Modulation of Early Neutrophil Granulation: The Circulating Tumor Cell-Extravesicular Connection in Pancreatic Ductal Adenocarcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Derived Tumor Cell Lines
2.2. EV Isolation
2.3. Transmission Electron Microscopy (TEM)
2.4. Western Blots
2.5. shRNA Mediated Silencing of TSG101
2.6. Antibody Arrays
2.7. Preparation of Samples for LC-MS
2.8. LC-MS/MS Analysis
2.9. Maxquant Label Free Quantitation
2.10. Neutrophil Isolation
2.11. EV-Neutrophil Co-Culture and Transcriptome Analyses
2.12. Neutrophil Granulation and NETosis Analysis
2.13. Real-Time PCR (qPCR) and Data Analyses
2.14. Enrichment Analyses
3. Results
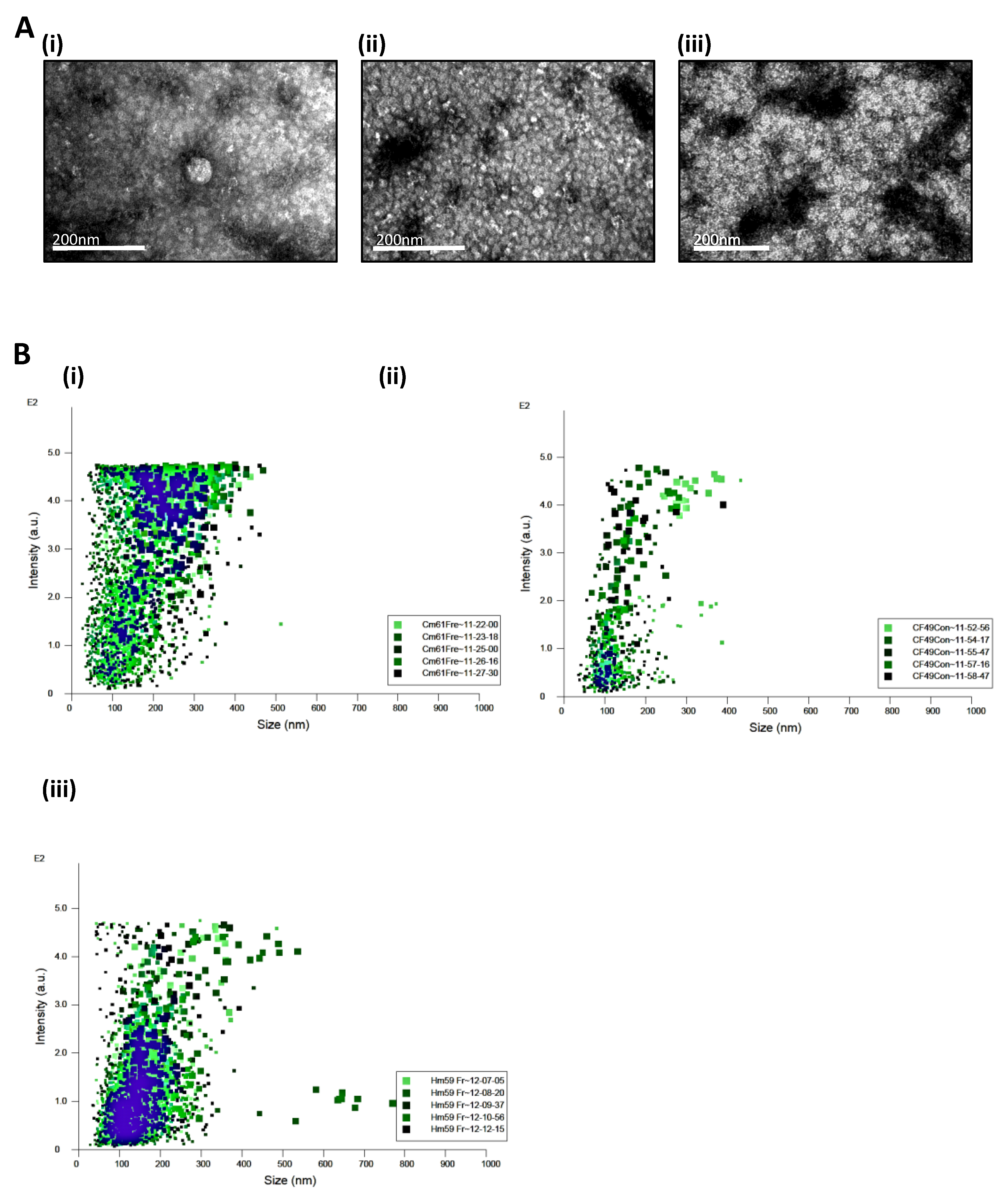
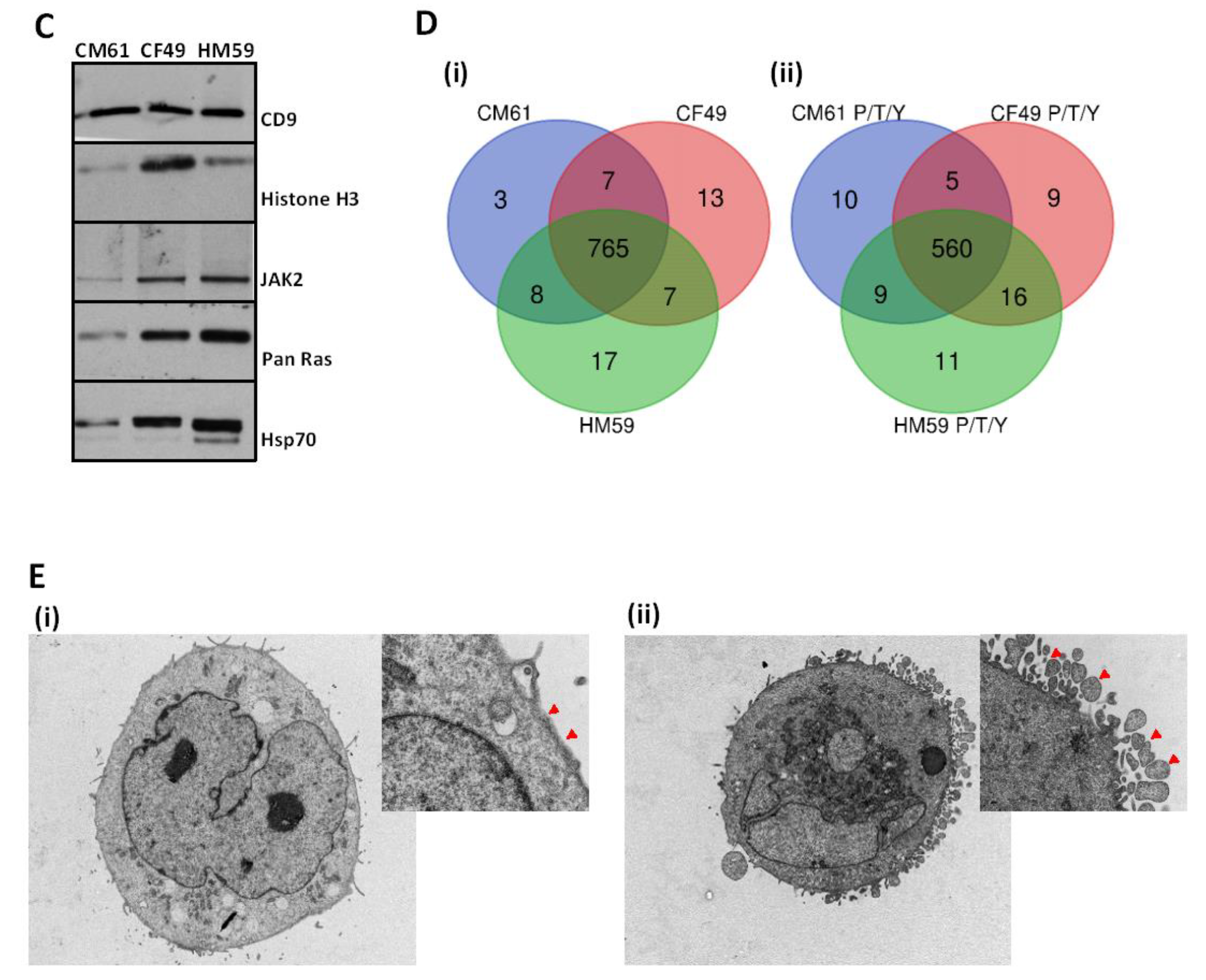
3.1. Circulating Tumor Cells Secrete Extracellular Vesicles
3.2. Secretion of EVs from the CTCs Is an Active Process
3.3. CTC EVome Contains Both a Protein and Phosphoprotein Cargo
3.4. Active Signaling Signature Is Conserved in the EV Cargo
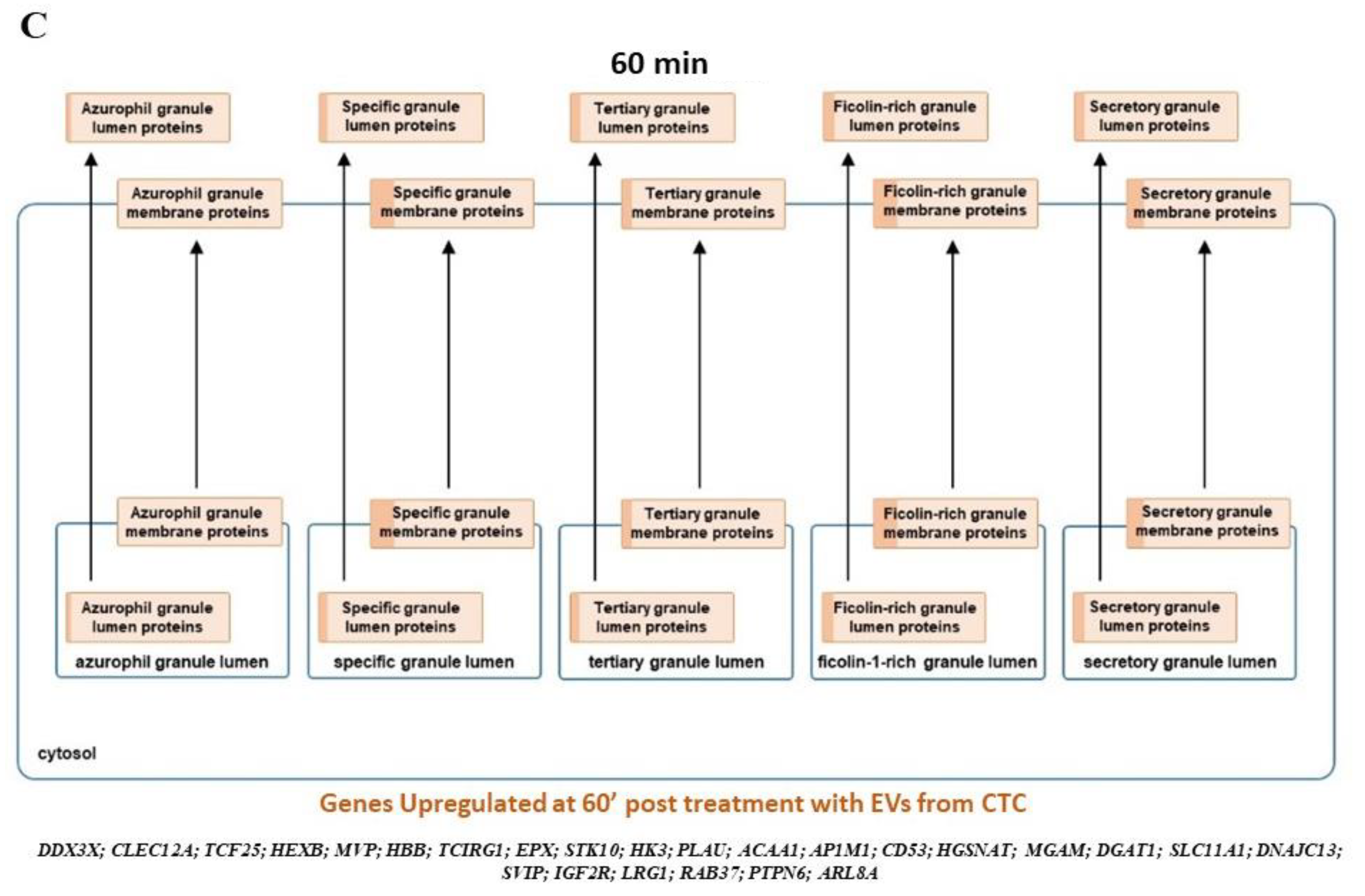
3.5. Neutrophil Degranulation and Reteplase Activity Pathways Are Modulated on Exposure to CTC EVs in Neutrophils
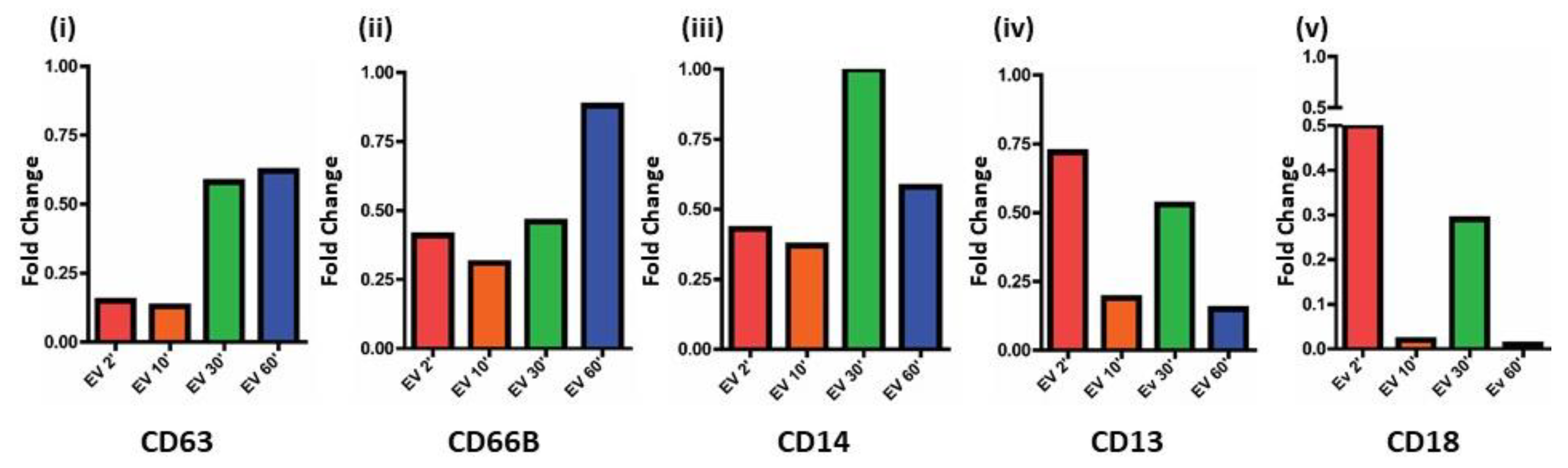
3.6. CTC EVs Modulate Granule Mobilization on Mature Human Neutrophils
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef]
- Bellon, E.; Gebauer, F.; Tachezy, M.; Izbicki, J.R.; Bockhorn, M. Pancreatic cancer and liver metastases: State of the art. Updates Surg. 2016, 68, 247–251. [Google Scholar] [CrossRef]
- Massague, J.; Obenauf, A.C. Metastatic colonization by circulating tumour cells. Nature 2016, 529, 298–306. [Google Scholar] [CrossRef]
- Aceto, N.; Toner, M.; Maheswaran, S.; Haber, D.A. En Route to Metastasis: Circulating Tumor Cell Clusters and Epithelial-to-Mesenchymal Transition. Trends Cancer 2015, 1, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Fang, F.; Zhang, Q. Circulating tumor cell clusters: What we know and what we expect (Review). Int. J. Oncol. 2016, 49, 2206–2216. [Google Scholar] [CrossRef] [PubMed]
- Szczerba, B.M.; Castro-Giner, F.; Vetter, M.; Krol, I.; Gkountela, S.; Landin, J.; Scheidmann, M.C.; Donato, C.; Scherrer, R.; Singer, J.; et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature 2019, 566, 553–557. [Google Scholar] [CrossRef]
- Aceto, N.; Bardia, A.; Miyamoto, D.T.; Donaldson, M.C.; Wittner, B.S.; Spencer, J.A.; Yu, M.; Pely, A.; Engstrom, A.; Zhu, H.; et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 2014, 158, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Cheung, K.J.; Padmanaban, V.; Silvestri, V.; Schipper, K.; Cohen, J.D.; Fairchild, A.N.; Gorin, M.A.; Verdone, J.E.; Pienta, K.J.; Bader, J.S.; et al. Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc. Natl. Acad. Sci. USA 2016, 113, E854–E863. [Google Scholar] [CrossRef]
