Simple Summary
The classical approach to study the immune response against a tumor was mixing immune cells with tumor cell suspensions in several experimental settings. These models lack the appropriate tissue architecture in which the immune response takes place and do not consider other cellular and extracellular players of the tumor microenvironment essential to understand the anti-tumor immune response. Thus, to confirm in vitro data, in vivo experiments have been extensively performed, using animal models that may not fully reproduce what happens in humans. Indeed, in animal-based studies, tumors are artificially generated in a short time, and immune cell subsets and receptor-ligands pairs, involved in tumor cells recognition by the immune system, are often different from human counterparts. To reduce the number of animals used, and possibly replace animal models, alternative methods of culture have been developed. Herein, some of these approaches will be described, highlighting their advantages and disadvantages, focusing on natural killer cells as the first line of anti-tumor effector cells able to contrast tumor growth.
Abstract
Several approaches have shown that the immune response against tumors strongly affects patients’ clinical outcome. Thus, the study of anti-tumor immunity is critical to understand and potentiate the mechanisms underlying the elimination of tumor cells. Natural killer (NK) cells are members of innate immunity and represent powerful anti-tumor effectors, able to eliminate tumor cells without a previous sensitization. Thus, the study of their involvement in anti-tumor responses is critical for clinical translation. This analysis has been performed in vitro, co-incubating NK with tumor cells and quantifying the cytotoxic activity of NK cells. In vivo confirmation has been applied to overcome the limits of in vitro testing, however, the innate immunity of mice and humans is different, leading to discrepancies. Different activating receptors on NK cells and counter-ligands on tumor cells are involved in the antitumor response, and innate immunity is strictly dependent on the specific microenvironment where it takes place. Thus, three-dimensional (3D) culture systems, where NK and tumor cells can interact in a tissue-like architecture, have been created. For example, tumor cell spheroids and primary organoids derived from several tumor types, have been used so far to analyze innate immune response, replacing animal models. Herein, we briefly introduce NK cells and analyze and discuss in detail the properties of 3D tumor culture systems and their use for the study of tumor cell interactions with NK cells.
1. Introduction
In late 1980s, the seminal findings of Rosenberg and colleagues on the so-called lymphokine activated killer (LAK) cells have shown that LAK-killing of tumor cells can eliminate both autologous and heterologous tumor cells in vitro, and cure mice from melanoma [1,2,3,4]. The transfer to the clinic of Rosenberg’s findings by the systemic administration of interleukin 2 (IL2) showed several drawbacks, such as the capillary leakage syndrome [5,6] leading to fatal outcome in some patients [6]. Indeed, IL2, essential for the generation of LAK cells, gave rise to relevant, unpredictable adverse effects in humans, not affecting murine models [1,2,3,4,5,6]. More recently, the key role of the immune response became evident testing immune-checkpoint (IC) blockers (B) to reactivate the anti-tumor immune response in host bearing tumors [7,8,9,10,11]. In this case, using appropriate tools such as humanized monoclonal antibodies (hmAb) to programmed cell death receptor 1 (PD1), programmed cell death receptor ligand 1 (PDL1) or cytotoxic activated T lymphocyte 4 receptor (CTLA4), it is possible to reactivate the adaptive anti-tumor-specific immune response [7,8,9,10,11]. This strategy is effective when IC-inhibited tumor-specific T cells are already present in the host, thus targeted hmAb can relieve the tumor microenvironment (TME)-mediated immunosuppression [11,12,13,14,15,16].
The study of the molecular mechanisms underlying IC-immunosuppression and patient-specific immune response is difficult in animal models [15,16]. Humanized murine models and patient-derived tumor xenografts (PDX) have been extensively used, with some success. However, it is conceivable that the complex cross-talk among the different cellular and extracellular matrix components of TME is not completely and adequately reconstructed in these hybrid animal models. One example among the others is the species-specificity of some fundamental immunomodulatory cytokines [17,18,19,20,21,22,23,24,25,26].
The innate immunity arm of the anti-tumor immune system has become more and more relevant to improve patient’s response to conventional anti-tumor therapies [27,28,29,30,31,32,33]. Unfortunately, innate cells such as natural killer (NK) cells do not display similar phenotypic and functional features in mice and humans [27,28,29]. To better understand how innate cells can be used to fight cancer, suitable and feasible 3D culture models composed of tumor cells, tumor stromal cells and immune effectors have been set up and used to evaluate the anti-tumor effect of NK cells.
2. Developing 3D Culture Models
The addition of appropriate scaffolds and flow-based systems could mimic the architecture of the tumor tissue and the dynamic conditions faced by immune cells approaching and invading the tumor [15,16]. It is evident that the molecular events detected in conventional culture systems, consisting of a mixture of different cell types, cannot be compared to what happens in a 3D-growing tumor mass. On the other side, human tumor cells injected in mice do not find the micro (cell and matrix component) and macro environment (vascular, lymphatic and nervous systems) in which the original tumor mass developed [15,16].
As a possible alternative, several 3D culture systems have been proposed and validated by the EU Reference Laboratories (EURL-ECVAM), as preclinical models, for the selection of anti-tumor drugs [15,16,25], starting from the simplest model of tumor cell homotypic spheroids, composed of a single cell type, through more complex spheroids with tumor and mesenchymal stromal cells (MSC), such as tumor associated fibroblasts (TAF) [16]. Patient-derived organoids, generated from tumors and usually composed of tumor cells at different stage of differentiation, have been used to study and improve the anti-tumor effect of immune cells [34,35,36,37,38,39,40,41]. Some of these developing systems are summarized in Figure 1.
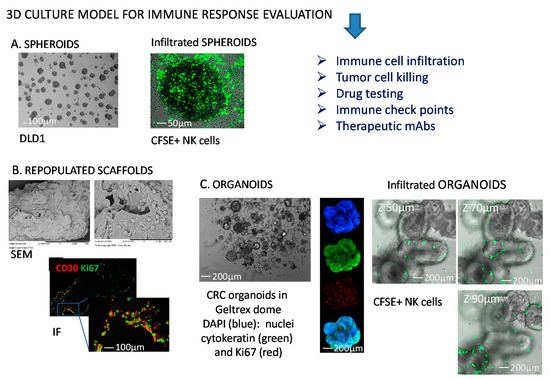
Figure 1.
Representative 3D models to study NK-tumor cell interactions. (A) Colorectal cancer (CRC) cell spheroids from DLD1 CRC cell line (left) infiltrated with NK cells (right) labelled with the green fluorescence probe carboxy fluorescein succinimidyl ester (CFSE). (B) Collagen scaffolds repopulated with mesenchymal stromal cells and Hodgkin’s lymphoma cells (scanning electron microscopy, SEM, upper) or analyzed by immunofluorescence (IF, lower) for the expression of CD30 antigen (HL cells, red) and Ki67 (proliferating antigen, green); (C) CRC organoids in a GeltrexTM dome (left), or labelled (middle) with 4′,6-diamidino-2-phenylindole (DAPI, blue), cytokeratin 2 (green), Ki67 (red), or merge (lower image); organoids analyzed by confocal microscopy upon NK cell (green labeled with CFSE) infiltration at different Z-stages (right). The dimension bars are shown in each panel.
