Simple Summary
There is limited information about genomic markers, especially for cervical adenocarcinoma treatment decisions. In this prospective study, it was found that nonsynonymous PIK3CA mutation detected in the patient’s circulating DNA collected before treatment or during follow-up was significantly associated with decreased progression-free survival or overall survival. It is the first indication of the predictive power of PIK3CA aberration in cervical adenocarcinoma. The work contributes to the development of liquid biopsies for the prolonged strategy of surveillance and indicates the possibility of tailoring management of this particular women’s cancer.
Abstract
Personalized treatment of genetically stratified subgroups has the potential to improve outcomes in many malignant tumors. This study distills clinically meaningful prognostic/predictive genomic marker for cervical adenocarcinoma using signature genomic aberrations and single-point nonsynonymous mutation-specific droplet digital PCR (ddPCR). Mutations in PIK3CA E542K, E545K, or H1047R were detected in 41.7% of tumors. PIK3CA mutation detected in the patient’s circulating DNA collected before treatment or during follow-up was significantly associated with decreased progression-free survival or overall survival. PIK3CA mutation in the circulating DNA during follow-up after treatment predicted recurrence with 100% sensitivity and 64.29% specificity. It is the first indication of the predictive power of PIK3CA mutations in cervical adenocarcinoma. The work contributes to the development of liquid biopsies for follow up surveillance and a possibility of tailoring management of this particular women’s cancer.
1. Introduction
Cervical cancer remains the fourth most common cancer in women worldwide [1]. The two major types of cervical cancer are squamous cell carcinoma and adenocarcinoma, which are morphologically distinct. While most of the decline in cervical cancer can be attributed to a reduction in cervical squamous cell carcinoma, the incidence of cervical adenocarcinoma, absolute and relative to cervical squamous cell carcinoma, has been rising dramatically over the past few decades, in particular in young women [2]. The prognosis for advanced cervical adenocarcinoma is especially poor. Cervical cancer is almost always associated with infection of oncogenic types of human papillomavirus (HPV); however, HPV infection alone is insufficient for malignant transformation. Other genetic events independent or in conjunction with HPV infection are required [3]. There is great value in the characterization of the genomic signatures of cervical adenocarcinoma for the development of new prognostic/predictive markers and targeted therapeutic regimes to improve the outcome of women suffering from this disease.
This study addresses combining the signature genomic aberrations and patient clinical course to explore whether DNA markers can predict outcomes for this specific subtype of cervical cancer using a droplet digital polymerase chain reaction (ddPCR) platform.
2. Materials and Methods
2.1. Study Population
Twenty-four patients with primary cervical adenocarcinoma managed in the Department of Obstetrics and Gynaecology, The Chinese University of Hong Kong, Prince of Wales Hospital were included this study (Table 1). Surgery was offered to patients with early-stage disease unless there was a contraindication. Chemo-irradiation or primary radiotherapy was offered to patients with late-stage disease, or to those with a contraindication to radical surgery. Pelvic lymphadenectomy was performed as part of the surgical treatment procedure. The research protocol was approved by the Research Ethics Committee of The Chinese University of Hong Kong. Patients were provided with written informed consent for the use of their biological material for research purposes.

Table 1.
Clinicopathological features of cervical adenocarcinoma.
2.2. Sample Collection
Prior to any radiation or chemotherapy, surgical or biopsy tumor tissue specimens were embedded in OCT compound, snap-frozen in liquid nitrogen within one hour after collection and stored at −80 °C. Cryo-sections of tumor specimens were manually or laser-capture micro-dissected to achieve a tumor cell purity of 90% or greater. Patient blood samples were collected from patients prior to treatment, and at 6, 12, and 18 months after treatment.
Genomic DNA from tumor cells and blood cell pellet as self-control was isolated using AllPrep kit (Qiagen, Germantown, MD, USA), and quantified using Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). Plasma cell-free DNA (cfDNA) was isolated from plasma using QIAmp Circulating Nucleic Acid Kit (Qiagen, Germantown, MD). The range of DNA amount isolated from 3 mL of plasma was 18.3 to 702.5 (median 31.3) ng.
