Differential Expression of BOC, SPOCK2, and GJD3 Is Associated with Brain Metastasis of ER-Negative Breast Cancers
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Sample Selection
2.2. Morphological Assessment
2.3. Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
2.4. Immunohistochemistry
2.5. Statistics
3. Results
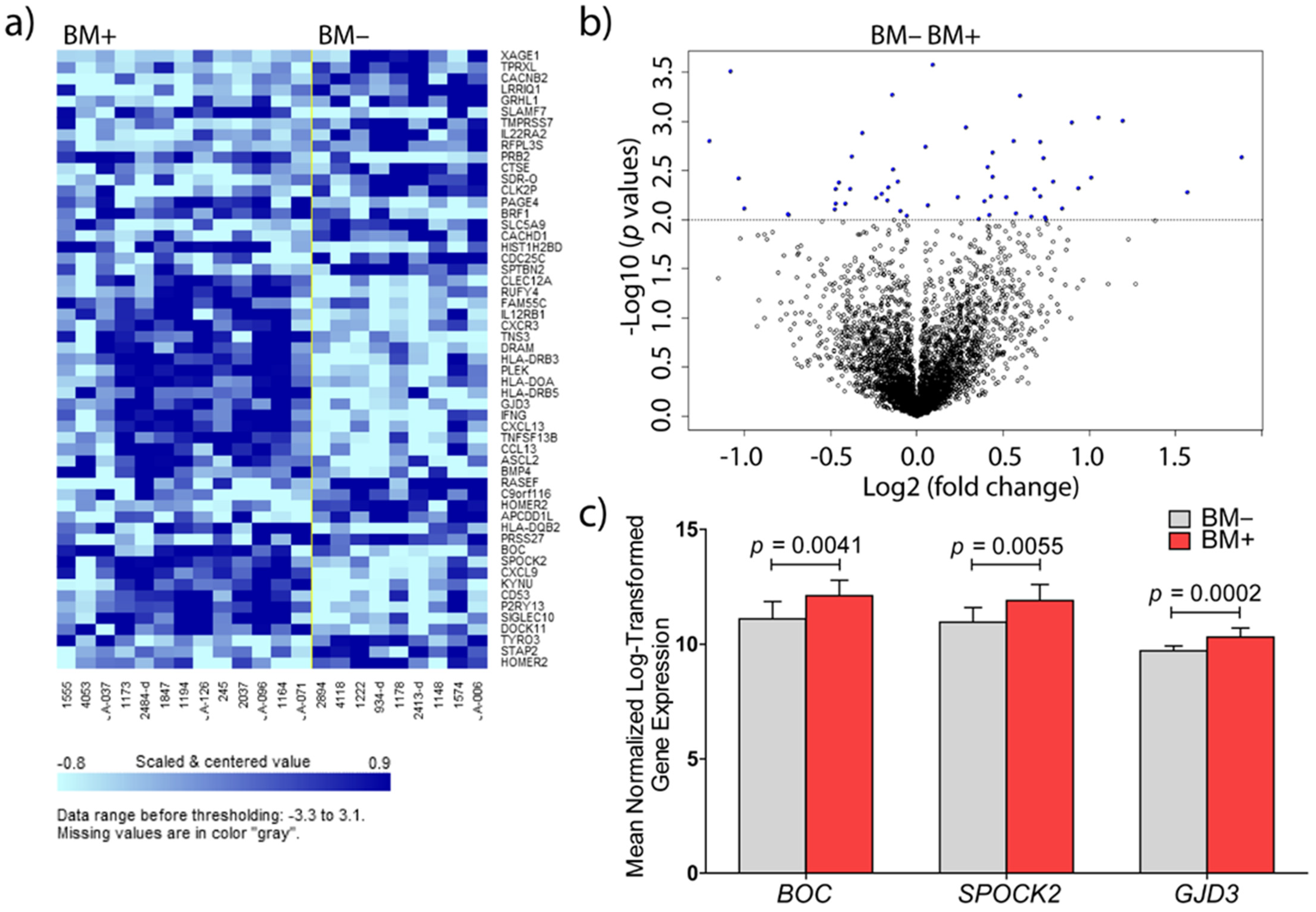
3.1. Identification of Genes Involved in Brain Metastasis of Primary Breast Cancer
3.2. Validations at mRNA and Protein Level
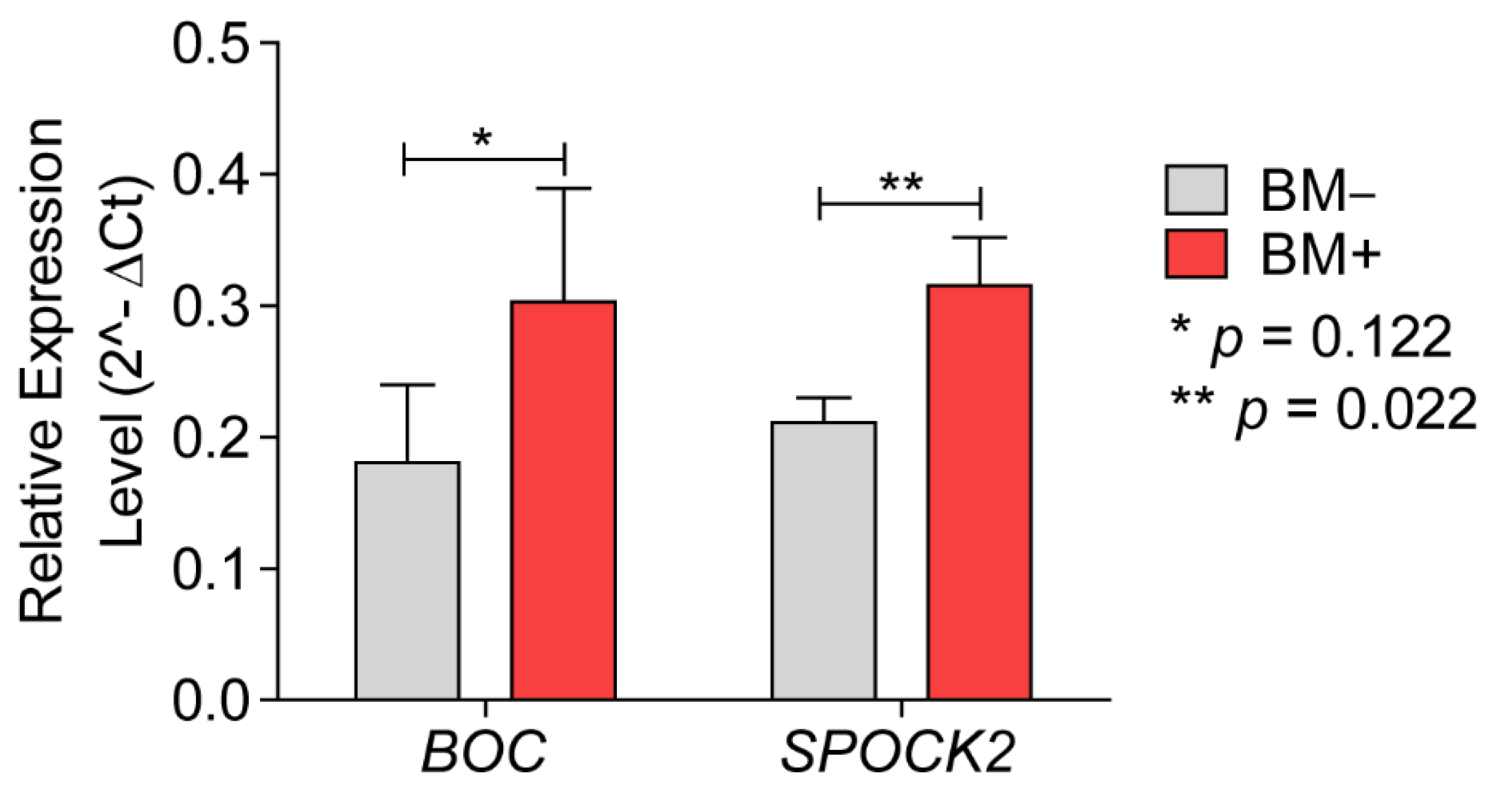
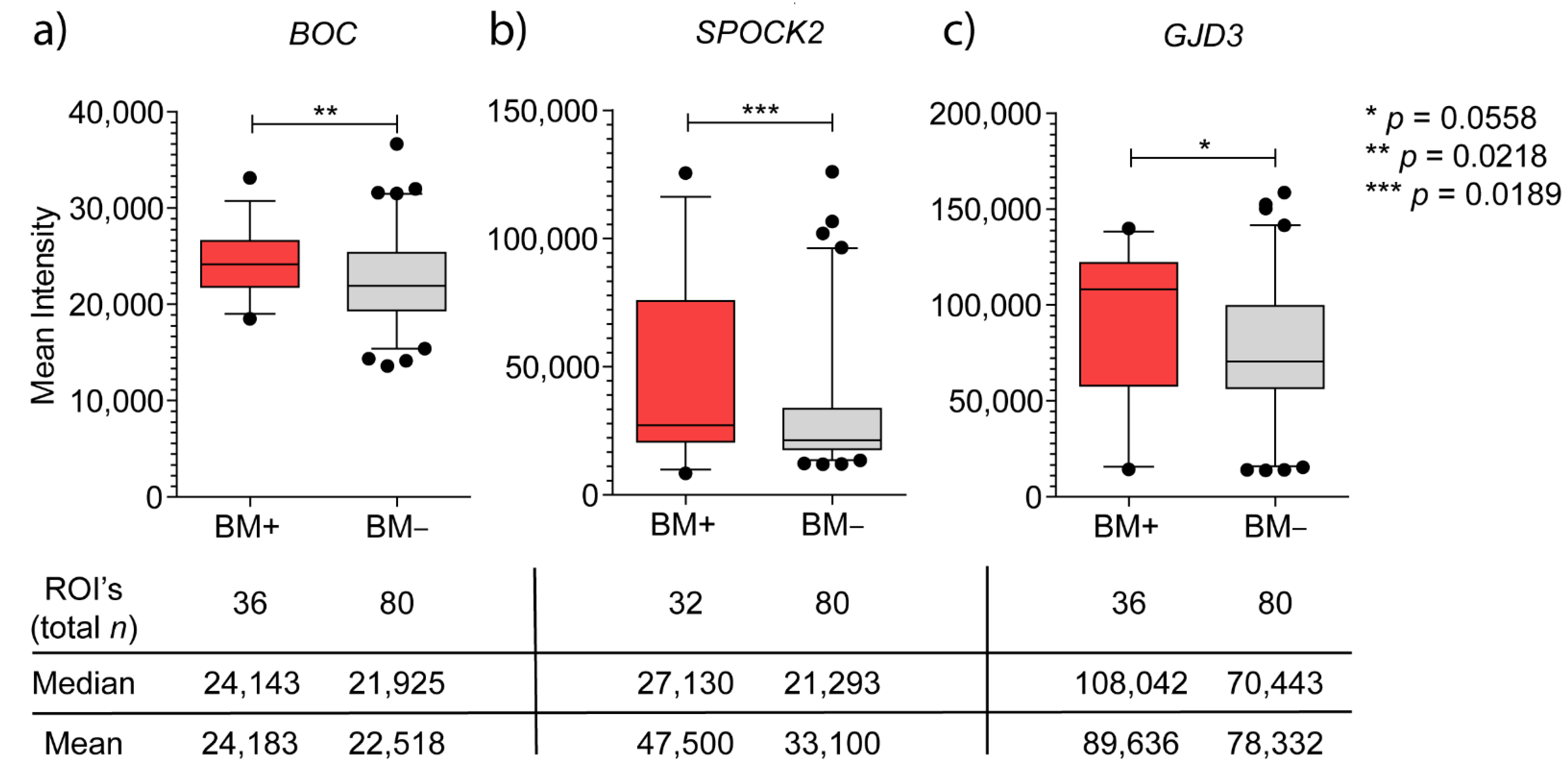
3.2.1. BOC mRNA Expression and Immunohistochemistry
3.2.2. SPOCK2 mRNA Expression and Immunohistochemistry
3.2.3. GJD3 mRNA Expression and Immunohistochemistry
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berghoff, A.S.; Bartsch, R.; Wöhrer, A.; Streubel, B.; Birner, P.; Kros, J.M.; Brastianos, P.K.; Von Deimling, A.; Preusser, M. Predictive molecular markers in metastases to the central nervous system: Recent advances and future avenues. Acta Neuropathol. 2014, 128, 879–891. [Google Scholar] [CrossRef]
- Morris, P.G.; Murphy, C.G.; Mallam, D.; Accordino, M.; Patil, S.; Howard, J.; Omuro, A.; Beal, K.; Seidman, A.D.; Hudis, C.A.; et al. Limited overall survival in patients with brain metastases from triple negative breast cancer. Breast J. 2012, 18, 345–350. [Google Scholar] [CrossRef]
- Heitz, F.; Harter, P.; Lueck, H.-J.; Fissler-Eckhoff, A.; Lorenz-Salehi, F.; Scheil-Bertram, S.; Traut, A.; du Bois, A. Triple-negative and HER2-overexpressing breast cancers exhibit an elevated risk and an earlier occurrence of cerebral metastases. Eur. J. Cancer 2009, 45, 2792–2798. [Google Scholar] [CrossRef] [PubMed]
- Sanna, G.; Franceschelli, L.; Rotmensz, N.; Botteri, E.; Adamoli, L.; Marenghi, C.; Munzone, E.; Rocca, M.C.; Verri, E.; Minchella, I.; et al. Brain metastases in patients with advanced breast cancer. Anticancer Res. 2007, 27, 2865–2869. [Google Scholar] [PubMed]