Herein, we will consider the 3D systems used to study NK-tumor cell cross-talk and the possible improvements of these models to better understand this interaction.
3. Natural Killer Cells as Anti-Tumor Effectors
Originally, NK cells have been identified as a lymphocyte subset able to recognize tumor cell targets, without priming [42,43,44,45]. This functional definition has been flanked by the identification of phenotypic features identifying NK cells like lymphocytes expressing CD16, the low affinity receptor for fragment crystallizable region of immunoglobulin G, FcγRIIIA [42,43,44,46,47]. Additionally, the neural cell adhesion molecule (NCAM), called CD56, is a typical marker of peripheral blood (PB) NK cells [46,47]. PBNK cells express CD56 at two detectable levels, the so called CD56low and CD56high NK cells, which display a high and a low anti-tumor potential, respectively [46,47]. Of note, CD56low and CD56high PBNK cells can produce low and high amounts of cytokines such as IFNγ and GM-CSF [46,47]. Thus, based on the expression of CD16 and CD56 surface molecules, different NK cell subsets can be distinguished (see Figure 2).
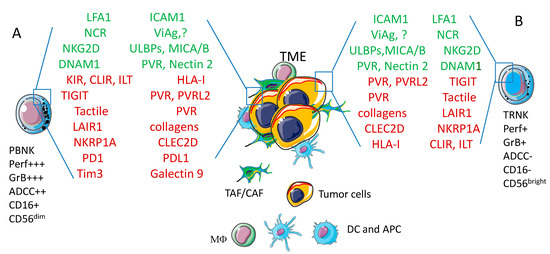
Figure 2.
Phenotype of peripheral blood (PBNK) and tissue resident (TRNK) NK cells. The large majority of PBNK (A) express CD16 and through this receptor for the Fc portion of immunoglobulins can activate the ADCC that induces the release of perforin and granzymes to kill target cells. A minority of PBNK does not express CD16, but displays high surface amounts of CD56 (CD56bright) antigen; these PBNK cells are similar to tissue TRNK (B). NK cells express a plethora of activating (green) or inhibiting (red) receptors that can interact with the corresponding ligands expressed on tumor cells within the tumor microenvironment (TME). Other cells of the TME, such as tumor/cancer associated fibroblasts (TAF/CAF), dendritic cells (DC) or antigen presenting cells (APC) and macrophages (MΦ) are also depicted. The engagement of activating receptors triggers cytolysis of tumor cells and/or the release of cytokines while the interaction of inhibiting receptors with corresponding ligands impair NK cell-mediated activities. The final net effect (triggering or inhibition) is determined by the balance of these positive and negative signals. The receptors and ligands mentioned in the figure are analyzed in detail in the text. Acronyms. LFA1: lymphocyte function antigen 1; ICAM1: intercellular cell adhesion molecule 1; NCR: Natural Cytotoxicity Receptor; ViAg: viral antigen; ?: not yet identified ligands; NKG2D: NK related group 2D; ULBPs: UL binding proteins; MICA/B: MHC-related molecule A/B; DNAM1: DNAX adhesion molecule 1; PVR: polio virus receptor; KIR: killer immunoglobulin receptor; CLIR: C-lectin inhibitory receptor; ILT: Immune lymphocyte transcript; HLA-I: human histocompatibility antigen; TIGIT: T cell immunoreceptor with Ig and ITIM domains; PVRL2: poliovirus receptor related 2; LAIR1: Leukocyte associated inhibitory receptor 1; NKRP1A: NK related protein 1A; CLEC2D: C-type lectin domain family 2 member D; PD1: programmed death receptor 1; PDL1: programmed death receptor ligand 1; Tim3:T-cell immunoglobulin and mucin-domain containing−3; Per: perforin; GrB: granzyme B; ADCC: antibody dependent cellular cytotoxicity.
Roughly, CD16+CD56low PBNK cells are potent antitumor effector cells and display a strong intracytoplasmic expression of cytotoxic molecules such as perforins and granzymes, whereas CD16-CD56high PBNK cells play a regulatory role due to the production of cytokines [46,47,48]. Of note, tissue-isolated NK cells display a surface phenotype more similar to CD16-CD56high than CD16+CD56low PBNK cells suggesting that tissue resident (TR), and thus tumor infiltrating NK cells are not prone to kill tumor cells. Thus, NK cells are surface CD3/TCR- with a different level of expression of CD16 and CD56 [49,50]. It is not clear to date whether resident NK cells are derived from PBNK cells and whether the two subsets described are different functional stage of the same NK cell at least in humans [51,52] (see Figure 2). Furthermore, several markers of NK cells are shared with the so-called innate lymphoid cells (ILC) including CD56, CD69, CD161, NKp30, NKp44 and NKp46 (these last three receptors called natural cytotoxic receptor, NCR) [49,50,51,52,53,54,55]. Indeed, NK cells have been considered among the different cells that belong to ILC1 subset of ILC [51,52], although it has been claimed that ILC1 and NK cells derive from different progenitor cells [53,54,55]. These molecules play a key role in NK cell-mediated activation and interaction with target cells (see Section 2).
4. Basic Interaction between Natural Killer Cells and Tumor Cells
A plethora of receptors expressed on NK cells, and the interaction with the corresponding ligands on tumor target cells, can lead to two main effect: activation or inhibition of NK cells [56,57,58,59]. These receptors–ligands pairs have been described extensively in several reviews that cover both murine and human NK cell features, and their detailed description is beyond the scope of this review. Recent reports can be consulted for a more comprehensive description of these characteristics [60,61,62,63,64,65,66,67,68]. Briefly, activating or inhibiting receptors can be found on NK cells [56,57,58,59], and their engagement eventually results in the killing or the protection from the killing of the target cell [56,57,58,59]. It is commonly accepted that NK cells do no kill self-healthy cells, because they recognize the self-MHC [56,57,58,59,64]. This recognition is mediated by receptors belonging to the Killer Ig like superfamily (KIR) and C-lectin type family (CLIR), that have been characterized in detail in other reviews [57,58,59]. When self-MHC or the receptors for MHC are covered in vitro by specific mAbs, NK cells can kill several self-cell types including tumor cells [57,58,59]. This killing is mediated by activating NK cell receptors whose engagement triggers intracellular calcium increase and activation of Akt kinase, inducing the release of cytotoxic granules at the NK-target cell interface [69,70,71,72,73]. Physiologically, the NK cell-mediated killing can occur when self-cells do not express MHC-class I, for instance when tumors downregulate these antigens to escape from the T cell-mediated antigen specific killing [74,75,76,77]. Nevertheless, several conflicting reports on the topic underline the complexity of self-recognition by NK cells and its regulation [56,57,58,59,78,79].