2.3. Digital Droplet Polymerase Chain Reaction (ddPCR)
A total of five single nucleotide variation (SNV) mutation sites from three genes were selected for ddPCR testing based on the cervical adenocarcinoma genomic landscape determined in previous studies, the yield of cfDNA, as well as the availability of established ddPCR mutation assays [4,5,6,7,8,9]. The ddPCR reactions contained 10 μL of 2×ddPCR supermix, 1 μL of 20× primer/probe mix, and 9 μL of diluted DNA (3 ng). The 20 μL reaction mix was transferred to a cartridge for a QX200 droplet generator (Bio-Rad, Hercules, CA, USA) followed by 70 μL of droplet generation oil into oil wells. After droplet generation, 40 μL of the reaction were then transferred to a 96-well plate and the plate was heat-sealed with sealing foil sheets. The optimized cycling conditions were 95 °C for 10 min, 40 cycles of 94 °C for 30 s, annealing temperature for 1 min, followed by 98 °C for 10 min and a hold at 4 °C using thermal cycler C1000 (Bio-Rad, Hercules, CA, USA). The annealing/extension temperature of PIK3CA E542K, PIK3CA E545K, PIK3CA H1047R, KRAS G12V and KRT6A F249L were 58.7 °C, 61 °C, 61 °C, 58.7 °C, and 57.9 °C, respectively. After PCR amplification, the plate was read by the QX200 droplet reader (Bio-Rad, Hercules, CA, USA) for analysis. The DNA targets were quantified using QuantaSoft Software (Bio-Rad, Hercules, CA, USA). The results are reported as copies of mutant allele per μL of reaction. To verify the ddPCR specificity, 10 g blocks gene fragments containing the mutant or wildtype sequence for the five targeted mutation sites were utilized as templates (Integrated DNA Technologies Inc., Coralville, IA, USA) (Supplementary Table S1), and 20,000 copies of the gene fragments were used for each assay.
2.4. Clinicopathological Data Collection and Statistical Analyses
Clinicopathological data from each patient were collected from medical records for statistical analysis. The associations between gene mutations and the clinicopathological features were analyzed by a Kaplan–Meier survival curve and log-rank test, Cox proportional hazards regression analysis, Fisher’s exact test, Binary logistic regression analysis, and receiver operating characteristic (ROC) curve analysis using MedCalc (Version 19.5.1) software. The results derived from the Kaplan–Meier survival curve and log-rank test in part was also verified using SPSS (Version 25) and SAS (University Edition) software. A two-sided p value < 0.05 was considered statistically significant.
3. Results
Using single-point mutation-specific ddPCR, a total of five nonsynonymous single nucleotide substitution mutation spots were measured in tumor DNA and circulating DNA (Supplementary Table S2). At least one of three PIK3CA missense mutation spots was detected in 10 (41.7%) of 24 tumor DNA samples, 7 (30.4%) of 23 circulating DNA samples collected before treatment, and 12 (29.3%) of 41 DNA samples collected during follow-up after treatment. There was no statistically significant difference among the three positions of the PIK3CA mutation detected in tumor DNA and in circulating DNA collected before treatment or during follow-up (p > 0.05). KRT6A F249L mutation was detected in 12 (50%) tumor DNA samples, seven (30.4%) circulating DNA samples collected before treatment, and 11(29.7%) circulating DNA samples collected during follow-up. KRAS G12V mutation was detected in four (25%) tumor DNA samples and not detected in circulating DNA samples collected before treatment or during follow-up.