- Cho, E.H.; Wendel, M.; Luttgen, M.; Yoshioka, C.; Marrinucci, D.; Lazar, D.; Schram, E.; Nieva, J.; Bazhenova, L.; Morgan, A.; et al. Characterization of circulating tumor cell aggregates identified in patients with epithelial tumors. Phys. Biol. 2012, 9, 016001. [Google Scholar] [CrossRef]
- Hou, J.M.; Krebs, M.G.; Lancashire, L.; Sloane, R.; Backen, A.; Swain, R.K.; Priest, L.J.; Greystoke, A.; Zhou, C.; Morris, K.; et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J. Clin. Oncol. 2012, 30, 525–532. [Google Scholar] [CrossRef]
- Jansson, S.; Bendahl, P.O.; Larsson, A.M.; Aaltonen, K.E.; Ryden, L. Prognostic impact of circulating tumor cell apoptosis and clusters in serial blood samples from patients with metastatic breast cancer in a prospective observational cohort. BMC Cancer 2016, 16, 433. [Google Scholar] [CrossRef] [PubMed]
- Molnar, B.; Floro, L.; Sipos, F.; Toth, B.; Sreter, L.; Tulassay, Z. Elevation in peripheral blood circulating tumor cell number correlates with macroscopic progression in UICC stage IV colorectal cancer patients. Dis. Markers 2008, 24, 141–150. [Google Scholar] [CrossRef]
- Kats-Ugurlu, G.; Roodink, I.; de Weijert, M.; Tiemessen, D.; Maass, C.; Verrijp, K.; van der Laak, J.; de Waal, R.; Mulders, P.; Oosterwijk, E.; et al. Circulating tumour tissue fragments in patients with pulmonary metastasis of clear cell renal cell carcinoma. J. Pathol. 2009, 219, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.C.; Chang, Y.T.; Chen, J.Y.; Jeng, Y.M.; Yang, C.Y.; Tien, Y.W.; Yang, S.H.; Chen, H.L.; Liang, T.Y.; Wang, C.F.; et al. Clinical Significance of Circulating Tumor Microemboli as a Prognostic Marker in Patients with Pancreatic Ductal Adenocarcinoma. Clin. Chem. 2016, 62, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Cools-Lartigue, J.; Spicer, J.; McDonald, B.; Gowing, S.; Chow, S.; Giannias, B.; Bourdeau, F.; Kubes, P.; Ferri, L. Neutrophil extracellular traps sequester circulating tumor cells and promote metastasis. J. Clin. Investig. 2013, 123, 3446–3458. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, M.J.; Radic, M. Neutrophil extracellular traps: Double-edged swords of innate immunity. J. Immunol. 2012, 189, 2689–2695. [Google Scholar] [CrossRef]
- Urban, C.F.; Ermert, D.; Schmid, M.; Abu-Abed, U.; Goosmann, C.; Nacken, W.; Brinkmann, V.; Jungblut, P.R.; Zychlinsky, A. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog. 2009, 5, e1000639. [Google Scholar] [CrossRef]