- Drolez, A.; Vandenhaute, E.; Delannoy, C.P.; Dewald, J.H.; Gosselet, F.; Cecchelli, R.; Julien, S.; Dehouck, M.-P.; Delannoy, P.; Mysiorek, C. ST6GALNAC5 expression decreases the interactions between breast cancer cells and the human blood-brain barrier. Int. J. Mol. Sci. 2016, 17, 1309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mustafa, D.A.M.; Pedrosa, R.M.S.M.; Smid, M.; Van Der Weiden, M.; De Weerd, V.; Nigg, A.L.; Berrevoets, C.; Zeneyedpour, L.; Priego, N.; Valiente, M.; et al. T lymphocytes facilitate brain metastasis of breast cancer by inducing Guanylate-Binding Protein 1 expression. Acta Neuropathol. 2018, 135, 581–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crowe, A.R.; Yue, W. Semi-quantitative determination of protein expression using immunohistochemistry staining and analysis: An integrated protocol. BIO-PROTOCOL 2019, 9, e3465. [Google Scholar] [CrossRef]
- Bos, P.D.; Zhang, X.H.-F.; Nadal, C.; Shu, W.; Gomis, R.R.; Nguyen, D.X.; Minn, A.J.; van de Vijver, M.J.; Gerald, W.L.; Foekens, J.A.; et al. Genes that mediate breast cancer metastasis to the brain. Nature 2009, 459, 1005–1009. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Mulieri, P.J.; Hu, Y.; Taliana, L.; Krauss, R.S. BOC, an Ig superfamily member, associates with CDO to positively regulate myogenic differentiation. EMBO J. 2002, 21, 114–124. [Google Scholar] [CrossRef]
- Allen, B.L.; Song, J.Y.; Izzi, L.; Althaus, I.W.; Kang, J.-S.; Charron, F.; Krauss, R.S.; McMahon, A.P. Overlapping roles and collective requirement for the coreceptors GAS1, CDO, and BOC in SHH pathway function. Dev. Cell 2011, 20, 775–787. [Google Scholar] [CrossRef] [Green Version]
- Okada, A.; Charron, F.; Morin, S.; Shin, D.S.; Wong, K.; Fabre, P.; Tessier-Lavigne, M.; McConnell, S.K. Boc is a receptor for sonic hedgehog in the guidance of commissural axons. Nat. Cell Biol. 2006, 444, 369–373. [Google Scholar] [CrossRef]
- Ingham, P.W.; McMahon, A.P. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 2001, 15, 3059–3087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, K.; Kaspar, B.K.; Gage, F.H.; Schaffer, D.V. Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nat. Neurosci. 