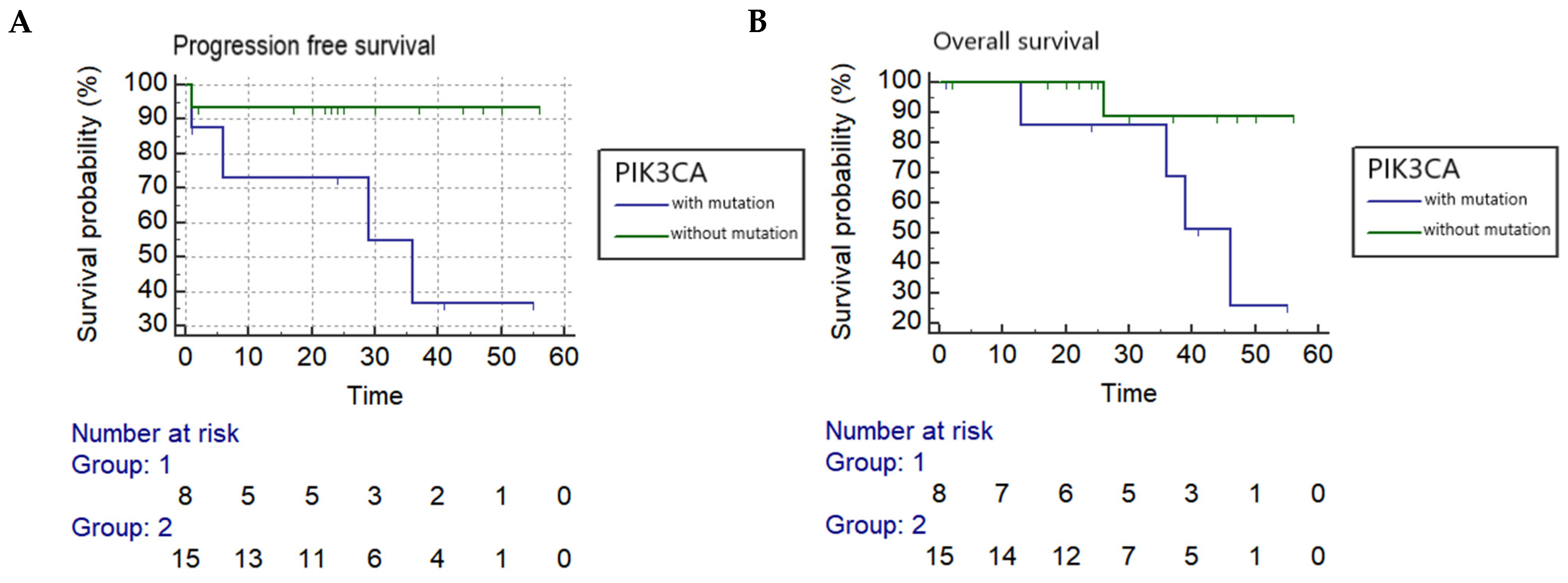
Kaplan–Meier log-rank analyses found a PIK3CA mutation in circulating DNA collected before treatment was associated with a statistically significant worse progression-free and overall survival with a hazard ratio of 7.7 and 6.6, respectively (PFS: p = 0.0291 and OS: p = 0.0499) (Figure 1), while reduced OS was associated with PIK3CA mutation in circulating DNA collected during follow-up after treatment (p = 0.0429). Tumor recurrence was also significantly correlated to both PFS and OS (p = 0.0001 and p = 0.0026, respectively). In addition, OS was significantly associated with pelvic LN metastasis (p = 0.0143), and reduced PFS and OS were significantly correlated to a late stage of cancer (p = 0.0036 and p = 0.0007, respectively) (Table 2).
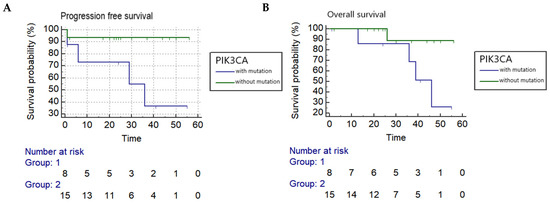
Figure 1.
Kaplan–Meier analysis (A) Progression-free survival correlated to PIK3CA in circulating DNA collected before treatment in cervical adenocarcinoma (p = 0.0291) (hazard ratio = 7.7068). (B) Overall survival correlated to PIK3CA in circulating DNA collected before treatment in cervical adenocarcinoma (p = 0.0499) (hazard ratio = 6.6105).

Table 2.
Survival analysis related to clinicopathological features and gene mutations in cervical adenocarcinoma using Kaplan–Meier survival curve and log-rank test.
In Cox proportional hazards regression analysis, the factors significantly associated with survival included the presence of PIK3CA mutation in circulating DNA collected before treatment, stage, tumor size and recurrence (p = 0.015, p = 0.0034, p = 0.016 and p < 0.0001, respectively) (Supplementary Table S3). The factors significantly correlated to recurrence included PIK3CA mutation in circulating DNA collected during follow-up and tumor size (p = 0.0114 and p = 0.0185, respectively) while late stage was marginally associated with recurrence (p = 0.0561) (Supplementary Table S4).
Correlation analysis using Fisher’s exact test showed that patient survival was significantly associated with stage, recurrence, and PIK3CA mutation status in circulating DNA collected before treatment (p = 0.0145, p = 0.005, and p = 0.0329, respectively), while recurrence was significantly associated with survival and PIK3CA mutation status in circulating DNA collected during follow-up (p = 0.005 and p = 0.0325, respectively) (Supplementary Table S5).
