390/cancers13112726
Fehrenbach U, Xin S, Hartenstein A, Auer TA, Dräger F, Froböse K, Jann H, Mogl M, Amthauer H, Geisel D, et al. Automatized Hepatic Tumor Volume Analysis of Neuroendocrine Liver Metastases by Gd-EOB MRI—A Deep-Learning Model to Support Multidisciplinary Cancer Conference Decision-Making. Cancers. 2021; 13(11):2726. https://doi.org/10.3390/cancers13112726
Chicago/Turabian StyleFehrenbach, Uli, Siyi Xin, Alexander Hartenstein, Timo Alexander Auer, Franziska Dräger, Konrad Froböse, Henning Jann, Martina Mogl, Holger Amthauer, Dominik Geisel, and et al. 2021. "Automatized Hepatic Tumor Volume Analysis of Neuroendocrine Liver Metastases by Gd-EOB MRI—A Deep-Learning Model to Support Multidisciplinary Cancer Conference Decision-Making" Cancers 13, no. 11: 2726. https://doi.org/10.3390/cancers13112726
APA StyleFehrenbach, U., Xin, S., Hartenstein, A., Auer, T. A., Dräger, F., Froböse, K., Jann, H., Mogl, M., Amthauer, H., Geisel, D., Denecke, T., Wiedenmann, B., & Penzkofer, T. (2021). Automatized Hepatic Tumor Volume Analysis of Neuroendocrine Liver Metastases by Gd-EOB MRI—A Deep-Learning Model to Support Multidisciplinary Cancer Conference Decision-Making. Cancers, 13(11), 2726. https://doi.org/10.3390/cancers13112726