2003, 6, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Kavran, J.M.; Ward, M.D.; Oladosu, O.O.; Mulepati, S.; Leahy, D.J. All mammalian hedgehog proteins interact with cell adhesion molecule, down-regulated by oncogenes (CDO) and brother of CDO (BOC) in a conserved manner. J. Biol. Chem. 2010, 285, 24584–24590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Magliano, M.P.; Hebrok, M. Hedgehog signalling in cancer formation and maintenance. Nat. Rev. Cancer 2003, 3, 903–911. [Google Scholar] [CrossRef]
- Jiang, J.; Hui, C.-C. Hedgehog signaling in development and cancer. Dev. Cell 2008, 15, 801–812. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.-S.; Feinleib, J.L.; Knox, S.; Ketteringham, M.A.; Krauss, R.S. Promyogenic members of the Ig and cadherin families associate to positively regulate differentiation. Proc. Natl. Acad. Sci. USA 2003, 100, 3989–3994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powell, D.R.; Williams, J.S.; Hernandez-Lagunas, L.; Salcedo, E.; O’Brien, J.H.; Artinger, K.B. Cdon promotes neural crest migration by regulating N-cadherin localization. Dev. Biol. 2015, 407, 289–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanders, T.A.; Llagostera, E.; Barna, M. Specialized filopodia direct long-range transport of SHH during vertebrate tissue patterning. Nat. Cell Biol. 2013, 497, 628–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Habib, J.G.; O’Shaughnessy, J.A. The hedgehog pathway in triple-negative breast cancer. Cancer Med. 2016, 5, 2989–3006. [Google Scholar] [CrossRef] [PubMed]
- Harris, L.G.; Pannell, L.K.; Singh, S.; Samant, R.S.; Shevde, L.A. Increased vascularity and spontaneous metastasis of breast cancer by hedgehog signaling mediated up-regulation of cyr61. Oncogene 2012, 31, 3370–3380. [Google Scholar] [CrossRef] [Green Version]
- Kwon, Y.J.; Hurst, D.R.; Steg, A.D.; Yuan, K.; Vaidya, K.S.; Welch, D.R.; Frost, A.R. Gli1 enhances migration and invasion via up-regulation of MMP-11 and promotes metastasis in ERalpha negative breast cancer cell lines. Clin. Exp. Metastasis 2011, 28, 437–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, Z.; Gibson, W.T.; Sexton, J.Z.; Aird, K.M.; Ingram, S.M.; Aldrich, A.J.; Lyerly, H.K.; Devi, G.R.; Williams, K.P. Targeting GLI1 expression in human inflammatory breast cancer cells enhances apoptosis and attenuates migration. Br. J. Cancer 2011, 104, 1575–1586. [Google Scholar] [CrossRef] [Green Version]
