Mutant p53L194F Harboring Luminal-A Breast Cancer Cells Are Refractory to Apoptosis and Cell Cycle Arrest in Response to MortaparibPlus, a Multimodal Small Molecule Inhibitor
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
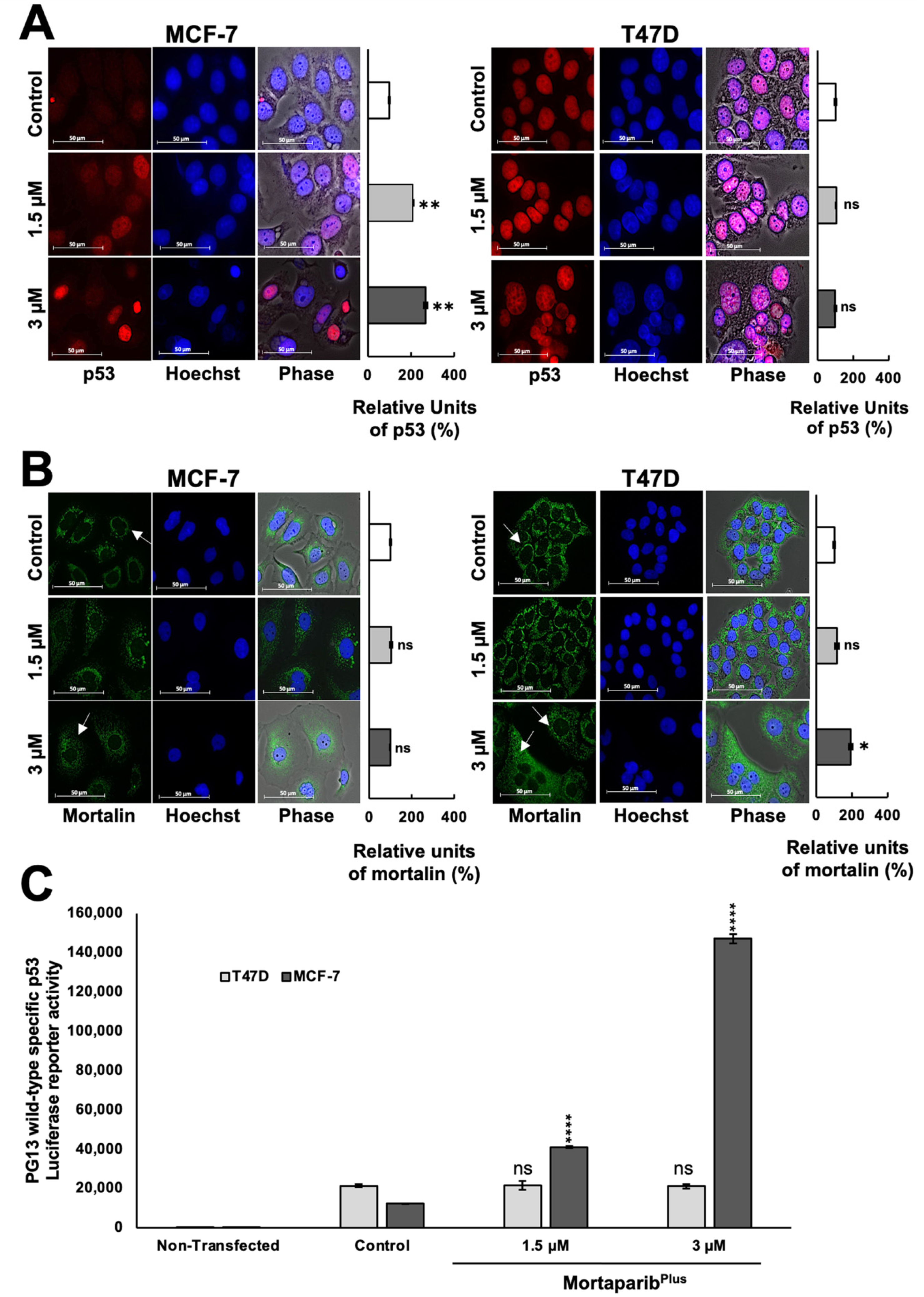
2.1. MortaparibPlus Blocked Mortalin–p53 Interaction in Luminal-A Breast Cancer Cells
2.2. MortaparibPlus Restored the Transcriptional Activation Function of the Wild-Type p53 in MCF-7 Cells Yielding Apoptosis and Cell Cycle Arrest
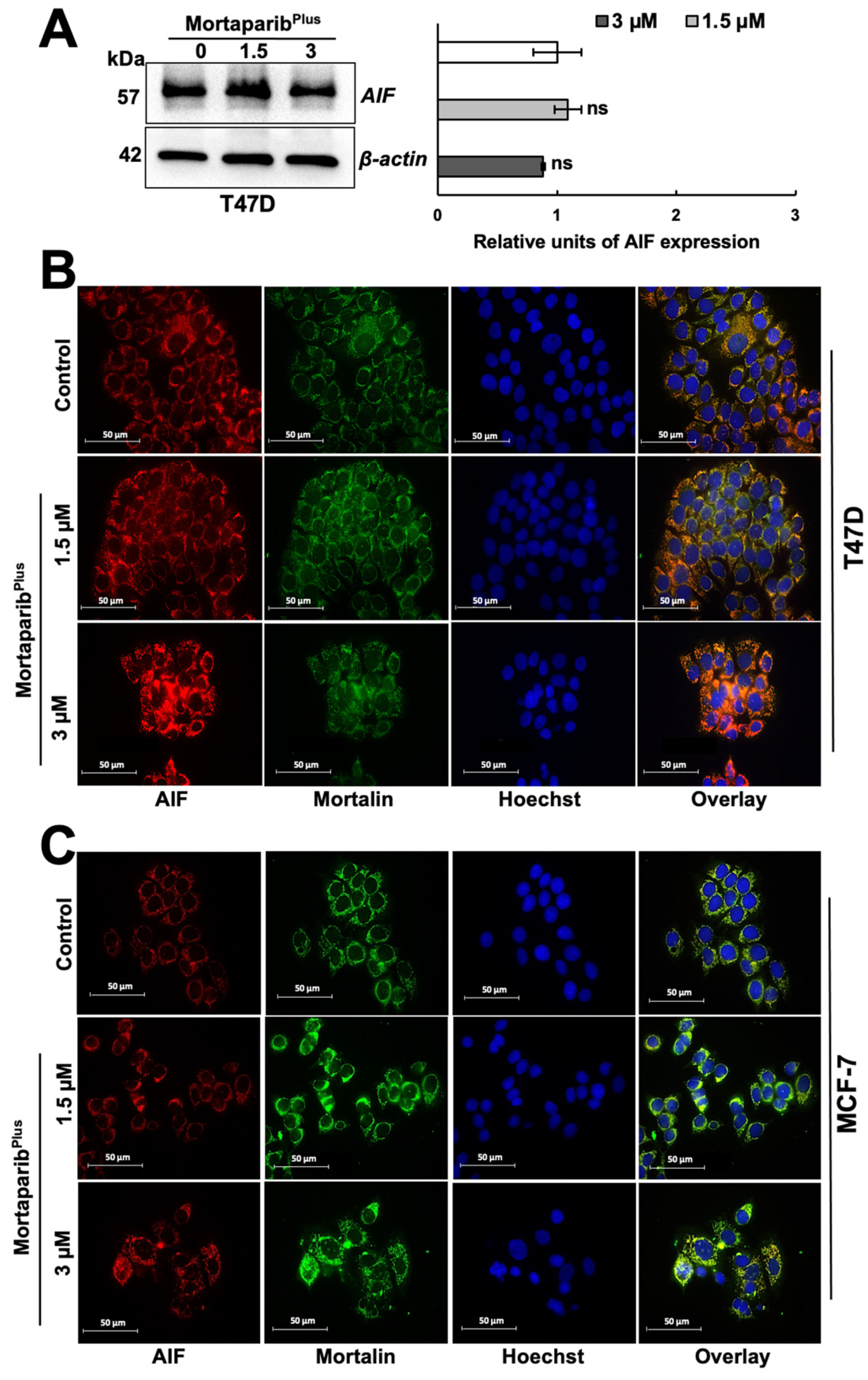
2.3. MortaparibPlus Induced Hyperactivation of PARP1s in T47D Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Drug Treatment
4.2. MTT Cytotoxicity Assay
4.3. Crystal Violet Assay
4.4. Lactate Dehydrogenase (LDH) Cytotoxicity Assay
4.5. Phase Contrast Light Microscopy
4.6. Western Blotting (WB)
4.7. Immunocytochemistry (ICC)
4.8. Luciferase Reporter Assay
4.9. Apoptosis Analysis
4.10. Cell Cycle Analysis
4.11. ATP Assay
4.12. ROS Assay
4.13. Immunoprecipitation
4.14. Mortalin Knockdown
4.15. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Szostakowska, M.; Trębińska-Stryjewska, A.; Grzybowska, E.A.; Fabisiewicz, A. Resistance to endocrine therapy in breast cancer: Molecular mechanisms and future goals. Breast Cancer Res. Treat. 2019, 173, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Eroles, P.; Bosch, A.; Pérez-Fidalgo, J.A.; Lluch, A. Molecular biology in breast cancer: Intrinsic subtypes and signaling pathways. Cancer Treat. Rev. 2012, 38, 698–707. [Google Scholar] [CrossRef] [PubMed]
- Ring, A.; Dowsett, M. Mechanisms of tamoxifen resistance. Endocrine-Related Cancer 2004, 11, 643–658. [Google Scholar] [CrossRef] [PubMed]
- Horlings, H.M.; Bergamaschi, A.; Nordgard, S.H.; Kim, Y.H.; Han, W.; Noh, N.-Y.; Salari, K.; Joosse, S.A.; Reyal, F.; Lingjaerde, O.C.; et al. ESR1 gene amplification in breast cancer: A common phenomenon? Nat. Genet. 2008, 40, 807–808. [Google Scholar] [CrossRef]
- Nielsen, K.V.; Ejlertsen, B.; Müller, S.; Møller, S.; Rasmussen, B.B.; Balslev, E.; Lænkholm, A.-V.; Christiansen, P.; Mouridsen, H.T. Amplification of ESR1 may predict resistance to adjuvant tamoxifen in postmenopausal patients with hormone receptor positive breast cancer. Breast Cancer Res. Treat. 2010, 127, 345–355. [Google Scholar] [CrossRef]
