NLRP7 Promotes Choriocarcinoma Growth and Progression through the Establishment of an Immunosuppressive Microenvironment
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Study
2.1.1. Normal and Pathological Human Tissue and Sera
2.1.2. Immunohistochemistry of the Human Placentas
2.1.3. ELISA
2.2. Cell Culture
2.2.1. HTR8/SV Neo Cell Line Culture
2.2.2. JEG3 Cell Line Culture
2.2.3. JEG3 Luc Cell Line Preparation
2.2.4. JEG3-Sh-NLRP7 Cell Line Preparation
2.3. 2D Culture System
Proliferation Assay
2.4. 3D Culture System
2.4.1. Anchorage-Independent Spheroids Formation
2.4.2. Anchorage-Independent Colony Formation Assay in Soft Agar
2.5. RNA Isolation and Real-Time PCR Analysis
2.6. Western Blot Analysis
2.7. Gene Sequencing
2.8. In Vivo Studies
2.8.1. Experimental Groups
2.8.2. Histology and Immunohistochemistry of Mouse Tissues
2.8.3. Bioluminescence Imaging
2.9. RNA-Seq Analysis
2.10. Antibody Cytokine Arrays
2.11. Statistical Analysis
3. Results
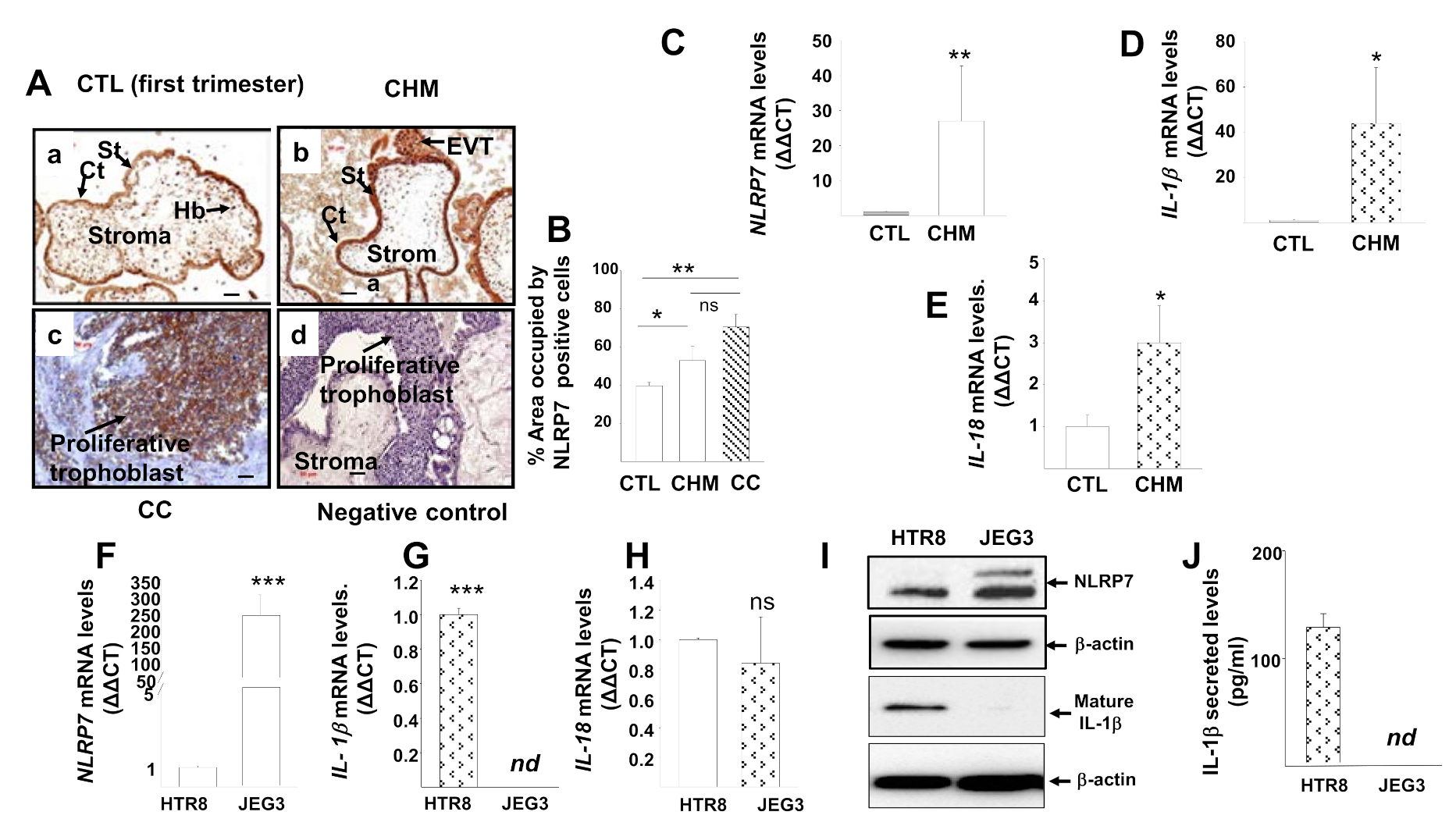
3.1. NLRP7 Inflammasome Is Highly Expressed in CHM and CC
3.2. NLRP7 Inflammasome Is Not Active in the Human Choriocarcinoma Cell Line, JEG3
3.3. NLRP7 Knockdown in JEG3 Cells
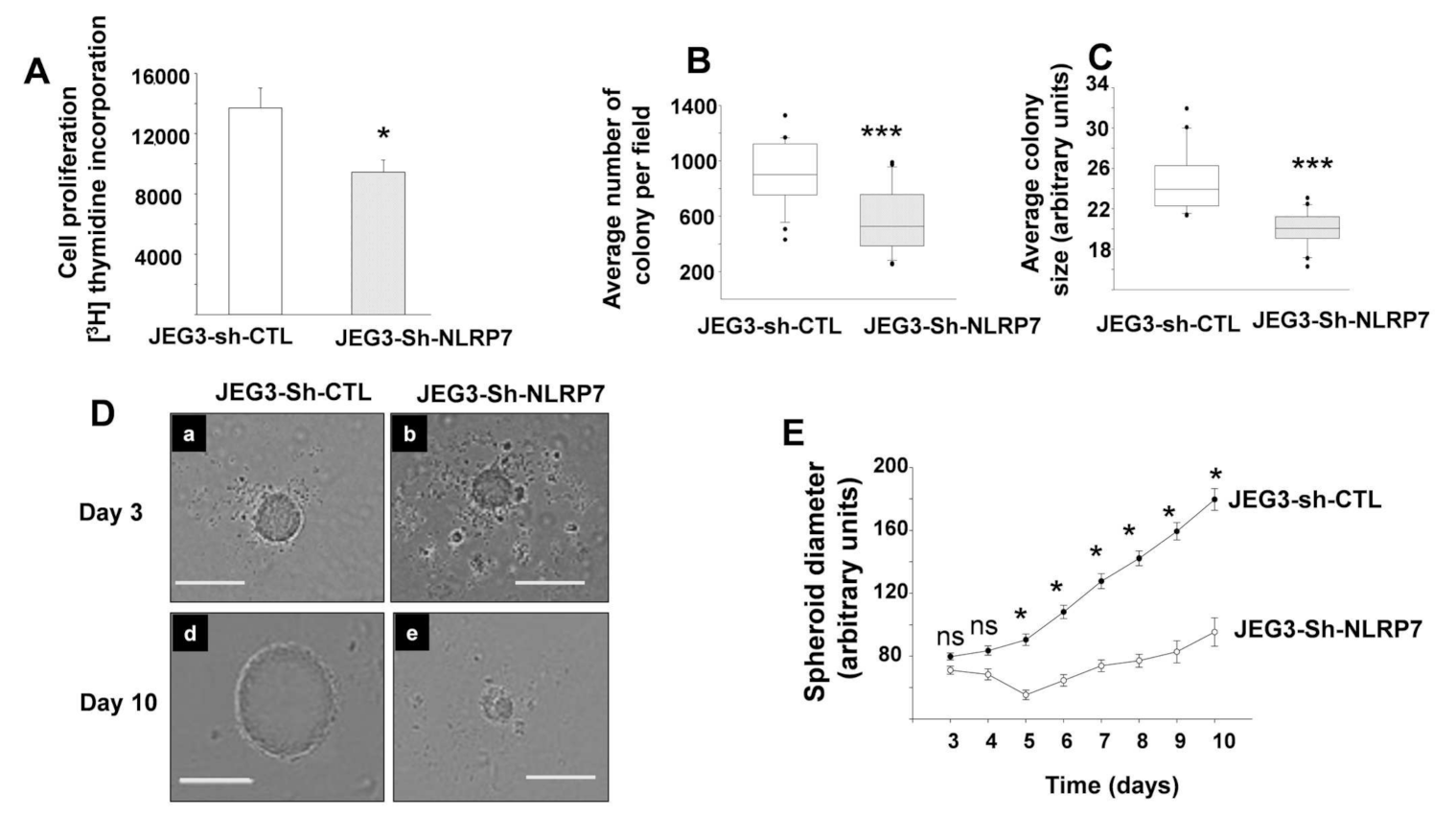
3.4. NLRP7 Knockdown Decreases the Proliferation of JEG3 Tumor Formation In Vitro
3.5. Role of NLRP7 in CC Development and Progression in Two Preclinical Mouse Models
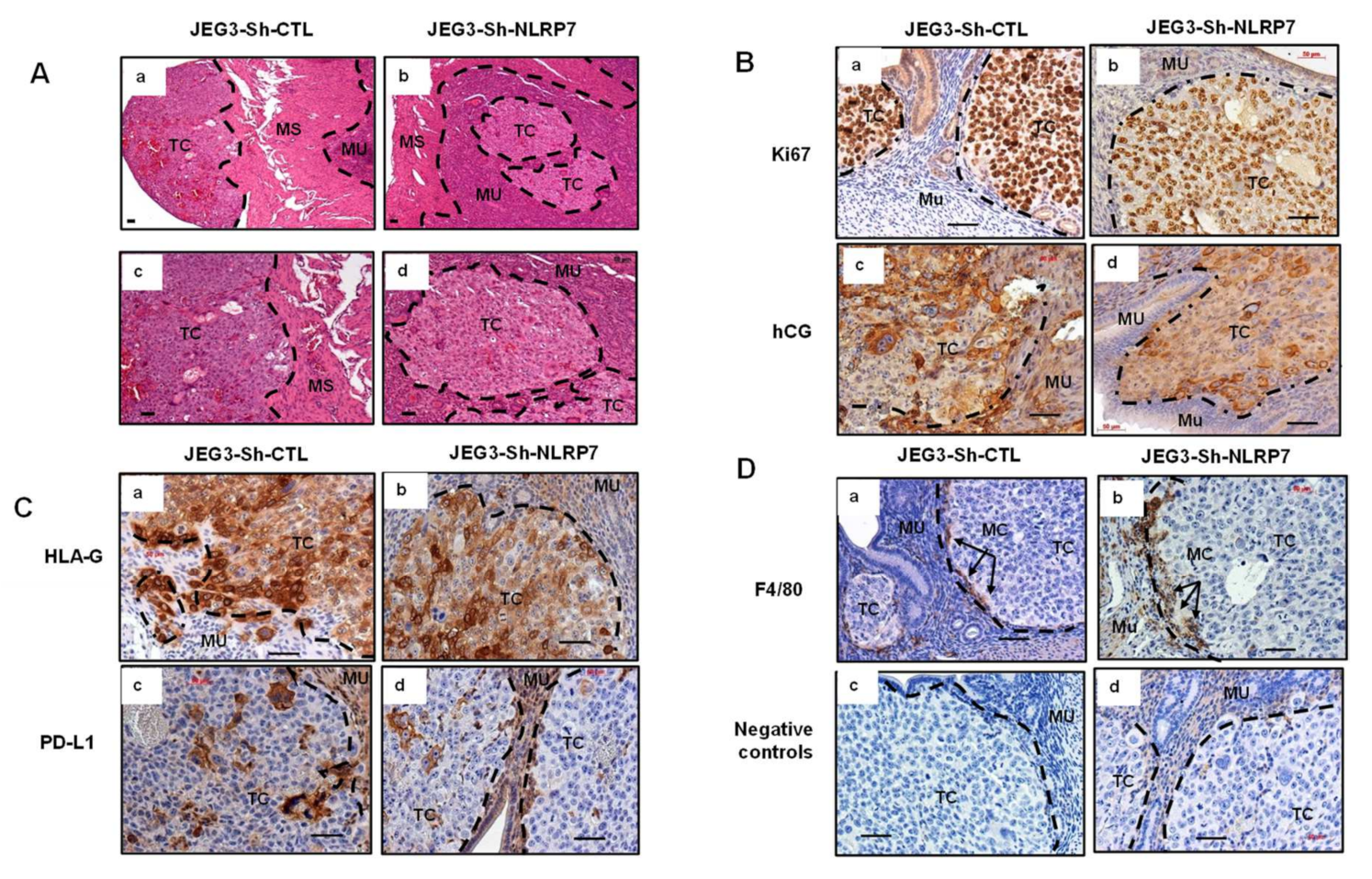
3.6. NLRP7 Knockdown Decreases CC Tumor Growth in the Mouse Placenta
3.7. NLRP7 Knockdown Decreases CC Tumor Growth in the Uterine Horn
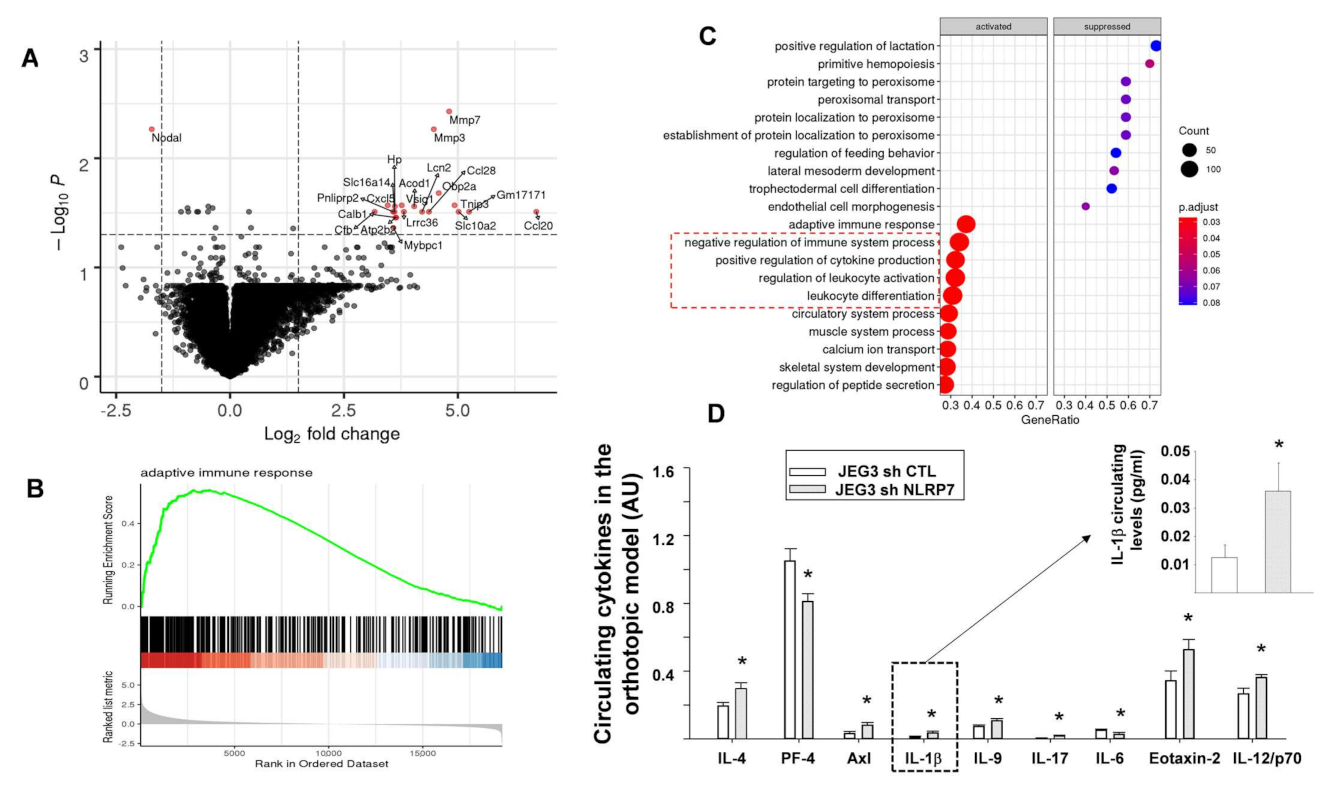
3.8. NLRP7 Knockdown Rescued Maternal Adaptive Immune Response
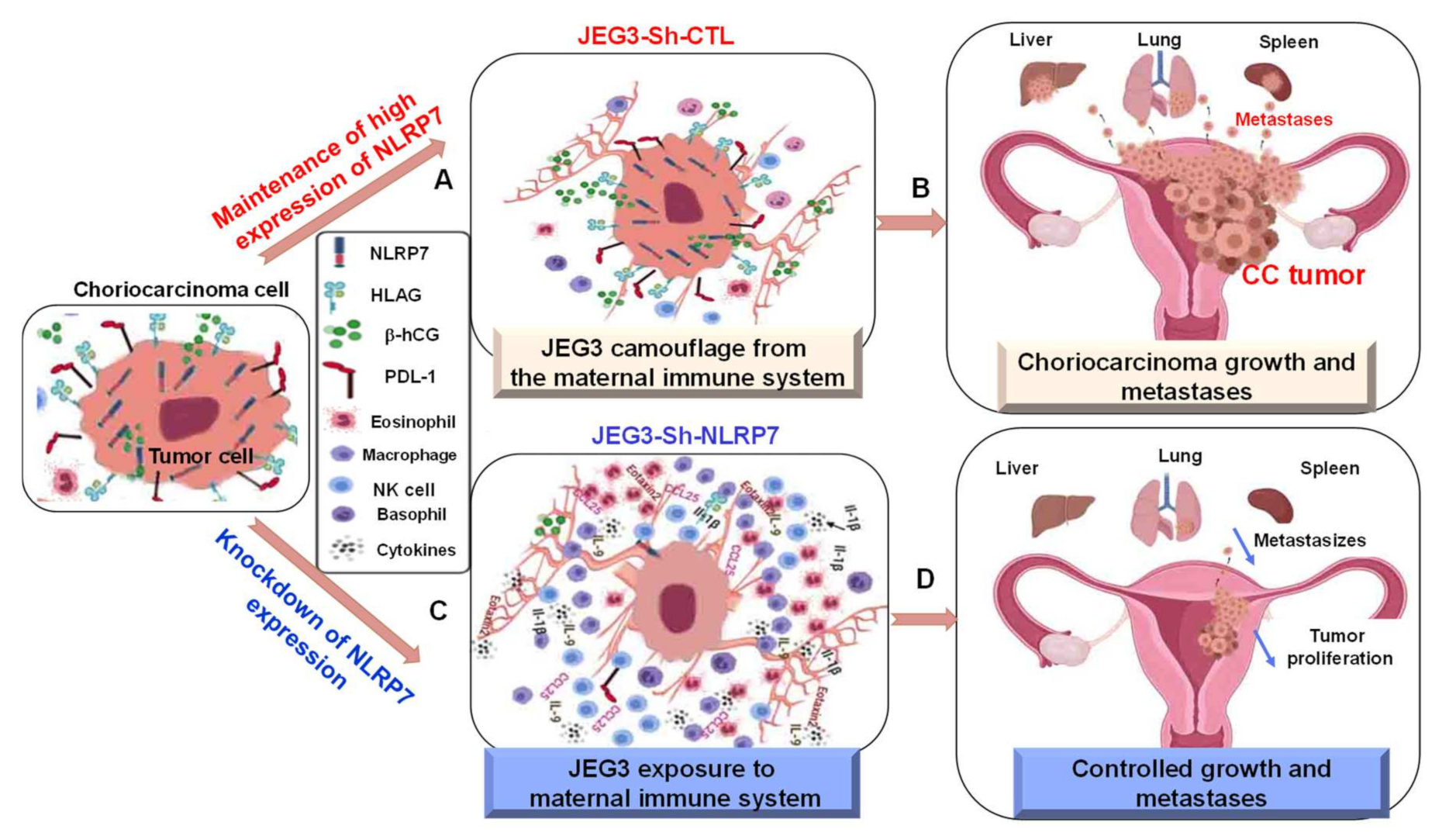
3.9. A Proposed Model for NLRP7 Contribution to the Development of CC In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bianchi, M.E. DAMPs, PAMPs and alarmins: All we need to know about danger. J. Leukoc. Biol. 2007, 81, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.Y.; Nunez, G. Sterile inflammation: Sensing and reacting to damage. Nat. Rev. Immunol. 2010, 10, 826–837. [Google Scholar] [CrossRef]