- Kameda, C.; Tanaka, H.; Yamasaki, A.; Nakamura, M.; Koga, K.; Sato, N.; Kubo, M.; Kuroki, S.; Tanaka, M.; Katano, M. The hedgehog pathway is a possible therapeutic target for patients with estrogen receptor-negative breast cancer. Anticancer Res. 2009, 29, 871–879. [Google Scholar] [PubMed]
- Nordgard, S.H.; Johansen, F.E.; Alnaes, G.I.G.; Bucher, E.; Syvänen, A.-C.; Naume, B.; Børresen-Dale, A.-L.; Kristensen, V.N. Genome-wide analysis identifies 16q deletion associated with survival, molecular subtypes, mRNA expression, and germline haplotypes in breast cancer patients. Genes Chromosom. Cancer 2008, 47, 680–696. [Google Scholar] [CrossRef] [PubMed]
- Huynh, M.-H.; Sage, E.H.; Ringuette, M. A calcium-binding motif in SPARC/osteonectin inhibits chordomesoderm cell migration during Xenopus laevis gastrulation: Evidence of counter-adhesive activity in vivo. Dev. Growth Differ. 1999, 41, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Nakada, M.; Yamada, A.; Takino, T.; Miyamori, H.; Takahashi, T.; Yamashita, J.; Sato, H. Suppression of membrane-type 1 matrix metalloproteinase (MMP)-mediated MMP-2 activation and tumor invasion by testican 3 and its splicing variant gene product, N-Tes. Cancer Res. 2001, 61, 8896–8902. [Google Scholar] [PubMed]
- Stetler-Stevenson, W.G.; Aznavoorian, S.; Liotta, L.A. Tumor cell interactions with the extracellular matrix during inva-sion and metastasis. Annu. Rev. Cell Biol. 1993, 9, 541–573. [Google Scholar] [CrossRef]
- Azzam, H.S.; Arand, G.; Lippman, M.E.; Thompson, E.W. Association of MMP-2 activation potential with metastatic progression in human breast cancer cell lines independent of MMP-2 production. J. Natl. Cancer Inst. 1993, 85, 1758–1764. [Google Scholar] [CrossRef]
- Nakada, M.; Nakamura, H.; Ikeda, E.; Fujimoto, N.; Yamashita, J.; Sato, H.; Seiki, M.; Okad, Y. Expression and tissue localization of membrane-type 1, 2, and 3 matrix metalloproteinases in human as-trocytic tumors. Am. J. Pathol. 1999, 154, 417–428. [Google Scholar] [CrossRef] [Green Version]