- Anurag, M.; Punturi, N.; Hoog, J.; Bainbridge, M.N.; Ellis, M.J.; Haricharan, S. Comprehensive Profiling of DNA Repair Defects in Breast Cancer Identifies a Novel Class of Endocrine Therapy Resistance Drivers. Clin. Cancer Res. 2018, 24, 4887–4899. [Google Scholar] [CrossRef]
- Wadhwa, R.; Kaul, S.; Ikawa, Y.; Sugimoto, Y. Identification of a novel member of mouse hsp70 family. Its association with cellular mortal phenotype. J. Biol. Chem. 1993, 268, 6615–6621. [Google Scholar] [CrossRef]
- Wadhwa, R.; Taira, K.; Kaul, S.C. Mortalin: A potential candidate for biotechnology and biomedicine. Histol. Histopathol. 2002, 17, 1173–1177. [Google Scholar]
- Wadhwa, R.; Taira, K.; Kaul, S.C. An Hsp70 family chaperone, mortalin/mthsp70/PBP74/Grp75: What, when, and where? Cell Stress Chaperones 2002, 7, 309–316. [Google Scholar] [CrossRef]
- Deocaris, C.C.; Kaul, S.C.; Wadhwa, R. The versatile stress protein mortalin as a chaperone therapeutic agent. Protein Pept. Lett. 2009, 16, 517–529. [Google Scholar] [CrossRef]
- Kaul, S.C.; Deocaris, C.C.; Wadhwa, R. Three faces of mortalin: A housekeeper, guardian and killer. Exp. Gerontol. 2007, 42, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Londono, C.; Osorio, C.; Gama, V.; Alzate, O. Mortalin, Apoptosis, and Neurodegeneration. Biomolecules 2012, 2, 143–164. [Google Scholar] [CrossRef] [PubMed]
- Dundas, S.R.; Lawrie, L.C.; Rooney, P.H.; Murray, G.I. Mortalin is over-expressed by colorectal adenocarcinomas and correlates with poor survival. J. Pathol. 2005, 205, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Wadhwa, R.; Takano, S.; Kaur, K.; Deocaris, C.C.; Pereira-Smith, O.M.; Reddel, R.R.; Kaul, S.C. Upregulation of mortalin/mthsp70/Grp75 contributes to human carcinogenesis. Int. J. Cancer 2006, 118, 2973–2980. [Google Scholar] [CrossRef]
- Na, Y.; Kaul, S.C.; Ryu, J.; Lee, J.-S.; Ahn, H.M.; Kaul, Z.; Kalra, R.S.; Li, L.; Widodo, N.; Yun, C.-O.; et al. Stress Chaperone Mortalin Contributes to Epithelial-to-Mesenchymal Transition and Cancer Metastasis. Cancer Res. 2016, 76, 2754–2765. [Google Scholar] [CrossRef]
- Yun, C.-O.; Bhargava, P.; Na, Y.; Lee, J.-S.; Ryu, J.; Kaul, S.C.; Wadhwa, R. Relevance of mortalin to cancer cell stemness and cancer therapy. Sci. Rep. 2017, 7, srep42016. [Google Scholar] [CrossRef] [PubMed]