- Kufer, T.A.; Sansonetti, P.J. NLR functions beyond pathogen recognition. Nat. Immunol. 2011, 12, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Shin, J.S.; Nahm, M.H. NOD-Like Receptors in Infection, Immunity, and Diseases. Yonsei Med. J. 2016, 57, 5–14. [Google Scholar] [CrossRef]
- Davis, B.K.; Wen, H.; Ting, J.P. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu. Rev. Immunol. 2011, 29, 707–735. [Google Scholar] [CrossRef] [PubMed]
- Burgener, S.S.; Schroder, K. Placental inflammasome signaling: Protection for mother and baby. J. Exp. Med. 2021, 218. [Google Scholar] [CrossRef]
- Kinoshita, T.; Wang, Y.; Hasegawa, M.; Imamura, R.; Suda, T. PYPAF3, a PYRIN-containing APAF-1-like protein, is a feedback regulator of caspase-1-dependent interleukin-1beta secretion. J. Biol. Chem. 2005, 280, 21720–21725. [Google Scholar] [CrossRef]
- Messaed, C.; Akoury, E.; Djuric, U.; Zeng, J.; Saleh, M.; Gilbert, L.; Seoud, M.; Qureshi, S.; Slim, R. NLRP7, a nucleotide oligomerization domain-like receptor protein, is required for normal cytokine secretion and co-localizes with Golgi and the microtubule-organizing center. J. Biol. Chem. 2011, 286, 43313–43323. [Google Scholar] [CrossRef]
- Murdoch, S.; Djuric, U.; Mazhar, B.; Seoud, M.; Khan, R.; Kuick, R.; Bagga, R.; Kircheisen, R.; Ao, A.; Ratti, B.; et al. Mutations in NALP7 cause recurrent hydatidiform moles and reproductive wastage in humans. Nat. Genet. 2006, 38, 300–302. [Google Scholar] [CrossRef] [PubMed]
- Akoury, E.; Zhang, L.; Ao, A.; Slim, R. NLRP7 and KHDC3L, the two maternal-effect proteins responsible for recurrent hydatidiform moles, co-localize to the oocyte cytoskeleton. Hum. Reprod. 2015, 30, 159–169. [Google Scholar] [CrossRef]
- Li, L.; Lu, X.; Dean, J. The maternal to zygotic transition in mammals. Mol. Asp. Med. 2013, 34, 919–938. [Google Scholar] [CrossRef]
- Cubellis, M.V.; Pignata, L.; Verma, A.; Sparago, A.; Del Prete, R.; Monticelli, M.; Calzari, L.; Antona, V.; Melis, D.; Tenconi, R.; et al. Loss-of-function maternal-effect mutations of PADI6 are associated with familial and sporadic Beckwith-Wiedemann syndrome with multi-locus imprinting disturbance. Clin. Epigenet. 2020, 12, 139. [Google Scholar] [CrossRef]