Binary logistic regression analysis showed that the stage (p = 0.018), tumor size (p = 0.039), and PIK3CA mutation in circulating DNA collected before treatment (p = 0.035), were significantly associated with survival.
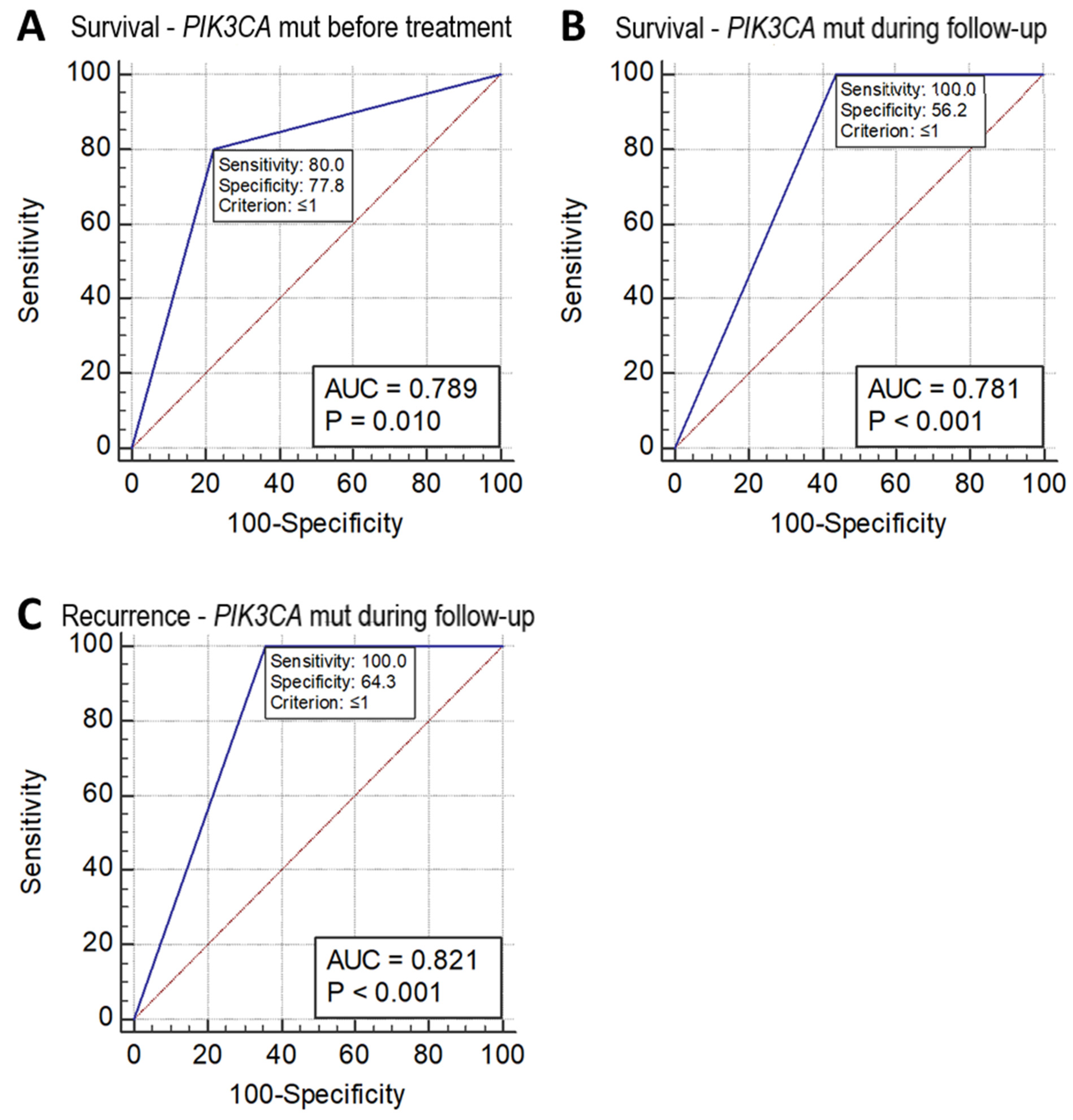
Receiver operating characteristic (ROC) analysis of sensitivity and specificity of these missense mutation measurements in the prediction of patient survival and recurrence showed that the mutation status of PIK3CA detected in circulating DNA collected before treatment and during follow-up after treatment could predict a reduction in survival with a sensitivity and specificity of 80.0% and 77.8% (p = 0.010, AUC = 0.789), and 100.0% and 56.2% (p = 0.0001, AUC 0.781), respectively. PIK3CA mutations detected in circulating DNA collected during follow-up after treatment could predict recurrence with a sensitivity and specificity of 100% and 64.3%, respectively (p = 0.001, AUC = 0.821) (Figure 2). However, the ROC analysis did not show p < 0.05 in the prediction of either the recurrence or survival status in the mutation of PIK3CA in tumor tissue DNA alone or of the other two SNVs (KRT6A F249L and KRAS G12V) in either tumor tissue DNA, or circulating DNA collected before treatment or during follow-up.