NK cells recognize and bind to the tumor target through several receptors [42,44,46,58]. The lymphocyte function associated antigen 1 (LFA1), a protein heterodimer composed of CD11a and CD18 chains, interacts with the intercellular adhesion molecule (ICAM) 1 expressed on target cells. ICAM1 is upregulated by IFNγ, induced by the inflammation that can accompany tumor growth [80,81,82,83,84,85]. Several other molecules, such as DNAM1, NKG2D and NCR, strengthen this binding and trigger the activation of killing [42,44,46,58]. DNAM1 and NKG2D receptors bind the corresponding counter-ligands on target cells, poliovirus receptor (PVR) and nectin 2 [86] or the UL binding proteins (ULBP1–6) and the MHC-related molecule A and B (MICA/B) [86,87,88,89,90,91]. Upon this binding NK cells can kill target cells through mainly two different mechanisms: release of lytic factors and enzymes (perforins and granzymes), and action of death inducing molecules such as FasL and tumor necrosis factor (TNF)α [92,93,94].
At the surface of NK cells are also expressed several receptors that deliver, upon engagement with counter-ligands, an inhibitory signal, blocking cytolysis and cytokine release [95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110]. These receptors, in addition to those involved in the recognition of self-MHC-class I mentioned above, include the leukocyte associated inhibitory receptor 1 (LAIR1) [95,96,97], some sialic acid-binding immunoglobulin-type lectins (siglecs 7 and 17) [98,99], the T cell immunoreceptor with Ig and ITIM domains (TIGIT) [100,101], the T cell activation increased late expression (Tactile, CD96) [102,103,104], the programmed cell death receptors 1 (PD1) [105,106,107], and the T-cell immunoglobulin and mucin-domain containing−3 (Tim 3) [108,109,110]. Thus, NK cells can kill a tumor target only when positive signals overwhelm negative ones [98]. This balance does not only rely on the expression of activating and inhibiting receptors on NK cells, but also on the expression of the corresponding counter ligands on target cells [95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110] (Figure 2).
The knowledge of the large majority of the molecular mechanisms involved in NK cell-mediated cytolysis has been derived from experiments in conventional culture systems, using tumor target cell suspensions labelled with radionuclide probes and incubated in V-bottomed well plates [111,112,113,114,115,116]. In these culture conditions, NK cells aggress carcinoma epithelial cells (the large majority of solid tumor tissues) as single cells with a spherical shape. These conditions are not similar to what happens in vivo, where tumor cells of epithelial origin grow attached to extracellular matrix in a compact 3D structure. Furthermore, the labelling with a radioactive probe limits the analysis of NK-tumor interactions to short-time assays, due to the toxicity of the probe itself. Thus, some of the findings found in conventional assays are far from the in vivo situation.
5. Tumor Cell Spheroids as a 3D Model to Study Tumor Cell Biology and Tumor-Natural Killer Cross-Talk
To transfer in a clinical setting the powerful anti-tumor immune activity of NK cells reported in vitro, several animal models have been used to confirm in vitro observations [18,19,20,21,22,23,24,25,26]. Unfortunately, these models are not fully appropriate, as human neoplastic cells are usually ectopically injected in immunocompromised animals to avoid xenogeneic reaction [18,19,20,21,22,23,24,25,26]. Furthermore, some side effects of the immunotherapy, such as capillary leakage syndrome, have not been detected in animals [1,2,3,4,5,6]. The implant of patient derived xenograft tissue (PDX) in non-obese diabetic severe combined immunodeficient gamma (NOD-SCIDγ or NSG) mice has been extensively used [18,22,24]. The anti-PDX effects of human immune cells, in these humanized mice, only partially resemble the development of a tumor in humans, as tumor microenvironment is complex and composed of different cellular and extracellular components that cannot be replaced by murine cells [18,19,20,21,22,23,24,25,26]. Recently, it has become evident that the replacement of animal experimentation, whenever possible, can be achieved by using 3D culture systems [15,16]. Thus, tumor cell spheroids (this paragraph) and organoids (next paragraph) have been created to study the biological behavior of tumor and immune cells [34,116,117]. Some of the advantages and disadvantages of the above-mentioned models to study immune response are summarized in Table 1.

Table 1.
Advantages and disadvantages of in vitro and in vivo tumor cell culture models to study interactions with immune cells.
Spheroids can be derived from either established cell lines or primary isolated tumor cells [35,118,119,120,121,122,123,124,125,126,127,128]. Usually, spheroids are obtained culturing tumor cells in ultra-low adherent plasticware or by hanging drop methods, forcing tumor cells to aggregate and generate tumor sphere in static or microfluidic systems [15,16,126,127,128,129,130,131,132,133,134,135,136]. The spheroids can be composed of a single (homotypic) or, less frequently, different (heterotypic) cell types [15,16,125,126]. Of note, it has been reported that spheroids can be obtained from patients’ tumor-specimens digestion, and self-immune cells can be challenged with self-spheroids to analyze the effect of different drugs and plan their application into the clinic [15,16,117,119,123]. By this way, the 3R (Reduce–Reuse–Recycle) policy has become more and more followed, trying to replace, reduce and refine the use of animal-based assays [134,135]. Herein, we will describe the experimental data derived from spheroid models where the cross-talk between NK and tumor cells have been analyzed.
5.1. NK Cells Interaction with Colorectal Carcinoma (CRC) Cells
5.1.1. CRC Biological Features and CRC Spheroids
Several reports have considered CRC spheroids as a good tool to study in a 3D system the features of tumor cells and their interactions with immune cells [125,126,127,136,137,138]. The interest on CRC is related to the strong clinical relevance of this tumor and the impact of immune infiltration on patient prognosis [139,140,141]. Indeed, CRC is the third cancer diagnosed in males, the second in females and it is the second cause of cancer-related death in USA [142,143,144]. CRC has been subdivided into four groups, the Consensus Molecular Subtypes (CMS), based on clinic, pathologic and molecular profiling data [145]. CRC is a very heterogeneous inter and intra patient disease [142,143,144,145]. Recent advances in treating tumors with ICB, are not effective in most CRC, except for microsatellite instability (MSI)-high cancers, showing high neoantigens load [143]. The studies of CRC biology in immunocompetent animal models are limited [146,147,148]. This prompts to generate models to study patients’ immune response variability and drug sensitivity, and to characterize the interaction of immune cells with 3D CRC-derived spheroids [119,120,125,126,127,136]. Tumor cell spheroids can be obtained applying different culture methods [125,126,127,128,129,130,131,132,133] (Figure 3).
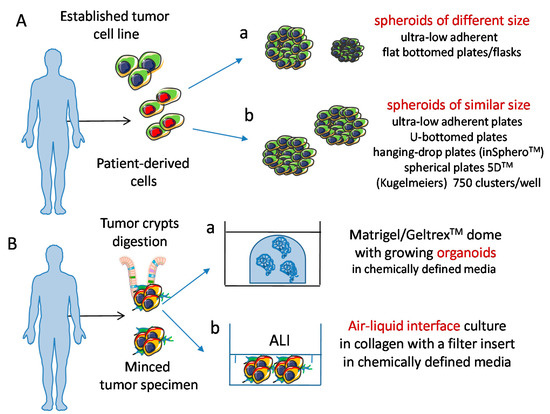
Figure 3.