- Ueno, H.; Nakamura, H.; Inoue, M.; Imai, K.; Noguchi, M.; Sato, H.; Seiki, M.; Okada, Y. Expression and tissue localization of membrane-types 1, 2, and 3 matrix metalloproteinases in human inva-sive breast carcinomas. Cancer Res. 1997, 57, 2055–2060. [Google Scholar]
- Nakada, M.; Miyamori, H.; Yamashita, J.; Sato, H. Testican 2 abrogates inhibition of membrane-type matrix metalloproteinases by other testican family pro-teins. Cancer Res. 2003, 63, 3364–3369. [Google Scholar] [PubMed]
- Groelz, D.; Sobin, L.; Branton, P.; Compton, C.; Wyrich, R.; Rainen, L. Non-formalin fixative versus formalin-fixed tissue: A comparison of histology and RNA quality. Exp. Mol. Pathol. 2013, 94, 188–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antonov, J.; Goldstein, D.R.; Oberli, A.; Baltzer, A.; Pirotta, M.; Fleischmann, A.; Altermatt, H.J.; Jaggi, R. Reliable gene expression measurements from degraded RNA by quantitative real-time PCR depend on short amplicons and a proper normalization. Lab. Investig. 2005, 85, 1040–1050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belluardo, N.; White, T.W.; Srinivas, M.; Trovato-Salinaro, A.; Ripps, H.; Mudò, G.; Bruzzone, R.; Condorelli, D.F. Identification and functional expression of HCx31.9, a novel gap junction gene. Cell Commun. Adhes. 2001, 8, 173–178. [Google Scholar] [CrossRef]
- Nielsen, P.A.; Beahm, D.L.; Giepmans, B.N.G.; Baruch, A.; Hall, J.E.; Kumar, N.M. Molecular cloning, functional expression, and tissue distribution of a novel human gap junction-forming protein, connexin-31.9. J. Biol. Chem. 2002, 277, 38272–38283. [Google Scholar] [CrossRef] [Green Version]
- Kumar, N.M.; Gilula, N.B. The gap junction communication channel. Cell 1996, 84, 381–388. [Google Scholar] [CrossRef] [Green Version]