- Wadhwa, R.; Takano, S.; Robert, M.; Yoshida, A.; Nomura, H.; Reddel, R.; Mitsui, Y.; Kaul, S.C. Inactivation of Tumor Suppressor p53 by Mot-2, a hsp70 Family Member. J. Biol. Chem. 1998, 273, 29586–29591. [Google Scholar] [CrossRef]
- Gestl, E.E.; Böttger, S.A. Cytoplasmic sequestration of the tumor suppressor p53 by a heat shock protein 70 family member, mortalin, in human colorectal adenocarcinoma cell lines. Biochem. Biophys. Res. Commun. 2012, 423, 411–416. [Google Scholar] [CrossRef]
- Walker, C.; Böttger, S.; Low, B. Mortalin-Based Cytoplasmic Sequestration of p53 in a Nonmammalian Cancer Model. Am. J. Pathol. 2006, 168, 1526–1530. [Google Scholar] [CrossRef]
- Lu, W.-J.; Lee, N.P.; Kaul, S.C.; Lan, F.; Poon, R.T.P.; Wadhwa, R.; Luk, J.M. Mortalin–p53 interaction in cancer cells is stress dependent and constitutes a selective target for cancer therapy. Cell Death Differ. 2011, 18, 1046–1056. [Google Scholar] [CrossRef] [PubMed]
- Grover, A.; Priyandoko, D.; Gao, R.; Shandilya, A.; Widodo, N.; Bisaria, V.S.; Kaul, S.C.; Wadhwa, R.; Sundar, D. Withanone binds to mortalin and abrogates mortalin–p53 complex: Computational and experimental evidence. Int. J. Biochem. Cell Biol. 2012, 44, 496–504. [Google Scholar] [CrossRef] [PubMed]
- Nigam, N.; Grover, A.; Goyal, S.; Katiyar, S.; Bhargava, P.; Wang, P.-C.; Sundar, D.; Kaul, S.C.; Wadhwa, R. Targeting Mortalin by Embelin Causes Activation of Tumor Suppressor p53 and Deactivation of Metastatic Signaling in Human Breast Cancer Cells. PLoS ONE 2015, 10, e0138192. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, P.; Grover, A.; Nigam, N.; Kaul, A.; Doi, M.; Ishida, Y.; Kakuta, H.; Kaul, S.C.; Terao, K.; Wadhwa, R. Anticancer activity of the supercritical extract of Brazilian green propolis and its active component, artepillin�C: Bioinformatics and experimental analyses of its mechanisms of action. Int. J. Oncol. 2018, 52, 925–932. [Google Scholar] [CrossRef]
- Widodo, N.; Kaur, K.; Shrestha, B.G.; Takagi, Y.; Ishii, T.; Wadhwa, R.; Kaul, S.C. Selective Killing of Cancer Cells by Leaf Extract of Ashwagandha: Identification of a Tumor-Inhibitory Factor and the First Molecular Insights to Its Effect. Clin. Cancer Res. 2007, 13, 2298–2306. [Google Scholar] [CrossRef]
- Wadhwa, R.; Sugihara, T.; Yoshida, A.; Nomura, H.; Reddel, R.; Simpson, R.; Maruta, H.; Kaul, S.C. Selective toxicity of MKT-077 to cancer cells is mediated by its binding to the hsp70 family protein mot-2 and reactivation of p53 function. Cancer Res. 2000, 60, 6818–6821. [Google Scholar]