Schematic representation of 3D culture models of tumor cells. (A) Spheroids can be obtained from tumor cell lines and tumor cell suspensions. Physical features of spheroids can change, depending on the culture conditions used, resulting in different (a) or similar (b) size. (B) Organoids can be derived from tumor cell suspensions seeded either in matrigel/geltrex domes (a) or in air liquid interface (ALI) cultures, (b) and can display a well-defined polarization of molecules like those localized at the basal or apical pole of cells. Importantly, spheroids and organoids can be cultured in chemically defined media; this is of relevance to avoid any influence during the experimental procedures of animal/viral derived factors; this leads to the generation of a more physiologic microenvironment.
5.1.2. Generation of CRC Spheroids and Molecules Involved in NK Cell Mediated Recognition of Tumor Cells
Usually, tumor cells are seeded onto ultra-low adherent culture wells [125,126,127]: this impairs adhesion of tumor cells to plastic substrate and favors the cell-to-cell aggregation. The size of spheroids can be controlled culturing the same number of cells in each well, and this is allowed when tumor cells are put in very little U or V bottomed wells or in hanging-drop plates [129,130,131,132,133]. Spheroids can be embedded into hydrogel drops that solidify at warm temperature, or by calcium addition, and be used as models of initial tumor growth [149,150,151,152,153,154]. This approach allows the contemporary assay of a large number of similar-sized spheroids, incubated with drugs and/or immune cells [154]. On the other hand, spheroids of different size can be randomly obtained when tumor cells are plated onto larger, flat-bottomed plates [125,126,127]. These different approaches can be used to standardize the results. Standardization is easier, having a lot of replicates of the immune-tumor cell interaction, when NK cells are faced with a spheroid of a defined size. Indeed, standardization is a key point to translate in vitro data to the clinic [155,156,157,158,159]. However, the use of spheroids of different size can characterize the behavior of NK cells when challenged with tumors showing different mass density, or inner hypoxic areas [126].
In this context, it has been shown that the NKG2D receptor plays a key role in the recognition of CRC spheroids derived from either established cell lines such as HT29 and DLD1, as well as primary cell suspension from tumor biopsies [119]. This finding is in line with previous observations obtained in conventional culture systems, supporting the notion that 3D culture can validate data from in vitro experiments in a more physiological microenvironment [119]. In this model, the contemporary targeting of NKG2D ligands MICA/B on tumor cells, and the inhibitory receptor NKG2A on NK cells, led to a synergistic effect in blocking tumor cell growth, even if it expressed the NKG2A ligand HLA-E [119]. The authors also originated patient-derived spheroids, mainly composed of a mixture of TAF (due to the initial 2D condition and the presence of serum) and a minor population of tumor epithelial cells. These spheroids were attacked by TIL (either NK or T cells) upon stimulation with IL15 [119]. The use of self-peripheral blood lymphocytes and self-TIL could aid to select appropriate drugs combinations targeting tumor cell growth without impairing the immune response [119].
5.1.3. Invasion and Killing of CRC Spheroids
Haploidentical allogeneic NK cells are an efficient tool to eliminate hematological cancer cells [160], and they have been evaluated as a therapeutic tool for killing CRC spheroids [120,161]. NK cells activated in vitro by the lymphoblastic R69 cell line, without IL2, can kill CRC spheroids derived from Caco−2, HT29 and HCT116 independently from the expression of PDL1 on tumor cells [162]. An inverse correlation between the effector/target ratio used in cytotoxicity assays against spheroids and NK cell infiltration has been shown [160]. At high E:T ratio allogeneic NK cell can kill CRC cells in spheroids without any apparent infiltration, this observation could be intuitive, as lots of NK cells covering the entire spheroid surface would kill all epithelial cells before being able to attempt any invasion of the disaggregating spheroid. On the other hand, the ability of NK cells to kill or invade a tumor spheroid could be triggered by a different balance of stimuli. In this perspective, it is important to define whether a so-called spheroid effectively displays a spherical 3D structure [126]. Indeed, the microscopic observation of a sphere as well as of a discoid is visualized as a circle in x–y axis. The spherical shape can be assessed by the use of microfluidic systems [126]. By this equipment [126], we characterized the exact shape and measured the mass density of CRC cell line spheroids. Mass density is the ratio between the weight and the volume of a spheroid, and it can be considered a parameter typical for each CRC cell line. Interestingly, this parameter changed during both spheroid infiltration by NK cells and NK cell-mediated killing of tumor cells [126]. These findings indicate that mass density can characterize a tumor cell spheroid, and its evaluation can provide new insights on how tumor cells respond to NK cell aggression. Of course, epithelial tumor spheroids lack many other components of TME, and only the analysis of complex heterotypic spheroids could complete the picture. The achievement of such a model is complex, as many paracrine stimuli would be activated and difficultly monitored. However, it is of note that mass density, and possibly other undefined biomechanical features, could give a deeper characterization of tumor behavior and responsiveness to immunotherapy.
5.2. NK Cells Interaction with Other Solid Tumor Spheroids
The cross-talk between NK cells and solid tumor cells has also been assessed using spheroids of neuroblastoma, ovarian, cervical and breast carcinomas [121,123,163,164,165]. Neuroblastoma spheroids have been used to assess NK-cell mediated cytotoxicity upon treatment with nutlin-3a, a small molecule that antagonizes the inhibitory effect of MDM2 on p53. The reactivation of p53 triggered the upregulation of DNAM1 ligands on neuroblastoma cells activating NK cell recognition and killing [165]. NK cells cultured with spheroids of ovarian cancer cells (IGROV1, SKOV3, OVCAR3) can downregulate the expression of DNAM1, the same happens co-culturing ovarian cancer specimens with NK cells [121]. The inhibitory effect mediated by DNAM1 downregulation was enforced by the upregulation of the inhibitory receptor TIGIT that can interact, like DNAM1, with PVR and nectin 2, while the levels of CD96, the third receptor for DNAM1 ligands, remained unaffected. These findings indicate that tumor cell line-derived spheroids are a suitable tool to analyze the response of NK cells. Triple negative breast carcinomas (TNBC, HCC1806 and MDA-MB231cell lines) spheroids have been used to assess the immunotherapeutic effect of the bispecific antibody targeting mesothelin and triggering CD16 by a Fab-like domain (MesobsFab) [164]. MesobsFab triggered NK cell-mediated killing and infiltration of tumor spheroids.
Finally, it has been reported that the expression of NKG2D ligands (NKG2DL) was partially downregulated in spheroids of the human cervix squamous cell carcinoma line SiHa and the cervical epidermoid carcinoma CaSki, compared to classical monolayer culture conditions [123]. This downregulation was due, at least in part, to the shedding of NKG2DL, that were detected in the culture medium. Though, these spheroids were infiltrated by NK cells that efficiently killed tumor cells through the activation of NKG2D-NKG2DL interaction. Indeed, anti-NKG2D-blocking antibodies strongly inhibited NK cell-mediated killing [123].