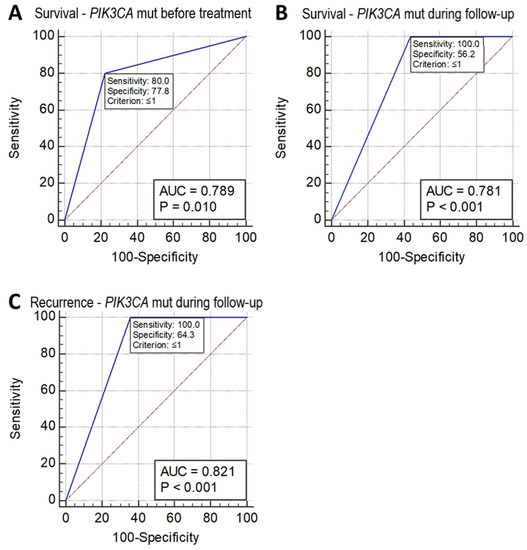
Figure 2.
ROC chart. (A) Survival of cervical adenocarcinoma predicted by PIK3CA mutation status as detected in circulating DNA collected before treatment. (B) Survival of cervical adenocarcinoma predicted by PIK3CA mutation status as detected in circulating DNA collected during follow-up after treatment. (C) Recurrence of cervical adenocarcinoma predicted by PIK3CA mutation status as detected in circulating DNA collected during follow-up after treatment.
4. Discussion
In a prior retrospective study, we identified mutations in FAT1, ARID1A, ERBB2, and PIK3CA in whole-exome sequencing of 15 patients with cervical adenocarcinoma [8]. We then assessed a second cohort of 24 patients from Hong Kong as a prospective validation set and verified recurrent genomic aberrations in this type of cervical cancer [9].
Due to the limited quantity of circulating DNA preparations, we performed the ddPCR analysis to five missense single nucleotide substitution mutation spots of three genes in the present study. The most significant finding in the study was that the measurement of three single-point nonsynonymous mutations in PIK3CA: E542K, E545K, and H1047R using ddPCR is predictive of survival in a subgroup of cervical adenocarcinoma. We noted that in six of seven (85.7%) cases with recurrence, there was a PIK3CA mutation detected in tumor tissue DNA or circulating cfDNA collected before the recurrence occurred clinically (Supplementary Table S6). One case had a recurrence without a PIK3CA mutation detected in her tumor DNA or circulating cfDNA collected before treatment; however, this case did not have cfDNA samples collected during follow-up for testing the PIK3CA mutation. Therefore, we could not exclude the possibility that a PIK3CA mutation had existed before recurrence occurred clinically. Although the numbers are small, the high frequency of PIK3CA variants identified in samples before recurrence is noteworthy. ddPCR is increasingly available in clinical care. Further study is warranted to validate our findings; however, ddPCR detection of PIK3CA could be implemented in clinical practice for the early identification of recurrent disease, which may have prognostic significance.