- Fatokun, A.A.; Dawson, V.L.; Dawson, T.M. Parthanatos: Mitochondrial-linked mechanisms and therapeutic opportunities. Br. J. Pharmacol. 2014, 171, 2000–2016. [Google Scholar] [CrossRef]
- Chaudhuri, A.R.; Nussenzweig, A.R.C.A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 610–621. [Google Scholar] [CrossRef]
- McCann, K.E.; Hurvitz, S.A. Advances in the use of PARP inhibitor therapy for breast cancer. Drugs Context 2018, 7, 1–30. [Google Scholar] [CrossRef]
- Abbotts, R.; Topper, M.J.; Biondi, C.; Fontaine, D.; Goswami, R.; Stojanovic, L.; Choi, E.Y.; McLaughlin, L.; Kogan, A.A.; Xia, L.; et al. DNA methyltransferase inhibitors induce a BRCAness phenotype that sensitizes NSCLC to PARP inhibitor and ionizing radiation. Proc. Natl. Acad. Sci. USA 2019, 116, 22609–22618. [Google Scholar] [CrossRef] [PubMed]
- Putri, J.F.; Bhargava, P.; Dhanjal, J.K.; Yaguchi, T.; Sundar, D.; Kaul, S.C.; Wadhwa, R. Mortaparib, a novel dual inhibitor of mortalin and PARP1, is a potential drug candidate for ovarian and cervical cancers. J. Exp. Clin. Cancer Res. 2019, 38, 499. [Google Scholar] [CrossRef]
- Sari, A.; Elwakeel, A.; Dhanjal, J.; Kumar, V.; Sundar, D.; Kaul, S.; Wadhwa, R. Identification and Characterization of MortaparibPlus—A Novel Triazole Derivative That Targets Mortalin-p53 Interaction and Inhibits Cancer-Cell Proliferation by Wild-Type p53-Dependent and -Independent Mechanisms. Cancers 2021, 13, 835. [Google Scholar] [CrossRef]
- Kao, J.; Salari, K.; Bocanegra, M.; Choi, Y.-L.; Girard, L.; Gandhi, J.; Kwei, K.A.; Hernandez-Boussard, T.; Wang, P.; Gazdar, A.F.; et al. Molecular Profiling of Breast Cancer Cell Lines Defines Relevant Tumor Models and Provides a Resource for Cancer Gene Discovery. PLoS ONE 2009, 4, e6146. [Google Scholar] [CrossRef] [PubMed]
- Poudel, P.; Nyamundanda, G.; Patil, Y.; Cheang, M.C.U.; Sadanandam, A. Heterocellular gene signatures reveal luminal-A breast cancer heterogeneity and differential therapeutic responses. npj Breast Cancer 2019, 5, 1–10. [Google Scholar] [CrossRef]
- Lim, L.Y.; Vidnovic, N.; Ellisen, L.W.; Leong, C.-O. Mutant p53 mediates survival of breast cancer cells. Br. J. Cancer 2009, 101, 1606–1612. [Google Scholar] [CrossRef]