- Beyer, E.C.; Berthoud, V.M. Gap junction gene and protein families: Connexins, innexins, and pannexins. Biochim. Biophys. Acta Biomembr. 2018, 1860, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Chen, C.; Li, Y.; Fu, X.; Xie, Y.; Li, Y.; Huang, Y. Cx31.1 acts as a tumour suppressor in non-small cell lung cancer (NSCLC) cell lines through inhibition of cell proliferation and metastasis. J. Cell. Mol. Med. 2012, 16, 1047–1059. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, P.; Saunders, M.M.; Li, Z.; Zhou, Z.; Sheaffer, N.; Kunze, E.L.; Samant, R.S.; Welch, D.R.; Donahue, H.J. Breast cancer metastatic potential: Correlation with increased heterotypic gap junctional intercellular com-munication between breast cancer cells and osteoblastic cells. Int. J. Cancer 2004, 111, 693–697. [Google Scholar] [CrossRef] [PubMed]
- Stoletov, K.; Strnadel, J.; Zardouzian, E.; Momiyama, M.; Park, F.D.; Kelber, J.A.; Pizzo, D.P.; Hoffman, R.; VandenBerg, S.R.; Klemke, R.L. Role of connexins in metastatic breast cancer and melanoma brain colonization. J. Cell Sci. 2013, 126, 904–913. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Zhou, Z.; Dou, K.; Wang, Y. Connexin evolution ameliorates the risk of various cancers. Eur. Rev. Med. Pharm. Sci. 2015, 19, 1662–1672. [Google Scholar]
- Czyż, J. The stage-specific function of gap junctions during tumourigenesis. Cell. Mol. Biol. Lett. 2008, 13, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Naus, C.C.; Laird, D.W. Implications and challenges of connexin connections to cancer. Nat. Rev. Cancer 2010, 10, 435–441. [Google Scholar] [CrossRef] [PubMed]
- Park, J.M.; Munoz, J.L.; Won, B.W.; Bliss, S.A.; Greco, S.J.; Patel, S.A.; Kandouz, M.; Rameshwar, P. Exogenous CXCL12 activates protein kinase C to phosphorylate connexin 43 for gap junctional intercellular communication among confluent breast cancer cells. Cancer Lett. 2013, 331, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Teleki, I.; Krenacs, T.; Szasz, M.A.; Kulka, J.; Wichmann, B.; Leo, C.; Papassotiropoulos, B.; Riemenschnitter, C.; Moch, H.; Varga, Z. The potential prognostic value of connexin 26 and 46 expression in neoadjuvant-treated breast cancer. BMC Cancer 2013, 13, 50. [Google Scholar] [CrossRef] [Green Version]
- Kanczuga-Koda, L.; Sulkowska, M.; Koda, M.; Rutkowski, R.; Sulkowski, S. Increased expression of gap junction protein—Connexin 32 in lymph node metastases of human ductal breast cancer. Folia Histochem. Cytobiol. 2007, 45, 175–180. [Google Scholar]