- Eggermann, T.; Kadgien, G.; Begemann, M.; Elbracht, M. Biallelic PADI6 variants cause multilocus imprinting disturbances and miscarriages in the same family. Eur. J. Hum. Genet. EJHG 2021, 29, 575–580. [Google Scholar] [CrossRef]
- Froeling, F.E.; Seckl, M.J. Gestational trophoblastic tumours: An update for 2014. Curr. Oncol. Rep. 2014, 16, 408. [Google Scholar] [CrossRef] [PubMed]
- Stevens, F.T.; Katzorke, N.; Tempfer, C.; Kreimer, U.; Bizjak, G.I.; Fleisch, M.C.; Fehm, T.N. Gestational Trophoblastic Disorders: An Update in 2015. Geburtshilfe Frauenheilkd 2015, 75, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Seckl, M.J.; Sebire, N.J.; Berkowitz, R.S. Gestational trophoblastic disease. Lancet 2010, 376, 717–729. [Google Scholar] [CrossRef]
- Sebire, N.J.; Savage, P.M.; Seckl, M.J.; Fisher, R.A. Histopathological features of biparental complete hydatidiform moles in women with NLRP7 mutations. Placenta 2013, 34, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Boufettal, H.; Khalkane, L.; Noun, M.; Hermas, S.; Samouh, N. Gestational choriocarcinoma at Ibn Rochd Hospital, Casablanca, 2004–2010. East. Mediterr. Health J. 2014, 19 (Suppl. 3), S208–S212. [Google Scholar] [PubMed]
- Cisse, C.T.; Lo, N.; Moreau, J.C.; Fall-Gaye, C.; Mendez, V.; Diadhiou, F. [Choriocarcinoma in Senegal: Epidemiology, prognosis and prevention]. Gynecol. Obs. Fertil. 2002, 30, 862–869. [Google Scholar] [CrossRef]
- Altieri, A.; Franceschi, S.; Ferlay, J.; Smith, J.; La Vecchia, C. Epidemiology and aetiology of gestational trophoblastic diseases. Lancet Oncol. 2003, 4, 670–678. [Google Scholar] [CrossRef]
- Smith, H.O. Gestational trophoblastic disease epidemiology and trends. Clin. Obs. Gynecol. 2003, 46, 541–556. [Google Scholar] [CrossRef]
- Berkowitz, R.S.; Goldstein, D.P. Chorionic tumors. N. Engl. J. Med. 1996, 335, 1740–1748. [Google Scholar] [CrossRef]
- Savage, P.; Kelpanides, I.; Tuthill, M.; Short, D.; Seckl, M.J. Brain metastases in gestational trophoblast neoplasia: An update on incidence, management and outcome. Gynecol. Oncol. 2015, 137, 73–76. [Google Scholar] [CrossRef]