- Jaillon, S.; Peri, G.; Delneste, Y.; Fremaux, I.; Doni, A.; Moalli, F.; Garlanda, C.; Romani, L.; Gascan, H.; Bellocchio, S.; et al. The humoral pattern recognition receptor PTX3 is stored in neutrophil granules and localizes in extracellular traps. J. Exp. Med. 2007, 204, 793–804. [Google Scholar] [CrossRef]
- Demers, M.; Krause, D.S.; Schatzberg, D.; Martinod, K.; Voorhees, J.R.; Fuchs, T.A.; Scadden, D.T.; Wagner, D.D. Cancers predispose neutrophils to release extracellular DNA traps that contribute to cancer-associated thrombosis. Proc. Natl. Acad. Sci. USA 2012, 109, 13076–13081. [Google Scholar] [CrossRef]
- Record, M.; Carayon, K.; Poirot, M.; Silvente-Poirot, S. Exosomes as new vesicular lipid transporters involved in cell-cell communication and various pathophysiologies. Biochim. Biophys. Acta 2014, 1841, 108–120. [Google Scholar] [CrossRef]
- Choi, D.S.; Kim, D.K.; Kim, Y.K.; Gho, Y.S. Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics 2013, 13, 1554–1571. [Google Scholar] [CrossRef]
- Batista, I.A.; Melo, S.A. Exosomes and the Future of Immunotherapy in Pancreatic Cancer. Int. J. Mol. Sci. 2019, 20, 567. [Google Scholar] [CrossRef]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.Y.; Xiao, F.; Li, J.; Zhou, Y.N.; Zhang, W.J.; Sun, B.; Wang, G. Exosomes and pancreatic diseases: Status, challenges, and hopes. Int. J. Biol. Sci. 2019, 15, 1846–1860. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef] [PubMed]
- Irizarry, R.A.; Bolstad, B.M.; Collin, F.; Cope, L.M.; Hobbs, B.; Speed, T.P. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003, 31, e15. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef]
- Kandasamy, K.; Mohan, S.S.; Raju, R.; Keerthikumar, S.; Kumar, G.S.; Venugopal, A.K.; Telikicherla, D.; Navarro, J.D.; Mathivanan, S.; Pecquet, C.; et al. NetPath: A public resource of curated signal transduction pathways. Genome Biol. 2010, 11, R3. [Google Scholar] [CrossRef]
- Nywening, T.M.; Belt, B.A.; Cullinan, D.R.; Panni, R.Z.; Han, B.J.; Sanford, D.E.; Jacobs, R.C.; Ye, J.; Patel, A.A.; Gillanders, W.E.; et al. Targeting both tumour-associated CXCR2+ neutrophils and CCR2+ macrophages disrupts myeloid recruitment and improves chemotherapeutic responses in pancreatic ductal adenocarcinoma. Gut 2018, 67, 1112–1123. [Google Scholar] [CrossRef]
- Huang, C.; Li, Z.; Li, N.; Li, Y.; Chang, A.; Zhao, T.; Wang, X.; Wang, H.; Gao, S.; Yang, S.; et al. Interleukin 35 Expression Correlates with Microvessel Density in Pancreatic Ductal Adenocarcinoma, Recruits Monocytes, and Promotes Growth and Angiogenesis of Xenograft Tumors in Mice. Gastroenterology 2018, 154, 675–688. [Google Scholar] [CrossRef]