- Finegold, D.N.; Baty, C.J.; Knickelbein, K.Z.; Perschke, S.; Noon, S.E.; Campbell, D.; Karlsson, J.M.; Huang, D.; Kimak, M.A.; Lawrence, E.C.; et al. Connexin 47 mutations increase risk for secondary lymphedema following breast cancer treatment. Clin. Cancer Res. 2012, 18, 2382–2390. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| # |
Age at Diagnosis (Years) | ER | PR | Her 2/neu | Lymph Node Status |
Neoadjuvant Therapy |
Adjuvant Therapy |
Metastasis Free-Period (Months) | Time to BM (Months) | 1st Metastatic Site | Other Metastatic Sites |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 32 | neg | neg | neg | pos | - | CT + RT | 6 | 11 | liver | lung, brain |
| 2 | 47 | neg | neg | neg | pos | - | CT + RT | 10 | - | skin | - |
| 3 | 53 | neg | neg | neg | pos | - | CT + RT | 48 | - | skin | - |
| 4 | 44 | neg | neg | neg | pos | - | CT + RT | 33 | - | lung | - |
| 5 | 42 | neg | neg | neg | pos | - | CT + RT | 24 | - | liver | - |
| 6 | 53 | neg | neg | neg | pos | - | CT + RT | 42 | 48 | skin | brain |
| 7 | 52 | neg | neg | neg | pos | - | CT + RT | 48 | 34 | lung | brain, lung |
| 8 | 59 | neg | neg | neg | pos | - | CT + RT | 35 | - | pleura | bone, liver, lung, meninges |
| 9 | 42 | neg | neg | neg | pos | - | CT + RT | 84 | - | bone | liver |
| 10 | 39 | neg | neg | neg | pos | - | CT + RT | 21 | 19 | skin | brain |
| 11 | 55 | neg | neg | neg | neg | - | CT + RT | 14 | - | liver | bone |
| 12 | 44 | neg | neg | neg | pos | - | CT + RT | 14 | 11 | brain | meninges, pleura |
| 13 | 61 | neg | neg | neg | pos | - | CT + RT | 26 | 56 | bone, lung | liver, brain |
| 14 | 64 | neg | neg | neg | pos | - | CT + RT | 7 | - | lung | liver, bone |
| 15 | 54 | neg | neg | neg | pos | - | CT + RT | 17 | - | liver | skin, bone, leptomengieal |
| 16 | 37 | neg | neg | neg | neg | - | CT + RT | 18 | - | lung, liver, bone | |
| 17 | 51 | neg | neg | neg | neg | - | CT + RT | 15 | - | bone | lung, skin, liver |
| 18 | 49 | neg | neg | neg | neg | - | CT + RT | 24 | - | bone | lung |
| 19 | 61 | neg | neg | neg | neg | - | CT + RT | 24 | 37 | lung | brain |