6. Tumor Cell Organoids: A More Reliable 3D Model to Study Tumor Cell Biology and Tumor-NK Cells Interactions?
Tumor organoids can be derived mainly by two procedures: using (a) an air–liquid interface (ALI) method, embedding the tissue sample en-bloc, without immersion in culture medium [166,167], or (b) matrix-embedded epithelial cells derived from tissue digestion, and continuously growing by the activity of Wnt and other mitogenic and stem factors [40,168,169] (Figure 3B). These approaches can be used to generate organoids from different tumor specimens [40,166,167,168,169] and both can be used to study immune system–tumor cell interactions [40,166,167,168,169]. The ALI method has been described quite recently, and allows to study tumor infiltrating lymphocyte (TIL) phenotype and functional activities for a discrete period and a few culture passages [166]. In this system, TIL can be expanded in the complex microenvironment of the original tumors [166]. Though, the complex structure of the tissue is progressively lost, along time [166]. Tumor associated fibroblasts are present, as smooth muscle alpha actin (SMA) and vimentin positive cells; this may be due to the non-enzymatically based procedure of generation of these organoids, and the presence of serum in the culture medium [166]. However, also myofibroblasts are progressively lost during culture. Thus, this model should be analyzed in short-term experiments for the evaluation of tumor and stroma patient-specific features. The survival of specific cell populations can be improved by the exogenous addition of growth factors. For example, CD3+ T lymphocytes (either CD4 or CD8), CD68+ macrophages, NKT cells and NK cells can be maintained for about two months in the presence of exogeneous IL2 [166].
On the other hand, matrix-embedded pure epithelial organoid cultures are generated by the proliferation and partial differentiation of epithelial stem cells, such as Lgr5+ cells. The presence of a very defined culture medium, allows to preserve the epithelial stem cell population by appropriate mitogenic stimuli and inhibition of differentiating signals, though it is not permissive for mesenchymal cell growth [170]. These organoids can be indefinitely expanded in culture and allowed to growth in different sizes. This model has been used to proof the possibility of deriving anti-tumor specific T lymphocytes in vitro, able to recognize and kill self-tumor cells [170]. In this case, while the generation of CD8 T cells was tumor antigen specific, the CD4+ T cell subset could respond to matrix mouse components. Thus, this system is more suitable to study cytotoxic T cell immune response, than CD4-mediated help. In the next paragraph, we discuss the reports regarding the interaction between organoids and NK cells, while the studies on T cell-mediated immune response are not considered, being treated in specific reviews [168,169,170,171].
6.1. How NK Cells Modify Their Behaviour upon Interaction with Organoids
6.1.1. Co-Culture Conditions of Organoids and Immune Cells
Data on how NK cells interact with organoids are scanty and it is relevant to clarify the exact culture conditions in which the experiments have been performed. Usually, immune cells are cultured in conventional RPMI1640 medium supplemented with fetal calf serum, while organoids should be cultured in chemically defined media, with specific growth factors and supplements (such as epidermal growth factor, EGF, fibroblast growth factor, FGF, Wnt3a, R-spondin1, noggin, B27, N-acetyl cysteine, and chemical inhibitors of the TGF-β and p38 pathways) [168,169,170,171]. It is not well-defined whether these different culture conditions may affect the cross-talk between immune and epithelial cells. Furthermore, immune cells should be supported by the addition of exogeneous IL2 or other immune cell stimulating triggers. It has been reported that unfractionated self-peripheral blood mononuclear cells (PBMC) can be co-cultured for a long time with epithelial cell organoids from CRC or non-small cell lung carcinoma (NSCLC) in the presence of IL2, IFNγ, anti-CD3 and anti-CD28 antibodies coated to plastic for 28 days. Anti-PD1 blocking antibodies were used to limit the inhibition of the immune response mediated by the interaction of T lymphocytes-expressed PD1, with PDL1 on epithelial organoids [168,169,170,171]. Under these complex experimental conditions, it has been shown for the first time to our knowledge, that an efficient T lymphocyte-specific immune response against self-tumor organoids can be elicited. This is the proof of principle that the presence of specific anti-tumor T lymphocytes in a given patient can be assessed in vitro [170].
6.1.2. Culture Requirements of NK Cells
NK cells require only IL2 (or another cytokine such as IL12, or IL7, or IL15, or IL21) to proliferate, differently from T lymphocytes, because they express the βγ chains of IL2 receptor [172,173,174]. These cytokines can enhance the basal cytolytic activity of NK cells and trigger an evident cytotoxic effect against carcinoma cells [172,173,174,175,176]. The expression of PD1 is limited to a minor population of polyclonal activated NK lymphocytes and mainly in selected donors [107,177]. Thus, the addition of anti-PD1 antibodies to a culture of NK cells with organoids is not needed. Peripheral blood NK cells, in pancreatic ductal adenocarcinoma (PDAC) patients, display CD16 and CD57 upregulation, while these markers are downregulated in the scanty NK cells infiltrating the tumor, showing also reduced NKG2D and NKp30 activating receptors expression [178]. The co-culture of PBMC from these patients, with self-organoids, determined a phenotype switch of NK cells, with downregulation of CD16, and low CD57, as observed in PDAC tumors. It is of note, that PDAC patients with a fraction of PB CD56low CD16- NK cells over 1% (the median level of this subset), showed less PDAC recurrence than patients with less than 1%. This would indicate that the TME of organoids co-cultured with self-PB NK cells in vitro could induce the selection of NK cell subsets mimicking patient’s situation. Thus, in vitro 3D systems can mimic pathophysiological conditions, supporting their role for a better understanding of the innate immune response against cancer [178].
6.1.3. Interactions of NK Cells and CRC-Derived Organoids
Patients-derived CRC organoids have been used for preclinical assessment of the chimeric antigen receptor (CAR) NK cells [179]. Using the standardized CAR-NK92 model, it was possible to quantify the CAR-mediated cytotoxicity against CRC organoids. First, it has been shown that the cytolytic activity of the CAR-NK92 cell line was strongly impaired by the presence of nicotinamide in the organoid medium, whereas the depletion of this compound did not affect the viability of organoids. Luciferase/GFP expressing CRC organoids were used as targets for EPCAM-specific CAR-NK92, in nicotinamide-free medium. These co-cultures were dynamically monitored, by confocal microscopy imaging and subsequent automatic analysis of recovered data. The cytotoxicity of CAR-NK92 cells against EPCAM+ organoids was quantified evaluating both the decrease of luciferase/GFP fluorescence and the area occupied by organoids [179]. In the same model, it has been shown that engineered EGFRvIII-CAR-NK92 cells can kill transduced EGFRvIII-organoids, but not non-transduced organoid controls. This indicates that organoids can be used as a tool to quantify CAR-NK92 cytotoxicity and that EGFRvIII-CAR-NK cells might be used as a therapeutic tool, as the EGFRvIII form of EGFR is a peculiar feature of several tumors [180,181].