- Pitarresi, J.R.; Liu, X.; Sharma, S.M.; Cuitino, M.C.; Kladney, R.D.; Mace, T.A.; Donohue, S.; Nayak, S.G.; Qu, C.; Lee, J.; et al. Stromal ETS2 Regulates Chemokine Production and Immune Cell Recruitment during Acinar-to-Ductal Metaplasia. Neoplasia 2016, 18, 541–552. [Google Scholar] [CrossRef]
- Bayne, L.J.; Beatty, G.L.; Jhala, N.; Clark, C.E.; Rhim, A.D.; Stanger, B.Z.; Vonderheide, R.H. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer Cell 2012, 21, 822–835. [Google Scholar] [CrossRef]
- Zhang, Y.F.; Zhou, Y.Z.; Zhang, B.; Huang, S.F.; Li, P.P.; He, X.M.; Cao, G.D.; Kang, M.X.; Dong, X.; Wu, Y.L. Pancreatic cancer-derived exosomes promoted pancreatic stellate cells recruitment by pancreatic cancer. J. Cancer 2019, 10, 4397–4407. [Google Scholar] [CrossRef] [PubMed]
- Rahbari, M.; Rahbari, N.; Reissfelder, C.; Weitz, J.; Kahlert, C. Exosomes: Novel implications in diagnosis and treatment of gastrointestinal cancer. Langenbeck’s Arch. Surg. 2016, 401, 1097–1110. [Google Scholar] [CrossRef]
- Colombo, M.; Moita, C.; van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Thery, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013, 126, 5553–5565. [Google Scholar] [CrossRef] [PubMed]
- Luyet, P.P.; Falguieres, T.; Pons, V.; Pattnaik, A.K.; Gruenberg, J. The ESCRT-I subunit TSG101 controls endosome-to-cytosol release of viral RNA. Traffic 2008, 9, 2279–2290. [Google Scholar] [CrossRef]
- Babst, M.; Odorizzi, G.; Estepa, E.J.; Emr, S.D. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic 2000, 1, 248–258. [Google Scholar] [CrossRef]
- Razi, M.; Futter, C.E. Distinct roles for Tsg101 and Hrs in multivesicular body formation and inward vesiculation. Mol. Biol. Cell 2006, 17, 3469–3483. [Google Scholar] [CrossRef]
- Lu, Q.; Hope, L.W.; Brasch, M.; Reinhard, C.; Cohen, S.N. TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation. Proc. Natl. Acad. Sci. USA 2003, 100, 7626–7631. [Google Scholar] [CrossRef]
- Guerreiro, E.M.; Ovstebo, R.; Thiede, B.; Costea, D.E.; Soland, T.M.; Kanli Galtung, H. Cancer cell line-specific protein profiles in extracellular vesicles identified by proteomics. PLoS ONE 2020, 15, e0238591. [Google Scholar] [CrossRef]
- Conway, J.R.; Herrmann, D.; Evans, T.J.; Morton, J.P.; Timpson, P. Combating pancreatic cancer with PI3K pathway inhibitors in the era of personalised medicine. Gut 2019, 68, 742–758. [Google Scholar] [CrossRef]
- Duluc, C.; Moatassim-Billah, S.; Chalabi-Dchar, M.; Perraud, A.; Samain, R.; Breibach, F.; Gayral, M.; Cordelier, P.; Delisle, M.B.; Bousquet-Dubouch, M.P.; et al. Pharmacological targeting of the protein synthesis mTOR/4E-BP1 pathway in cancer-associated fibroblasts abrogates pancreatic tumour chemoresistance. EMBO Mol. Med. 2015, 7, 735–753. [Google Scholar] [CrossRef] [PubMed]