Over 80% of somatic missense mutations in PIK3CA are found in the kinase and helical domains of the PIK3CA subunit, and cluster in three “hotspots”: E542K, E545K (in kinase domain), and H1047R (in helical domain) [10]. Mutational activation of PIK3CA detected in tumor tissue has been found in association with both an adverse clinical outcome of patients in multiple solid tumors and can predict a favorable response to PIK3CA inhibitors [10]. However, there is disagreement as to whether the PIK3CA mutation status correlates with patient survival in cervical cancer in previous reports [11,12,13,14,15,16,17,18,19] (Supplemental Table S7). The discrepancy of the PIK3CA mutation detection rate in tumor tissue of cervical adenocarcinoma among these reports were attributed to the differing purity of tumor cells in a sample, differing sensitivity of the measurement, or differing populations and regions as well as differing sample sizes. It is desirable to conduct a collaborative study in multiple centers in different regions with different races using a standardized mutation detection method in the future. These studies only tested PIK3CA mutations in tumor DNA but not circulating DNA and did not clearly analyze different sub-types of cervical cancer separately, while our work contributes novel data to this assessment. The correlation between the findings of mutations detected in cfDNA in other malignant tumors and negative prognosis was also reported. In patients of advanced non-small-cell lung cancer (NSCLC) with tissue epidermal growth factor receptor (EGFR) T790M-positive, an absence of detectable plasma T790M at baseline is associated with longer progression-free survival, which may be attributed to a lower disease burden [20]. In patients with estrogen receptor-positive advanced metastatic breast cancer (ER+ mBC) in the BEECH study, it was shown that early on-treatment ctDNA dynamics are a surrogate for progression-free survival. Dynamic ctDNA assessment has the potential to substantially enhance early drug development [21].
In the present study, somatic point mutations in KRAS appeared in a low frequency in both tumor and circulating DNA samples. KRAS is a dominant oncogene with alterations found in both COSMIC and cBioPortal. There are multiple hotspot mutations in KRAS, but only one spot, p.G12V, was examined in the present study due to the limited amount of circulating DNA available in the sample pool. A KRT6A point substitution mutation was detected in 12 of 24 (50%) patients in this study. Keratin 6A is a type II cytokeratin hyperexpressed keratin 6A in lung adenocarcinoma that promotes lung cancer proliferation and metastasis via epithelial–mesenchymal transition and cancer stem cells transformation [22]. Although the mutation was not associated with clinicopathological features, it may be worthwhile to explore its pathologic functions in cervical adenocarcinoma and the potential to be a therapeutic target in this disease.
One of the limitations of the current work is the small sample size. While five major medical centers were included in this study, the final results were only able to be obtained from Prince of Wales Hospital (PWH), Hong Kong, resulting in a smaller case series.
However, despite the small sample size, the results were significant and thus warrant further investigation. Some questions remain: (1) are the associations valid across multiple cancer centers in different regions and different demographics, (2) can ddPCR of the PIK3CA mutation be implemented for accurate and affordable disease monitoring, and (3) can an appropriate PIK3CA inhibitor improve outcomes for a late stage of cancer with PIK3CA mutation detectable? Hopefully, a subgroup of particularly advanced cervical adenocarcinoma patients can benefit from this specialized prognosis, treatment and management. When designing continuing prospective validation studies, a priori power analysis and sample calculation are suggested to achieve the most reliable output.
5. Conclusions
This translational study identified that the detection of PIK3CA mutations in three hotspots and combining tumor and circulating DNA assessment using robust ddPCR was predictive of a reduction in both progression-free and overall survival in a clinically important sub-group of cervical adenocarcinoma. The stratification of patients with cervical adenocarcinoma, regardless of stage, based on the PIK3CA mutation could be implemented in a clinical setting able to perform ddPCR assays on liquid biopsies, and may offer the possibility of tailoring management.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cancers13133218/s1. Table S1. DNA sequences of gBlock gene fragments used as controls for each mutation site. Table S2. Gene mutation copy (Copies/ng) detected in tumor tissue DNA, and circulating DNA collected before treatment and during the follow-up period. Table S3. Cox regression analysis of survival related to clinicopathological features and gene mutations shown as p-value in cervical adenocarcinoma. Table S4. Cox regression analysis of recurrence related to clinicopathological features and gene mutations shown as p-value in cervical adenocarcinoma. Table S5. Association of survival and with clinicopathological features and gene mutations in cervical adenocarcinoma analyzed using Fisher’s exact test shown as p-value. Table S6. PIK3CA mutation detection in cervical adenocarcinoma with recurrence. Table S7. PIK3CA mutation and clinical outcome in cervical adenocarcinoma in previous reports.