6.2. Improvements of 3D Culture Systems and Therapeutic Relevance of Studies in 3D Models
6.2.1. Limitations of Spheroids and Organoids as 3D Culture Models
Both tumor spheroids and tumor organoids have limitations in mimicking the physiological conditions where NK cells interact with tumor cells. First, immune cells should reach a tumor by extravasation, interacting with the endothelial cell layer [182,183,184,185]. Afterwards, NK cells need to degrade the extracellular matrix, to reach tumor tissue [186,187,188]. In some instances, they can encounter mesenchymal stromal cells (MSC) that are in close proximity to tumor cells. MSC themselves can be influenced by the growing tumor and can be activated. MSC proximal to the tumor mass do not show altered markers, compared to healthy MSC, resembling reactive MSC instead of tumor-infiltrating MSC [189,190,191]. Thus, MSC close to the tumor have some features of repairing tissue MSC [189,190,191]. Additionally, NK cells come across with several healthy cells before reaching tumor cells [192,193,194]. It is conceivable that NK cells may interact with a tumor-budding spheroid during the progressive growth of the neoplastic mass [195,196,197,198]. The infiltration of the tumor will be different if NK cells reach the tumor from its periphery, or from a vessel generated inside the tumor mass. In this context, the study of the NK-tumor cell interaction has to consider the dynamic conditions that accompany tumor growth and its vascularization [195,196,197,198]. It is evident that, within a tumor mass, a strong heterogeneity is present due to the differential expression of altered genes (mutations, up and down regulation of genes, chromosomal/copy number alterations) in different cell populations, and the heterogeneity of the metabolic state of these cells [199,200,201,202,203]. The expression of mutated genes can be dependent on the metabolic state and the primary source of energy used by tumor cells, and vice versa, some mutations trigger metabolic responses [204,205,206,207].
6.2.2. Microfluidic Model to Study the NK Cell Distribution in Tumor Spheroids
The influence of metabolism on tumor growth/mutation and the immune response could be tested by the generation of microfluidic models [126,208,209,210,211]. For example, two parallel capillary-like structures layered with endothelial cells have been plated in an extracellular matrix hydrogel; in the middle space, between these capillaries, a breast cancer spheroid was co-plated. This model was used to study a therapeutic antibody and NK cells distributions starting from the vessel lumen [212]. NK cells could sense the tumor, actively approaching and infiltrating the spheroid, apparently better than the passive diffusion of the therapeutic antibody. Indeed, antibodies diffused in the hydrogel, but they initially localized only on the outer surface of the spheroid, next to the artificial vessel. Only at 24 hrs the whole periphery of spheroid was covered by the antibody, though it could not penetrate inside the spheroid. NK cells sensed spheroids through several chemokine receptors and penetrated into the inner portion of the spheroid. These NK cells can kill tumor cells both at the periphery and inside the tumor mass [212]. This model is of great interest because the differential penetration of therapeutic antibody and NK cells in an artificial system that resembles the TME can provide information on how the anti-tumor immune response can be regulated. However, it does not take into consideration the plethora of activating and inhibiting signals that NK cells can receive during their localization, as mentioned in Section 3.
6.2.3. Heterotypic Spheroids to Better Mimic TME
The use of heterotypic spheroids would be essential to better understand the complex interaction between anti-tumor effector cells and TME [122,125,213]. Recently, several reports have demonstrated the possibility to reconstitute in vitro the conditions to generate heterotypic spheroid [122,125,213]. The combination of carcinoma cells, fibroblasts and immune cells was used to create 3D heterotypic spheroids with the hanging drop method [122]. In particular, the co-culture of the CRC cell line LS174T and fetal fibroblasts MRC-5 led to the generation of spheroids with a highly compact core of fibroblasts surrounded by carcinoma cells [122]. This kind of spheroids were infiltrated by monocytes and to a lesser extent by CD3+ T cells. This model was used to assess the localization and functional effects of IgG-IL2v, that is a variant of IL2 linked to IgG, able to interact only with lymphocytes expressing βγ chains of IL2R such as NK, CD8+ cytolytic effector cells and CD4+ helper T cells, but not regulatory T cells expressing high levels of CD25. IgG-IL2v could increase T cells and CD3-CD56+CD69+ NK cells infiltration of spheroids, and the killing of the peripheral layer of epithelial carcinoma cells. In this model, the use of T cell bispecific [TCB) antibodies, targeting carcinoembryonic antigen (CEA) or fibroblast activating protein (FAP), showed that T cells could be selectively activated to localize and kill either carcinoma cells or fibroblasts. This heterotypic model could be suitable to assess the efficiency of different therapeutic tools [123].
6.3. Future Perspectives and Applications of NK-Tumor Cell Interaction in 3D Models
As reported in the previous paragraphs, 3D models are a suitable tool to dissect how patient-derived NK cells may influence the growth of a tumor mass (Figure 4).
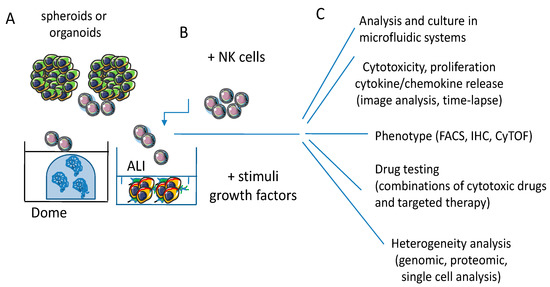
Figure 4.
3D culture models for NK/effector-tumor cell interaction. (A) Spheroids and organoids allow a more holistic approach to study the interactions between tumor and effector cells, provided the use of chemically defined media with components that do not alter the phenotypic and functional features of epithelial cells and lymphocytes and avoid factors that can metabolically inhibit one or another of these two cell populations. (B) The use of specific stimuli such as growth factors or cytokines can mimic in vivo conditions like in conventional cultures. The 3D structure of tumor mass forces the lymphocytes to interact with tumor cells and penetrate into spheroids and/or organoids. This interaction should trigger mechanisms of lymphocyte invasion, while also tumor cells respond to this trigger. (C) Endpoints and assays involved in effector lymphocyte/tumor cell 3D cultures analysis. NK/effector cell-mediated cytotoxicity and cytokine release through imaging software tools and time-lapse recording, including computerized microfluidic systems. NK/effector cell phenotype can be assessed using different methodological approaches, such as fluorescence activated cell sorting (FACS), immunohistochemistry (IHC) or mass cytofluorimetry (CyTOF). Drug testing, including combinatorial drug regimens, can be performed evaluating spheroid/organoid size and the differences of metabolism and oxygenation of the tumor mass, as it happens in vivo. The response of tumor cells, as well as that of NK cells, can be analyzed in detail applying genomic and proteomic analysis at the single cell level upon the disaggregation of the 3D culture model.
Tumor spheroids and organoids allow the study of the molecular mechanisms mediating lymphocyte and NK cell infiltration of a tumor mass [126]. During this process, it is relevant to analyze the immune synapse between activating and/or inhibiting NK cells receptors and their counter-ligands on tumor cells. NK cell cytoskeleton rearrangements could be analyzed to define if the simultaneous interaction with more than one single tumor cell may influence the redistribution of the cytotoxic granules. During the infiltration of spheroids from the periphery to the center, NK cells probably activate/deactivate different molecular mechanisms allowing migration, protein digestion and matrix/cell interactions. These mechanisms do not play a role in conventional 2D or cell suspension-based culture system. This would suggest that 3D culture models could shed new light on the NK cell signal transduction triggered during infiltration.