- Kaneda, M.M.; Cappello, P.; Nguyen, A.V.; Ralainirina, N.; Hardamon, C.R.; Foubert, P.; Schmid, M.C.; Sun, P.; Mose, E.; Bouvet, M.; et al. Macrophage PI3Kgamma Drives Pancreatic Ductal Adenocarcinoma Progression. Cancer Discov. 2016, 6, 870–885. [Google Scholar] [CrossRef] [PubMed]
- Chi, H. Regulation and function of mTOR signalling in T cell fate decisions. Nat. Rev. Immunol. 2012, 12, 325–338. [Google Scholar] [CrossRef] [PubMed]
- Simpson, D.; Siddiqui, M.A.; Scott, L.J.; Hilleman, D.E. Reteplase: A review of its use in the management of thrombotic occlusive disorders. Am. J. Cardiovasc. Drugs 2006, 6, 265–285. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, E.; Seyedhosseini-Ghaheh, H.; Mahnam, K.; Jahanian-Najafabadi, A.; Mir Mohammad Sadeghi, H. Reteplase: Structure, Function, and Production. Adv. Biomed. Res. 2019, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Placke, T.; Orgel, M.; Schaller, M.; Jung, G.; Rammensee, H.G.; Kopp, H.G.; Salih, H.R. Platelet-derived MHC class I confers a pseudonormal phenotype to cancer cells that subverts the antitumor reactivity of natural killer immune cells. Cancer Res. 2012, 72, 440–448. [Google Scholar] [CrossRef]
- Palumbo, J.S.; Kombrinck, K.W.; Drew, A.F.; Grimes, T.S.; Kiser, J.H.; Degen, J.L.; Bugge, T.H. Fibrinogen is an important determinant of the metastatic potential of circulating tumor cells. Blood 2000, 96, 3302–3309. [Google Scholar] [CrossRef] [PubMed]
- Biggerstaff, J.P.; Seth, N.; Amirkhosravi, A.; Amaya, M.; Fogarty, S.; Meyer, T.V.; Siddiqui, F.; Francis, J.L. Soluble fibrin augments platelet/tumor cell adherence in vitro and in vivo, and enhances experimental metastasis. Clin. Exp. Metastasis 1999, 17, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Wong, K.H.K.; Khankhel, A.H.; Zeinali, M.; Reategui, E.; Phillips, M.J.; Luo, X.; Aceto, N.; Fachin, F.; Hoang, A.N.; et al. Microfluidic isolation of platelet-covered circulating tumor cells. Lab Chip 2017, 17, 3498–3503. [Google Scholar] [CrossRef]
- Neutrophils Help Circulating Tumor Cells Metastasize. Cancer Discov. 2019, 9, 458–459. [CrossRef]
- Mollinedo, F. Neutrophil Degranulation, Plasticity, and Cancer Metastasis. Trends Immunol. 2019, 40, 228–242. [Google Scholar] [CrossRef]
- Borregaard, N.; Sorensen, O.E.; Theilgaard-Monch, K. Neutrophil granules: A library of innate immunity proteins. Trends Immunol. 2007, 28, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Mollinedo, F.; Nakajima, M.; Llorens, A.; Barbosa, E.; Callejo, S.; Gajate, C.; Fabra, A. Major co-localization of the extracellular-matrix degradative enzymes heparanase and gelatinase in tertiary granules of human neutrophils. Biochem. J. 1997, 327 Pt 3, 917–923. [Google Scholar] [CrossRef]