| 20 | 39 | neg | neg | pos | neg | - | CT | 51 | 95 | lung | brain, bone |
| 21 | 33 | neg | neg | neg | pos | - | CT + RT | 14 | - | lung, | liver, adrenal |
| 22 | 46 | neg | neg | neg | pos | - | CT + RT | 17 | - | bone | bone, lung, liver |
| 23 | 42 | neg | neg | neg | pos | - | CT | 51 | - | lung | liver |
| 24 | 63 | neg | neg | neg | pos | - | CT + RT | 42 | - | bone | - |
| 25 | 40 | neg | neg | neg | neg | - | CT + RT | 4 | 66 | lung | brain |
| 26 | 34 | neg | neg | neg | pos | - | CT + RT | 9 | - | liver | bone |
| 27 | 32 | neg | neg | neg | pos | N.A. | N.A. | 12 | - | skin | - |
| 28 | 25 | neg | neg | - | neg | - | CT + RT | 115 | 130 | bone | liver, brain |
| 29 | 53 | neg | neg | neg | pos | - | CT + RT | 15 | - | pleura | - |
| 30 | 33 | neg | neg | neg | pos | - | CT + RT | 34 | - | lung | liver, adrenal |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pedrosa, R.M.S.M.; Wismans, L.V.; Sinke, R.; van der Weiden, M.; van Eijck, C.H.J.; Kros, J.M.; Mustafa, D.A.M. Differential Expression of BOC, SPOCK2, and GJD3 Is Associated with Brain Metastasis of ER-Negative Breast Cancers. Cancers 2021, 13, 2982. https://doi.org/10.3390/cancers13122982
Pedrosa RMSM, Wismans LV, Sinke R, van der Weiden M, van Eijck CHJ, Kros JM, Mustafa DAM. Differential Expression of BOC, SPOCK2, and GJD3 Is Associated with Brain Metastasis of ER-Negative Breast Cancers. Cancers. 2021; 13(12):2982. https://doi.org/10.3390/cancers13122982
Chicago/Turabian StylePedrosa, Rute M. S. M., Leonoor V. Wismans, Renata Sinke, Marcel van der Weiden, Casper H. J. van Eijck, Johan M. Kros, and Dana A. M. Mustafa. 2021. "Differential Expression of BOC, SPOCK2, and GJD3 Is Associated with Brain Metastasis of ER-Negative Breast Cancers" Cancers 13, no. 12: 2982. https://doi.org/10.3390/cancers13122982
APA StylePedrosa, R. M. S. M., Wismans, L. V., Sinke, R., van der Weiden, M., van Eijck, C. H. J., Kros, J. M., & Mustafa, D. A. M. (2021). Differential Expression of BOC, SPOCK2, and GJD3 Is Associated with Brain Metastasis of ER-Negative Breast Cancers. Cancers, 13(12), 2982. https://doi.org/10.3390/cancers13122982