- Grummer, R.; Donner, A.; Winterhager, E. Characteristic growth of human choriocarcinoma xenografts in nude mice. Placenta 1999, 20, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Abi Nahed, R.; Reynaud, D.; Borg, A.J.; Traboulsi, W.; Wetzel, A.; Sapin, V.; Brouillet, S.; Dieudonne, M.N.; Dakouane-Giudicelli, M.; Benharouga, M.; et al. NLRP7 is increased in human idiopathic fetal growth restriction and plays a critical role in trophoblast differentiation. J. Mol. Med. 2019, 97, 355–367. [Google Scholar] [CrossRef]
- Ohno, S.; Kinoshita, T.; Ohno, Y.; Minamoto, T.; Suzuki, N.; Inoue, M.; Suda, T. Expression of NLRP7 (PYPAF3, NALP7) protein in endometrial cancer tissues. Anticancer Res. 2008, 28, 2493–2497. [Google Scholar]
- Okada, K.; Hirota, E.; Mizutani, Y.; Fujioka, T.; Shuin, T.; Miki, T.; Nakamura, Y.; Katagiri, T. Oncogenic role of NALP7 in testicular seminomas. Cancer Sci. 2004, 95, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Bolze, P.A.; Massardier, J.; Buenerd, A.; Thivolet Bejui, F.; Perrin, C.; Rouvet, I.; Sanlaville, D.; Maze, M.C.; Dufay, N.; Gaucherand, P.; et al. [Elaboration of a national biobank for the study of gestational trophoblastic diseases]. J. Gynecol. Obs. Biol. Reprod. 2016, 45, 559–562. [Google Scholar] [CrossRef]
- Alfaidy, N.; Chauvet, S.; Donadio-Andrei, S.; Salomon, A.; Saoudi, Y.; Richaud, P.; Aude-Garcia, C.; Hoffmann, P.; Andrieux, A.; Moulis, J.M.; et al. Prion protein expression and functional importance in developmental angiogenesis: Role in oxidative stress and copper homeostasis. Antioxid. Redox Signal. 2013, 18, 400–411. [Google Scholar] [CrossRef]
- Hoffmann, P.; Saoudi, Y.; Benharouga, M.; Graham, C.H.; Schaal, J.P.; Mazouni, C.; Feige, J.J.; Alfaidy, N. Role of EG-VEGF in human placentation: Physiological and pathological implications. J. Cell Mol. Med. 2009, 13, 2224–2235. [Google Scholar] [CrossRef] [PubMed]
- Kohler, P.O.; Bridson, W.E. Isolation of hormone-producing clonal lines of human choriocarcinoma. J. Clin. Endocrinol. Metab. 1971, 32, 683–687. [Google Scholar] [CrossRef]
- Brouillet, S.; Hoffmann, P.; Benharouga, M.; Salomon, A.; Schaal, J.P.; Feige, J.J.; Alfaidy, N. Molecular characterization of EG-VEGF-mediated angiogenesis: Differential effects on microvascular and macrovascular endothelial cells. Mol. Biol. Cell 2010, 21, 2832–2843. [Google Scholar] [CrossRef]