- Mollinedo, F.; Perez-Sala, D.; Gajate, C.; Jimenez, B.; Rodriguez, P.; Lacal, J.C. Localization of rap1 and rap2 proteins in the gelatinase-containing granules of human neutrophils. FEBS Lett. 1993, 326, 209–214. [Google Scholar] [CrossRef]
- Lacal, P.; Pulido, R.; Sanchez-Madrid, F.; Mollinedo, F. Intracellular location of T200 and Mo1 glycoproteins in human neutrophils. J. Biol. Chem. 1988, 263, 9946–9951. [Google Scholar] [CrossRef]
- Mollinedo, F.; Schneider, D.L. Subcellular localization of cytochrome b and ubiquinone in a tertiary granule of resting human neutrophils and evidence for a proton pump ATPase. J. Biol. Chem. 1984, 259, 7143–7150. [Google Scholar] [CrossRef]
- Mollinedo, F.; Pulido, R.; Lacal, P.M.; Sanchez-Madrid, F. Mobilization of gelatinase-rich granules as a regulatory mechanism of early functional responses in human neutrophils. Scand. J. Immunol. 1991, 34, 33–43. [Google Scholar] [CrossRef]
- Naegelen, I.; Plancon, S.; Nicot, N.; Kaoma, T.; Muller, A.; Vallar, L.; Tschirhart, E.J.; Brechard, S. An essential role of syntaxin 3 protein for granule exocytosis and secretion of IL-1alpha, IL-1beta, IL-12b, and CCL4 from differentiated HL-60 cells. J. Leukoc. Biol. 2015, 97, 557–571. [Google Scholar] [CrossRef]
- Monti, M.; De Rosa, V.; Iommelli, F.; Carriero, M.V.; Terlizzi, C.; Camerlingo, R.; Belli, S.; Fonti, R.; Di Minno, G.; Del Vecchio, S. Neutrophil Extracellular Traps as an Adhesion Substrate for Different Tumor Cells Expressing RGD-Binding Integrins. Int. J. Mol. Sci. 2018, 19, 2350. [Google Scholar] [CrossRef] [PubMed]
- Goetzl, E.J.; Goetzl, L.; Karliner, J.S.; Tang, N.; Pulliam, L. Human plasma platelet-derived exosomes: Effects of aspirin. FASEB J. 2016, 30, 2058–2063. [Google Scholar] [CrossRef] [PubMed]
- Bdeir, K.; Gollomp, K.; Stasiak, M.; Mei, J.; Papiewska-Pajak, I.; Zhao, G.; Worthen, G.S.; Cines, D.B.; Poncz, M.; Kowalska, M.A. Platelet-Specific Chemokines Contribute to the Pathogenesis of Acute Lung Injury. Am. J. Respir. Cell Mol. Biol. 2017, 56, 261–270. [Google Scholar] [CrossRef]
- Brown, A.J.; Sepuru, K.M.; Sawant, K.V.; Rajarathnam, K. Platelet-Derived Chemokine CXCL7 Dimer Preferentially Exists in the Glycosaminoglycan-Bound Form: Implications for Neutrophil-Platelet Crosstalk. Front. Immunol. 2017, 8, 1248. [Google Scholar] [CrossRef]