- Soldani, C.; Scovassi, A.I. Poly(ADP-ribose) polymerase-1 cleavage during apoptosis: An update. Apoptosis 2002, 7, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Imamura, H.; Sakamoto, S.; Yoshida, T.; Matsui, Y.; Penuela, S.; Laird, D.W.; Mizukami, S.; Kikuchi, K.; Kakizuka, A. Single-cell dynamics of pannexin-1-facilitated programmed ATP loss during apoptosis. eLife 2020, 9, e61960. [Google Scholar] [CrossRef] [PubMed]
- Aredia, F.; Scovassi, A.I. Poly(ADP-ribose): A signaling molecule in different paradigms of cell death. Biochem. Pharmacol. 2014, 92, 157–163. [Google Scholar] [CrossRef]
- Andrabi, S.A.; Kim, N.S.; Yu, S.-W.; Wang, H.; Koh, D.W.; Sasaki, M.; Klaus, J.A.; Otsuka, T.; Zhang, Z.; Koehler, R.C.; et al. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc. Natl. Acad. Sci. USA 2006, 103, 18308–18313. [Google Scholar] [CrossRef]
- Andrabi, S.A.; Dawson, T.M.; Dawson, V.L. Mitochondrial and Nuclear Cross Talk in Cell Death. Ann. N. Y. Acad. Sci. 2008, 1147, 233–241. [Google Scholar] [CrossRef]
- Ravagnan, L.; Gurbuxani, S.; Susin, S.A.; Maisse, C.; Daugas, E.; Zamzami, N.; Mak, T.; Jäättelä, M.; Penninger, J.M.; Garrido, C.; et al. Heat-shock protein 70 antagonizes apoptosis-inducing factor. Nat. Cell Biol. 2001, 3, 839–843. [Google Scholar] [CrossRef]
- Gurbuxani, S.; Schmitt, E.; Cande, C.; Parcellier, A.; Hammann, A.; Daugas, E.; Kouranti, I.; Spahr, C.; Pance, A.; Kroemer, G.; et al. Heat shock protein 70 binding inhibits the nuclear import of apoptosis-inducing factor. Oncogene 2003, 22, 6669–6678. [Google Scholar] [CrossRef]
- McKeage, M.J.; Berners-Price, S.; Galettis, P.; Bowen, R.J.; Brouwer, W.; Ding, L.; Zhuang, L.; Baguley, B.C. Role of lipophilicity in determining cellular uptake and antitumour activity of gold phosphine complexes. Cancer Chemother. Pharmacol. 2000, 46, 343–350. [Google Scholar] [CrossRef]
- Modak, R.; Basha, J.; Bharathy, N.; Maity, K.; Mizar, P.; Bhat, A.V.; Vasudevan, M.; Rao, V.K.; Kok, W.K.; Natesh, N.; et al. Probing p300/CBP Associated Factor (PCAF)-Dependent Pathways with a Small Molecule Inhibitor. ACS Chem. Biol. 2013, 8, 1311–1323. [Google Scholar] [CrossRef]
- Harbeck, N.; Thomssen, C.; St. Gnant, M. Gallen 2013: Brief Preliminary Summary of the Consensus Discussion. Breast Care 2013, 8, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Gasco, M.; Shami, S.; Crook, T. The p53 pathway in breast cancer. Breast Cancer Res. 2002, 4, 1–7. [Google Scholar] [CrossRef]