- Brouillet, S.; Hoffmann, P.; Chauvet, S.; Salomon, A.; Chamboredon, S.; Sergent, F.; Benharouga, M.; Feige, J.J.; Alfaidy, N. Revisiting the role of hCG: New regulation of the angiogenic factor EG-VEGF and its receptors. Cell Mol. Life Sci. 2012, 69, 1537–1550. [Google Scholar] [CrossRef]
- Alfaidy, N.; Hoffmann, P.; Gillois, P.; Gueniffey, A.; Lebayle, C.; Garcin, H.; Thomas-Cadi, C.; Bessonnat, J.; Coutton, C.; Villaret, L.; et al. PROK1 Level in the Follicular Microenvironment: A New Noninvasive Predictive Biomarker of Embryo Implantation. J. Clin. Endocrinol. Metab. 2016, 101, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Murthi, P.; Brouillet, S.; Pratt, A.; Borg, A.; Kalionis, B.; Goffin, F.; Tsatsaris, V.; Munaut, C.; Feige, J.; Benharouga, M.; et al. An EG-VEGF-dependent decrease in homeobox gene NKX3.1 contributes to cytotrophoblast dysfunction: A possible mechanism in human fetal growth restriction. Mol. Med. 2015. [Google Scholar] [CrossRef]
- Nguyen, N.M.; Zhang, L.; Reddy, R.; Dery, C.; Arseneau, J.; Cheung, A.; Surti, U.; Hoffner, L.; Seoud, M.; Zaatari, G.; et al. Comprehensive genotype-phenotype correlations between NLRP7 mutations and the balance between embryonic tissue differentiation and trophoblastic proliferation. J. Med. Genet. 2014, 51, 623–634. [Google Scholar] [CrossRef] [PubMed]
- Traboulsi, W.; Sergent, F.; Boufettal, H.; Brouillet, S.; Slim, R.; Hoffmann, P.; Benlahfid, M.; Zhou, Q.Y.; Balboni, G.; Onnis, V.; et al. Antagonism of EG-VEGF Receptors as Targeted Therapy for Choriocarcinoma Progression In Vitro and In Vivo. Clin. Cancer Res. 2017, 23, 7130–7140. [Google Scholar] [CrossRef]
- Fournier, T. Human chorionic gonadotropin: Different glycoforms and biological activity depending on its source of production. Ann. D’endocrinologie 2016, 77, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.F.; Pan, X.; Qian, B.; Shi, M.H. [Expression of PD-1 and PD-L1 in the peripheral blood of advanced non-small-cell lung cancer patients and its implications]. Zhonghua Yi Xue Za Zhi 2019, 99, 111–114. [Google Scholar] [CrossRef]
- Axelrod, H.D.; Valkenburg, K.C.; Amend, S.R.; Hicks, J.L.; Parsana, P.; Torga, G.; DeMarzo, A.M.; Pienta, K.J. AXL Is a Putative Tumor Suppressor and Dormancy Regulator in Prostate Cancer. Mol. Cancer Res. 2019, 17, 356–369. [Google Scholar] [CrossRef]