Author Contributions
Y.-F.W., T.K.H.C., G.D. and R.S.B. conceived and designed the study. T.-H.C., S.-F.Y., J.H.S.L., S.-Y.Y. and L.W. collected clinical data, specimens and samples. C.S.P., A.I.O., M.D.D., A.N., B.M.W. and A.D., contributed to whole exome sequencing data generation and related data analyses. M.-Y.Y. contributed to histopathology data generation and performed pathology review of tumor specimens. R.R.Y.W., G.D., Y.-F.W. and Y.M. performed computational and statistical data analyses. K.-M.L. contributed to ddPCR data generation, and method descriptions. M.J.W.J., K.M.E., A.R.T., C.S.P., V.W.W., N.S.H., M.R.D., S.-O.A.L., S.C.M., P.K.S.C., M.S.L., C.P.C. and R.W.K.C. contributed expertise, checked the data and edited the final version of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Hong Kong Research Grant Council (Reference Number 14101315).
Institutional Review Board Statement
The protocol was approved by the Ethics Committee of The Chinese University of Hong Kong, Hong Kong (Project Code 2014.514). The investigations were carried out following the rules of the Declaration of Helsinki.
Informed Consent Statement
All subjects gave their informed consent for inclusion before they participated in the study.
Data Availability Statement
Data is contained within the article or Supplementary Materials.
Acknowledgments
We sincerely thank Jin-he Lang, Matthew Meyerson, David Smith and Gad Getz for strong support in the project funding application. We very much thank Jin-he Lang, Xiu-gui Sheng, Yi-le Chen, Guonan Zhang, Ge Lou and their team members who made a lot of efforts in the preparation of subject recruitment. We also greatly thank Bernard Rosner, Robert Glynn and Frank Schoonjans for their important expert advice in statistical analysis.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Available online: https://www.iarc.fr/featured-news/media-centre-iarc-news-wcd (accessed on 2 February 2017).
- del Carmen, M.G.; Schorge, J.O. Invasive Cervical Adenocarcinoma; UpToDate: Waltham, MA, USA, 2019. [Google Scholar]
- Wang, S.S.; Hildesheim, A. Chapter 5: Viral and host factors in human papillomaviruspersistence and progression. JNCI Monogr. 2003, 31, 35–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ojesina, A.I.; Lichtenstein, L.; Freeman, S.S.; Pedamallu, C.S.; Imaz-Rosshandler, I.; Pugh, T.J.; Cherniack, A.D.; Ambrogio, L.; Cibulskis, K.; Bertelsen, B.; et al. Landscape of genomic alterations in cervical carcinomas. Nature 2014, 506, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Integrated genomic and molecular characterization of cervical cancer. Nature 2017, 543, 378–384. [Google Scholar] [CrossRef] [PubMed]
- Halle, M.K.; Ojesina, A.I.; Engerud, H.; Woie, K.; Tangen, I.L.; Holst, F.; Høivik, E.; Kusonmano, K.; Haldorsen, I.S.; Vintermyr, O.K.; et al. Clinicopathologic and molecular markers in cervical carcinoma: A prospective cohort study. Am. J. Obstet. Gynecol. 2017, 217, 432.e1–432.e17. [Google Scholar] [CrossRef] [PubMed]
- Lachkar, B.; Minaguchi, T.; Akiyama, A.; Liu, S.; Zhang, S.; Xu, C.; Shikama, A.; Tasaka, N.; Sakurai, M.; Nakao, S.; et al. Prognostic significance of PIK3CA mutation in stage IIB to IVA cervical cancers treated by concurrent chemoradiotherapy with weekly cisplatin. Med. Baltim. 2018, 97, e11392. [Google Scholar] [CrossRef] [PubMed]
- Chung, T.K.H.; Van Hummelen, P.; Chan, P.K.S.; Cheung, T.H.; Yim, S.F.; Yu, M.Y.; Ducar, M.D.; Thorner, A.R.; MacConaill, L.E.; Doran, G.; et al. Genomic aberrations in cervical adenocarcinomas in Hong Kong Chinese women. Int. J. Cancer 2015, 136, 776–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, T.K.H.; Cheung, T.H.; Yim, S.F.; Thorner, A.R.; Ducar, M.D.; Lau, K.K.M.; Wong, R.R.Y.; Berkowitz, R.S.; Wong, Y.F. Recurrent mutations and liquid biopsy in cervical adenocarcinoma. In Proceedings of the Second AACR International Conference: Translational Cancer Medicine, Cancer Discoveries for Clinical Application, Sao Paulo, Brazil, 27–29 September 2018; p. B45. [Google Scholar]