- Berger, C.; Qian, Y.; Chen, X. The p53-Estrogen Receptor Loop in Cancer. Curr. Mol. Med. 2013, 13, 1229–1240. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Bergman, J.; Wiman, K.G.; Bykov, V.J. Role of Thiol Reactivity for Targeting Mutant p53. Cell Chem. Biol. 2018, 25, 1219–1230.e3. [Google Scholar] [CrossRef] [PubMed]
- Ramraj, S.K.; Elayapillai, S.P.; Pelikan, R.C.; Zhao, Y.D.; Isingizwe, Z.R.; Kennedy, A.L.; Lightfoot, S.A.; Benbrook, D.M. Novel ovarian cancer maintenance therapy targeted at mortalin and mutant p53. Int. J. Cancer 2020, 147, 1086–1097. [Google Scholar] [CrossRef] [PubMed]
- Cavanaugh, F.P., Jr.; Moskwa, P.S.; Donish, W.H.; Pera, P.J.; Richardson, D.; Andrese, A.P. A semi-automated neutral red based chemosensitivity assay for drug screening. Investig. New Drugs 1990, 8, 347–354. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elwakeel, A.; Sari, A.N.; Dhanjal, J.K.; Meidinna, H.N.; Sundar, D.; Kaul, S.C.; Wadhwa, R. Mutant p53L194F Harboring Luminal-A Breast Cancer Cells Are Refractory to Apoptosis and Cell Cycle Arrest in Response to MortaparibPlus, a Multimodal Small Molecule Inhibitor. Cancers 2021, 13, 3043. https://doi.org/10.3390/cancers13123043
Elwakeel A, Sari AN, Dhanjal JK, Meidinna HN, Sundar D, Kaul SC, Wadhwa R. Mutant p53L194F Harboring Luminal-A Breast Cancer Cells Are Refractory to Apoptosis and Cell Cycle Arrest in Response to MortaparibPlus, a Multimodal Small Molecule Inhibitor. Cancers. 2021; 13(12):3043. https://doi.org/10.3390/cancers13123043
Chicago/Turabian StyleElwakeel, Ahmed, Anissa Nofita Sari, Jaspreet Kaur Dhanjal, Hazna Noor Meidinna, Durai Sundar, Sunil C. Kaul, and Renu Wadhwa. 2021. "Mutant p53L194F Harboring Luminal-A Breast Cancer Cells Are Refractory to Apoptosis and Cell Cycle Arrest in Response to MortaparibPlus, a Multimodal Small Molecule Inhibitor" Cancers 13, no. 12: 3043. https://doi.org/10.3390/cancers13123043
APA StyleElwakeel, A., Sari, A. N., Dhanjal, J. K., Meidinna, H. N., Sundar, D., Kaul, S. C., & Wadhwa, R. (2021). Mutant p53L194F Harboring Luminal-A Breast Cancer Cells Are Refractory to Apoptosis and Cell Cycle Arrest in Response to MortaparibPlus, a Multimodal Small Molecule Inhibitor. Cancers, 13(12), 3043. https://doi.org/10.3390/cancers13123043