- Chen, H.J.; Edwards, R.; Tucci, S.; Bu, P.; Milsom, J.; Lee, S.; Edelmann, W.; Gumus, Z.H.; Shen, X.; Lipkin, S. Chemokine 25-induced signaling suppresses colon cancer invasion and metastasis. J. Clin. Investig. 2012, 122, 3184–3196. [Google Scholar] [CrossRef] [PubMed]
- Fabre, J.A.S.; Giustinniani, J.; Garbar, C.; Merrouche, Y.; Antonicelli, F.; Bensussan, A. The Interleukin-17 Family of Cytokines in Breast Cancer. Int. J. Mol. Sci. 2018, 19, 3880. [Google Scholar] [CrossRef]
- Ghiringhelli, F.; Apetoh, L.; Tesniere, A.; Aymeric, L.; Ma, Y.; Ortiz, C.; Vermaelen, K.; Panaretakis, T.; Mignot, G.; Ullrich, E.; et al. Activation of the NLRP3 inflammasome in dendritic cells induces IL-1beta-dependent adaptive immunity against tumors. Nat. Med. 2009, 15, 1170–1178. [Google Scholar] [CrossRef]
- Li, Z.; Chen, L.; Qin, Z. Paradoxical roles of IL-4 in tumor immunity. Cell Mol. Immunol. 2009, 6, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sun, M.; Zhao, H.; Huang, Y.; Li, D.; Mao, D.; Zhang, Z.; Zhu, X.; Dong, X.; Zhao, X. IL-9 Exerts Antitumor Effects in Colon Cancer and Transforms the Tumor Microenvironment In Vivo. Technol. Cancer Res. Treat. 2019, 18, 1533033819857737. [Google Scholar] [CrossRef]
- Babic, A.; Shah, S.M.; Song, M.; Wu, K.; Meyerhardt, J.A.; Ogino, S.; Yuan, C.; Giovannucci, E.L.; Chan, A.T.; Stampfer, M.J.; et al. Soluble tumour necrosis factor receptor type II and survival in colorectal cancer. Br. J. Cancer 2016, 114, 995–1002. [Google Scholar] [CrossRef]
- Lombardo, G.; Gili, M.; Grange, C.; Cavallari, C.; Dentelli, P.; Togliatto, G.; Taverna, D.; Camussi, G.; Brizzi, M.F. IL-3R-alpha blockade inhibits tumor endothelial cell-derived extracellular vesicle (EV)-mediated vessel formation by targeting the beta-catenin pathway. Oncogene 2018, 37, 1175–1191. [Google Scholar] [CrossRef] [PubMed]
- Masjedi, A.; Hashemi, V.; Hojjat-Farsangi, M.; Ghalamfarsa, G.; Azizi, G.; Yousefi, M.; Jadidi-Niaragh, F. The significant role of interleukin-6 and Its signaling pathway in the immunopathogenesis and treatment of breast cancer. Biomed. Pharm. 2018, 108, 1415–1424. [Google Scholar] [CrossRef]