- Wetterholm, E.; Linders, J.; Merza, M.; Regner, S.; Thorlacius, H. Platelet-derived CXCL4 regulates neutrophil infiltration and tissue damage in severe acute pancreatitis. Transl. Res. 2016, 176, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Hwaiz, R.; Rahman, M.; Zhang, E.; Thorlacius, H. Platelet secretion of CXCL4 is Rac1-dependent and regulates neutrophil infiltration and tissue damage in septic lung damage. Br. J. Pharmacol. 2015, 172, 5347–5359. [Google Scholar] [CrossRef] [PubMed]
- Rosales, C. Neutrophil: A Cell with Many Roles in Inflammation or Several Cell Types? Front. Physiol. 2018, 9, 113. [Google Scholar] [CrossRef]
- Yamada, M.; Kubo, H.; Kobayashi, S.; Ishizawa, K.; He, M.; Suzuki, T.; Fujino, N.; Kunishima, H.; Hatta, M.; Nishimaki, K.; et al. The increase in surface CXCR4 expression on lung extravascular neutrophils and its effects on neutrophils during endotoxin-induced lung injury. Cell. Mol. Immunol. 2011, 8, 305–314. [Google Scholar] [CrossRef]
- Rodrigues, P.; Vanharanta, S. Circulating Tumor Cells: Come Together, Right Now, Over Metastasis. Cancer Discov. 2019, 9, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Teijeira, A.; Garasa, S.; Gato, M.; Alfaro, C.; Migueliz, I.; Cirella, A.; de Andrea, C.; Ochoa, M.C.; Otano, I.; Etxeberria, I.; et al. CXCR1 and CXCR2 Chemokine Receptor Agonists Produced by Tumors Induce Neutrophil Extracellular Traps that Interfere with Immune Cytotoxicity. Immunity 2020, 52, 856–871.e8. [Google Scholar] [CrossRef] [PubMed]
- Adrover, J.M.; Aroca-Crevillen, A.; Crainiciuc, G.; Ostos, F.; Rojas-Vega, Y.; Rubio-Ponce, A.; Cilloniz, C.; Bonzon-Kulichenko, E.; Calvo, E.; Rico, D.; et al. Programmed ‘disarming’ of the neutrophil proteome reduces the magnitude of inflammation. Nat. Immunol. 2020, 21, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Genschmer, K.R.; Russell, D.W.; Lal, C.; Szul, T.; Bratcher, P.E.; Noerager, B.D.; Abdul Roda, M.; Xu, X.; Rezonzew, G.; Viera, L.; et al. Activated PMN Exosomes: Pathogenic Entities Causing Matrix Destruction and Disease in the Lung. Cell 2019, 176, 113–126.e15. [Google Scholar] [CrossRef] [PubMed]
- Stefanius, K.; Servage, K.; de Souza Santos, M.; Gray, H.F.; Toombs, J.E.; Chimalapati, S.; Kim, M.S.; Malladi, V.S.; Brekken, R.; Orth, K. Human pancreatic cancer cell exosomes, but not human normal cell exosomes, act as an initiator in cell transformation. eLife 2019, 8, e40226. [Google Scholar] [CrossRef] [PubMed]
- Castillo, J.; Bernard, V.; San Lucas, F.A.; Allenson, K.; Capello, M.; Kim, D.U.; Gascoyne, P.; Mulu, F.C.; Stephens, B.M.; Huang, J.; et al. Surfaceome profiling enables isolation of cancer-specific exosomal cargo in liquid biopsies from pancreatic cancer patients. Ann. Oncol. 2018, 29, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Tang, P.; Tao, L.; Yuan, C.; Zhang, L.; Xiu, D. Serum Derived Exosomes From Pancreatic Cancer Patients Promoted Metastasis: An iTRAQ-Based Proteomic Analysis. Onco Targets Ther. 2019, 12, 9329–9339. [Google Scholar] [CrossRef] [PubMed]