Another challenge in a 3D culture models is the quantitative analysis of the cytotoxicity of NK cells in tumor cells. This can be achieved evaluating different parameters in tumor cells: caspase activation, increment/reduction of fluorescent probes (passive markers, active substrates, or reporter constructs), and residual living cells quantification [119,121,123,128,160,163,165,166,168,170,171,178,179]. The quantitative analysis of killing has been assessed mainly by image analysis [119,126,128,212,214,215]. This procedure needs appropriate equipment, a long time for the acquisition of large sets of images at different zeta planes of the 3D structure, and the complex informatic analysis with advanced software [214,215]. Furthermore, the accuracy and precision of results depends on the sensitivity of the equipment and the algorithms of the software [214,215]. The development of easier and standardized quantification assays dedicated to these 3D models is a must, as the conventional cytotoxic tests are clearly insufficient to define the behavior of tumor cells during the interaction with effector cells [126,128].
7. Conclusions
The immune system can deeply interact with tumor cells in a hostile TME. While the reactivation of the adaptive immunity by ICB is an additional therapeutic tool against cancer, several solid tumors do not show relevant responses to ICB therapy [7,8,9,13,77,98,143,160,177]. NK cells can successfully eliminate tumor cells, but are regulated by a complex balance of positive and negative signals, delivered from the receptor ligands expressed on tumor cells. The role of MHC class I molecules in shaping NK cell self-reactivity is essential. Thus, to fully understand NK cell interaction with self-tumor cells, appropriate culture models should be developed. 3D culture systems, such as spheroids and organoids derived from patients’ specimens, would allow a deeper analysis of the complex structural architecture of NK-tumor cell recognition/killing. Moreover, they can represent a tool to test therapeutic approaches and novel drugs in an autologous setting. These dynamic models, associated with microfluidic chips, can give new insights on the phenotypic and functional features of NK cells, and their application in cancer therapy.
Author Contributions
Conceptualization and data curation, A.P., F.V., J.L.C.F., D.C., R.B. and M.R.Z.; writing—original draft preparation, A.P.; writing—review and editing all the authors; supervision, A.P. and M.R.Z.; project administration, A.P.; funding acquisition, A.P., R.B. and M.R.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by grants from the Associazione Italiana per la Ricerca sul Cancro (AIRC) IG 21648, Compagnia di San Paolo (ROL 32567), 5xmille 2015 and 2016 and Ricerca Corrente from Italian Ministry of Health to A.P.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Atkins, M.B.; Lotze, M.T.; Dutcher, J.P.; Fisher, R.I.; Weiss, G.; Margolin, K.; Abrams, J.; Sznol, M.; Parkinson, D.; Hawkins, M.; et al. High-Dose Recombinant Interleukin 2 Therapy for Patients With Metastatic Melanoma: Analysis of 270 Patients Treated Between 1985 and 1993. J. Clin. Oncol. 1999, 17, 2105. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, S.A. Immersion in the search for effective cancer immunotherapies. Mol. Med. 2021, 27, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, S.A. IL-2: The First Effective Immunotherapy for Human Cancer. J. Immunol. 2014, 192, 5451–5458. [Google Scholar] [CrossRef] [PubMed]
- Andriole, G.L.; Mulé, J.J.; Hansen, C.T.; Linehan, W.M.; Rosenberg, S.A. Evidence that lymphokine-activated killer cells and natural killer cells are distinct based on an analysis of congenitally immunodeficient mice. J. Immunol. 1985, 135, 2911–2913. [Google Scholar] [PubMed]
- Dhupkar, P.; Gordon, N. Interleukin-2: Old and New Approaches to Enhance Immune-Therapeutic Efficacy. Adv. Exp. Med. Biol. 2017, 995, 33–51. [Google Scholar] [CrossRef]
- Jeong, G.H.; Lee, K.H.; Lee, I.R.; Oh, J.H.; Kim, D.W.; Shin, J.W.; Kronbichler, A.; Eisenhut, M.; Van Der Vliet, H.J.; Abdel-Rahman, O.; et al. Incidence of Capillary Leak Syndrome as an Adverse Effect of Drugs in Cancer Patients: A Systematic Review and Meta-Analysis. J. Clin. Med. 2019, 8, 143. [Google Scholar] [CrossRef] [Green Version]
- Nishino, M.; Ramaiya, N.H.; Hatabu, H.; Hodi, F.S. Monitoring immune-checkpoint blockade: Response evaluation and biomarker development. Nat. Rev. Clin. Oncol. 2017, 14, 655–668. [Google Scholar] [CrossRef]
- Xiao, Q.; Nobre, A.; Piñeiro, P.; Berciano-Guerrero, M.Á.; Alba, E.; Cobo, M.; Lauschke, V.M.; Barragán, I. Genetic and Epigenetic Biomarkers of Immune Checkpoint Blockade Response. J. Clin. Med. 2020, 9, 286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siozopoulou, V.; Domen, A.; Zwaenepoel, K.; Van Beeck, A.; Smits, E.; Pauwels, P.; Marcq, E. Immune Checkpoint Inhibitory Therapy in Sarcomas: Is There Light at the End of the Tunnel? Cancers 2021, 13, 360. [Google Scholar] [CrossRef]
- De Santis, F.; Fucà, G.; Schadendorf, D.; Mantovani, A.; Magnani, L.; Lisanti, M.; Pettitt, S.; Bellone, M.; Del Sal, G.; Minucci, S.; et al. Anticancer innovative therapy congress: Highlights from the 10th anniversary edition. Cytokine Growth Factor Rev. 2021, 59, 1–8. [Google Scholar] [CrossRef]
- Zhou, L.; Xu, N.; Shibata, H.; Saloura, V.; Uppaluri, R. Epigenetic modulation of immunotherapy and implications in head and neck cancer. Cancer Metastasis Rev. 2021, 40, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Kaymak, I.; Williams, K.S.; Cantor, J.R.; Jones, R.G. Immunometabolic Interplay in the Tumor Microenvironment. Cancer Cell 2021, 39, 28–37. [Google Scholar] [CrossRef]
- Jung, G.; Benítez-Ribas, D.; Sánchez, A.; Balaguer, F. Current Treatments of Metastatic Colorectal Cancer with Immune Checkpoint Inhibitors—2020 Update. J. Clin. Med. 2020, 9, 3520. [Google Scholar] [CrossRef] [PubMed]
- Menter, T.; Tzankov, A.; Dirnhofer, S. The tumor microenvironment of lymphomas: Insights into the potential role and modes of actions of checkpoint inhibitors. Hematol. Oncol. 2021, 39, 3–10. [Google Scholar] [CrossRef]