- Pucci, F.; Rickelt, S.; Newton, A.P.; Garris, C.; Nunes, E.; Evavold, C.; Pfirschke, C.; Engblom, C.; Mino-Kenudson, M.; Hynes, R.O.; et al. PF4 Promotes Platelet Production and Lung Cancer Growth. Cell Rep. 2016, 17, 1764–1772. [Google Scholar] [CrossRef]
- Christgen, S.; Kanneganti, T.D. Inflammasomes and the fine line between defense and disease. Curr. Opin. Immunol. 2020, 62, 39–44. [Google Scholar] [CrossRef]
- Mahadevan, S.; Wen, S.; Wan, Y.W.; Peng, H.H.; Otta, S.; Liu, Z.; Iacovino, M.; Mahen, E.M.; Kyba, M.; Sadikovic, B.; et al. NLRP7 affects trophoblast lineage differentiation, binds to overexpressed YY1 and alters CpG methylation. Hum. Mol. Genet. 2014, 23, 706–716. [Google Scholar] [CrossRef] [PubMed]
- Nilkaeo, A.; Bhuvanath, S. Interleukin-1 modulation of human placental trophoblast proliferation. Mediat. Inflamm. 2006, 2006, 79359. [Google Scholar] [CrossRef]
- Prutsch, N.; Fock, V.; Haslinger, P.; Haider, S.; Fiala, C.; Pollheimer, J.; Knofler, M. The role of interleukin-1beta in human trophoblast motility. Placenta 2012, 33, 696–703. [Google Scholar] [CrossRef]
- Yu, N.; Yan, W.; Yin, T.; Wang, Y.; Guo, Y.; Zhou, D.; Xu, M.; Ding, J.; Yang, J. HCG-Activated Human Peripheral Blood Mononuclear Cells (PBMC) Promote Trophoblast Cell Invasion. PLoS ONE 2015, 10, e0125589. [Google Scholar] [CrossRef] [PubMed]
- Chow, M.T.; Sceneay, J.; Paget, C.; Wong, C.S.; Duret, H.; Tschopp, J.; Moller, A.; Smyth, M.J. NLRP3 suppresses NK cell-mediated responses to carcinogen-induced tumors and metastases. Cancer Res. 2012, 72, 5721–5732. [Google Scholar] [CrossRef]
- Karan, D.; Tawfik, O.; Dubey, S. Expression analysis of inflammasome sensors and implication of NLRP12 inflammasome in prostate cancer. Sci. Rep. 2017, 7, 4378. [Google Scholar] [CrossRef] [PubMed]
- Karki, R.; Kanneganti, T.D. Diverging inflammasome signals in tumorigenesis and potential targeting. Nat. Rev. Cancer 2019, 19, 197–214. [Google Scholar] [CrossRef]
- Radian, A.D.; de Almeida, L.; Dorfleutner, A.; Stehlik, C. NLRP7 and related inflammasome activating pattern recognition receptors and their function in host defense and disease. Microbes Infect. 2013, 15, 630–639. [Google Scholar] [CrossRef]