| Protein | Phosphorylation Site(s) | Biological Significance |
|---|---|---|
| CRAF | S621 | Essential for preventing protein degradation (Activation Site) |
| BRAF | S365; S729 | 14-3-3 Binding; BRAF-CRAF heterodimer formation (Activation Site) |
| MEK1 | S222 | Activation Site |
| AKT1 | S122; T308 | Activation Site |
| ERK2 | T185; Y187 | Activation Site |
| RSK2 | S227; S369 | Activation Site |
| YAP1 | S109 | Activation Site |
| Time Point | Differentially Regulated Genes | Family of Genes Upregulated (%) | Family of Genes Downregulated (%) |
|---|---|---|---|
| 10′ EV treated vs. 2′ Naïve Untreated | Upregulated: 1676 Downregulated: 1234 | Multicomplex: 62.53 Coding: 24.82 Non Coding: 8.89 Unassigned: 3.34 Pseudogenes: 0.42 | Multicomplex: 88.01 Coding: 8.35 Non Coding: 3.08 Unassigned: 0.57 |
| 30′ EV treated vs. 2′ Naïve Untreated | Upregulated: 638 Downregulated: 511 | Multicomplex: 86.36 Coding: 8.46 Non Coding: 4.08 Unassigned: 1.1 | Multicomplex: 78.28 Coding: 8.41 Non Coding: 8.02 Unassigned: 5.28 |
| 60′ EV treated vs. 2′ Naïve Untreated | Upregulated: 428 Downregulated: 462 | Multicomplex: 86.45 Coding: 9.58 Non Coding: 3.27 Unassigned: 0.7 | Multicomplex: 77.49 Coding: 8.87 Non Coding: 7.58 Unassigned: 6.06 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Charles Jacob, H.K.; Charles Richard, J.L.; Signorelli, R.; Kashuv, T.; Lavania, S.; Vaish, U.; Boopathy, R.; Middleton, A.; Boone, M.M.; Sundaram, R.; et al. Modulation of Early Neutrophil Granulation: The Circulating Tumor Cell-Extravesicular Connection in Pancreatic Ductal Adenocarcinoma. Cancers 2021, 13, 2727. https://doi.org/10.3390/cancers13112727
Charles Jacob HK, Charles Richard JL, Signorelli R, Kashuv T, Lavania S, Vaish U, Boopathy R, Middleton A, Boone MM, Sundaram R, et al. Modulation of Early Neutrophil Granulation: The Circulating Tumor Cell-Extravesicular Connection in Pancreatic Ductal Adenocarcinoma. Cancers. 2021; 13(11):2727. https://doi.org/10.3390/cancers13112727
Chicago/Turabian StyleCharles Jacob, Harrys Kishore, John Lalith Charles Richard, Rossana Signorelli, Tyler Kashuv, Shweta Lavania, Utpreksha Vaish, Ranjitha Boopathy, Ashley Middleton, Melinda Minucci Boone, Ramakrishnan Sundaram, and et al. 2021. "Modulation of Early Neutrophil Granulation: The Circulating Tumor Cell-Extravesicular Connection in Pancreatic Ductal Adenocarcinoma" Cancers 13, no. 11: 2727. https://doi.org/10.3390/cancers13112727