- Ligresti, G.; Militello, L.; Steelman, L.S.; Cavallaro, A.; Basile, F.; Nicoletti, F.; Stivala, F.; McCubrey, J.A.; Libra, M. PIK3CA mutations in human solid tumors. Cell Cycle 2009, 8, 1352–1358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McIntyre, J.B.; Wu, J.S.; Craighead, P.S.; Phan, T.; Köbel, M.; Lees-Miller, S.P.; Ghatage, P.; Magliocco, A.M.; Doll, C.M. PIK3CA mutational status and overall survival in patients with cervical cancer treated with radical chemoradiotherapy. Gynecol. Oncol. 2014, 128, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.A.; Howitt, B.E.; Myers, A.P.; Dahlberg, S.E.; Palescandolo, E.; van Hummelen, P.; MacConaill, L.E.; Shoni, M.; Wagle, N.; Jones, R.T.; et al. Oncogenic mutations in cervical cancer: Enomic differences between adenocarcinomas and squamous cell carcinomas of the cervix. Cancer 2013, 119, 3776–3783. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.; Li, J.; Jiang, W.; Shen, X.; Yang, W.; Wu, X.; Yang, H. Comprehensive analysis of targetable oncogenic mutations in chinese cervical cancers. Oncotarget 2015, 6, 4968–4975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, L.; Jiang, W.; Li, J.; Shen, X.; Yang, W.; Yang, G.; Wu, X.; Yang, H. PIK3CA mutation analysis in Chinese patients with surgically resected cervical cancer. Sci. Rep. 2015, 5, 14035. [Google Scholar] [CrossRef] [PubMed]
- Akbarov, K.; Isayev, I.; Melikova, L.; Guliyev, E.; Aliyeva, N. PIK3CA gene mutation frequency among cervical cancer patients in Azerbajian. Ann. Oncol. 2017, 28 (Suppl. S10), 86–93. [Google Scholar]
- Hodgson, A.; Amemiya, Y.; Seth, A.; Cesari, M.; Djordjevic, B.; Parra-Herran, C. Genomic abnormalities in invasive endocervical adenocarcinoma correlate with pattern of invasion: Biologic and clinical implications. Mod. Pathol. 2017, 30, 1633–1641. [Google Scholar] [CrossRef] [PubMed]
- Razia, S.; Nakayama, K.; Nokamura, K.; Ishibashi, T.; Ishikawa, M.; Minamoto, T.; Iida, K.; Otsuki, Y.; Nakayama, S.; Ishikawa, N. Clinicopathological and biological analysis of PIK3CA mutation and amplification in cervical carcinomas. Exp. Ther. Med. 2019, 18, 2278–2284. [Google Scholar] [CrossRef] [Green Version]
- Scholl, S.; Popovic, M.; de la Rochefordiere, A.; Girard, E.; Dureau, S.; Mandic, A.; Koprivsek, K.; Samet, N.; Craina, M.; Margan, M.; et al. Clinical and genetic landscape of treatment naive cervical cancer: Alterations in PIK3CA and in epigenetic modulators associated with sub-optimal outcome. EBioMedicine 2019, 43, 253–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arafeh, R.; Samules, Y. PIK3CA in caner: The past 30 years. Semin. Cancer Biol. 2019, 59, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Papadimitrakopoulou, V.A.; Han, J.; Ahn, M.; Ramalingam, S.S.; Delmonte, A.; Hsia, T.; Laskin, J.; Kim, S.; He, Y.; Tsai, C.; et al. Epidermal growth factor receptor mutation analysis in tissue and plasma from the AURA3 trial: Osimertinib versus platinum-pemetrexed for T790M mutation-positive advanced non–small cell lung cancer. Cancer 2020, 126, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Hrebien, S.; Citi, V.; Garcia-Murillas, I.; Cutts, R.; Fenwick, K.; Kozarewa, I.; McEwen, R.; Ratnayake, J.; Maudsley, R.; Carr, T.H.; et al. Early ctDNA dynamics as a surrogate for progression-free survival in advanced breast cancer in the BEECH trial. Ann. Oncol. 2019, 30, 945–952. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Zhang, W.; Zhang, M.; Wang, X.; Peng, S.; Zhang, R. KRT6A Promotes EMT and Cancer Stem Cell Transformation in Lung Adenocarcinoma. Technol. Cancer Res. Treat. 2020, 19, 15330338209212. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).