- Kallenbach, L.R.; Bianchi, D.W.; Peter, I.; Stroh, H.; Johnson, K.L. Maternal background strain influences fetal-maternal trafficking more than maternal immune competence in mice. J. Reprod. Immunol. 2011, 90, 188–194. [Google Scholar] [CrossRef]
- Schumacher, A.; Heinze, K.; Witte, J.; Poloski, E.; Linzke, N.; Woidacki, K.; Zenclussen, A.C. Human chorionic gonadotropin as a central regulator of pregnancy immune tolerance. J. Immunol. 2013, 190, 2650–2658. [Google Scholar] [CrossRef]
- Schumacher, A.; Poloski, E.; Sporke, D.; Zenclussen, A.C. Luteinizing hormone contributes to fetal tolerance by regulating adaptive immune responses. Am. J. Reprod. Immunol. 2014, 71, 434–440. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reynaud, D.; Abi Nahed, R.; Lemaitre, N.; Bolze, P.-A.; Traboulsi, W.; Sergent, F.; Battail, C.; Filhol, O.; Sapin, V.; Boufettal, H.; et al. NLRP7 Promotes Choriocarcinoma Growth and Progression through the Establishment of an Immunosuppressive Microenvironment. Cancers 2021, 13, 2999. https://doi.org/10.3390/cancers13122999
Reynaud D, Abi Nahed R, Lemaitre N, Bolze P-A, Traboulsi W, Sergent F, Battail C, Filhol O, Sapin V, Boufettal H, et al. NLRP7 Promotes Choriocarcinoma Growth and Progression through the Establishment of an Immunosuppressive Microenvironment. Cancers. 2021; 13(12):2999. https://doi.org/10.3390/cancers13122999
Chicago/Turabian StyleReynaud, Deborah, Roland Abi Nahed, Nicolas Lemaitre, Pierre-Adrien Bolze, Wael Traboulsi, Frederic Sergent, Christophe Battail, Odile Filhol, Vincent Sapin, Houssine Boufettal, and et al. 2021. "NLRP7 Promotes Choriocarcinoma Growth and Progression through the Establishment of an Immunosuppressive Microenvironment" Cancers 13, no. 12: 2999. https://doi.org/10.3390/cancers13122999
APA StyleReynaud, D., Abi Nahed, R., Lemaitre, N., Bolze, P.-A., Traboulsi, W., Sergent, F., Battail, C., Filhol, O., Sapin, V., Boufettal, H., Hoffmann, P., Aboussaouira, T., Murthi, P., Slim, R., Benharouga, M., & Alfaidy, N. (2021). NLRP7 Promotes Choriocarcinoma Growth and Progression through the Establishment of an Immunosuppressive Microenvironment. Cancers, 13(12), 2999. https://doi.org/10.3390/cancers13122999