- Rodrigues, J.; Heinrich, M.A.; Teixeira, L.M.; Prakash, J. 3D In Vitro Model (R)evolution: Unveiling Tumor–Stroma Interactions. Trends Cancer 2021, 7, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Colombo, E.; Cattaneo, M.G. Multicellular 3D Models to Study Tumour-Stroma Interactions. Int. J. Mol. Sci. 2021, 22, 1633. [Google Scholar] [CrossRef]
- De La Rochère, P.; Guil-Luna, S.; Decaudin, D.; Azar, G.; Sidhu, S.S.; Piaggio, E. Humanized Mice for the Study of Immuno-Oncology. Trends Immunol. 2018, 39, 748–763. [Google Scholar] [CrossRef]
- Zitvogel, L.; Pitt, J.M.; Daillère, R.; Smyth, M.J.; Kroemer, G. Mouse models in oncoimmunology. Nat. Rev. Cancer 2016, 16, 759–773. [Google Scholar] [CrossRef]
- Morton, J.J.; Bird, G.; Refaeli, Y.; Jimeno, A. Humanized Mouse Xenograft Models: Narrowing the Tumor–Microenvironment Gap. Cancer Res. 2016, 76, 6153–6158. [Google Scholar] [CrossRef] [Green Version]
- Rosato, R.R.; Dávila-González, D.; Choi, D.S.; Qian, W.; Chen, W.; Kozielski, A.J.; Wong, H.; Dave, B.; Chang, J.C. Evaluation of anti-PD-1-based therapy against triple-negative breast cancer patient-derived xenograft tumors engrafted in humanized mouse models. Breast Cancer Res. 2018, 20, 108. [Google Scholar] [CrossRef]
- Shultz, L.D.; Brehm, M.A.; Garcia-Martinez, J.V.; Greiner, D.L. Humanized mice for immune system investigation: Progress, promise and challenges. Nat. Rev. Immunol. 2012, 12, 786–798. [Google Scholar] [CrossRef] [PubMed]
- Ito, R.; Takahashi, T.; Ito, M. Humanized mouse models: Application to human diseases. J. Cell. Physiol. 2018, 233, 3723–3728. [Google Scholar] [CrossRef]
- Martinov, T.; McKenna, K.M.; Tan, W.H.; Collins, E.J.; Kehret, A.R.; Linton, J.D.; Olsen, T.M.; Shobaki, N.; Rongvaux, A. Building the Next Generation of Humanized Hemato-Lymphoid System Mice. Front. Immunol. 2021, 12, 643852. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.W.; Miljanic, M.; Triplett, T.; Ramirez, C.; Aung, K.L.; Eckhardt, S.G.; Capasso, A. Current methods in translational cancer research. Cancer Metastasis Rev. 2021, 40, 7–30. [Google Scholar] [CrossRef]
- Hahn, W.C.; Bader, J.S.; Braun, T.P.; Califano, A.; Clemons, P.A.; Druker, B.J.; Ewald, A.J.; Fu, H.; Jagu, S.; Kemp, C.J.; et al. Cancer Target Discovery and Development Network. An expanded universe of cancer targets. Cell 2021, 184, 1142–1155. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Bockorny, B.; Paul, I.; Akshinthala, D.; Frappart, P.-O.; Gandarilla, O.; Bose, A.; Sanchez-Gonzalez, V.; Rouse, E.E.; Lehoux, S.D.; et al. PDX-derived organoids model in vivo drug response and secrete biomarkers. JCI Insight 2020, 5, 32990680. [Google Scholar] [CrossRef] [PubMed]
- Mestas, J.; Hughes, C.C. Of mice and not men: Differences between mouse and human immunology. J. Immunol. 2004, 172, 2731–2738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vacca, P.; Chiossone, L.; Mingari, M.C.; Moretta, L. Heterogeneity of NK Cells and Other Innate Lymphoid Cells in Human and Murine Decidua. Front. Immunol. 2019, 10, 170. [Google Scholar] [CrossRef] [Green Version]
- Anderson, D.A., 3rd; Dutertre, C.-A.; Ginhoux, F.; Murphy, K.M. Genetic models of human and mouse dendritic cell development and function. Nat. Rev. Immunol. 2021, 21, 101–115. [Google Scholar] [CrossRef]
- Mian, S.A.; Anjos-Afonso, F.; Bonnet, D. Advances in Human Immune System Mouse Models for Studying Human Hematopoiesis and Cancer Immunotherapy. Front. Immunol. 2021, 11, 9236. [Google Scholar] [CrossRef]
- Rameshbabu, S.; Labadie, B.W.; Argulian, A.; Patnaik, A. Targeting Innate Immunity in Cancer Therapy. Vaccines 2021, 9, 138. [Google Scholar] [CrossRef]
- Wang, F.; Lau, J.K.C.; Yu, J. The role of natural killer cell in gastrointestinal cancer: Killer or helper. Oncogene 2021, 40, 717–730. [Google Scholar] [CrossRef]
- Morandi, F.; Yazdanifar, M.; Cocco, C.; Bertaina, A.; Airoldi, I. Engineering the Bridge between Innate and Adaptive Immunity for Cancer Immunotherapy: Focus on gammadelta T and NK Cells. Cells 2020, 9, 1757. [Google Scholar] [CrossRef] [PubMed]
- Sherman, H.; Gitschier, H.J.; Rossi, A.E. A Novel Three-Dimensional Immune Oncology Model for High-Throughput Testing of Tumoricidal Activity. Front. Immunol. 2018, 9, 857. [Google Scholar] [CrossRef] [PubMed]
- Dangles-Marie, V.; Richon, S.; EL Behi, M.; Echchakir, H.; Dorothée, G.; Thiery, J.; Validire, P.; Vergnon, I.; Menez, J.; Ladjimi, M.; et al. A three-dimensional tumor cell defect in activating autologous CTLs is associated with inefficient antigen presentation correlated with heat shock protein-70 down-regulation. Cancer Res. 2003, 63, 3682–3687. [Google Scholar]
- Jenkins, R.W.; Aref, A.R.; Lizotte, P.; Ivanova, E.; Stinson, S.; Zhou, C.W.; Bowden, M.; Deng, J.; Liu, H.; Miao, D.; et al. Ex Vivo Profiling of PD-1 Blockade Using Organotypic Tumor Spheroids. Cancer Discov. 2018, 8, 196–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujii, M.; Sato, T. Somatic cell-derived organoids as prototypes of human epithelial tissues and diseases. Nat. Mater. 2021, 20, 156–169. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Zhou, X.; Wang, S.; Trinkle, C. Tumor organoid models in precision medicine and investigating cancer-stromal interactions. Pharmacol. Ther. 2021, 218, 107668. [Google Scholar] [CrossRef]
- Schutgens, F.; Clevers, H. Human Organoids: Tools for Understanding Biology and Treating Diseases. Annu. Rev. Pathol. Mech. Dis. 2020, 15, 211–234. [Google Scholar] [CrossRef] [Green Version]
- Bar-Ephraim, Y.E.; Kretzschmar, K.; Clevers, H. Organoids in immunological research. Nat. Rev. Immunol. 2020, 20, 279–293. [Google Scholar] [CrossRef]
- Kim, J.; Koo, B.-K.; Knoblich, J.A. Human organoids: Model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Trinchieri, G. Biology of Natural Killer Cells. Adv. Immunol. 1989, 47, 187–376. [Google Scholar] [PubMed]
- Bellone, G.; Valiante, N.M.; Viale, O.; Ciccone, E.; Moretta, L.; Trinchieri, G. Regulation of hematopoiesis in vitro by alloreactive natural killer cell clones. J. Exp. Med. 1993, 177, 1117–1125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Myers, J.A.; Miller, J.S. Exploring the NK cell platform for cancer immunotherapy. Nat. Rev. Clin. Oncol. 2021, 18, 85–100. [Google Scholar] [CrossRef] [PubMed]
- Smith-Garvin, J.E.; Koretzky, G.A.; Jordan, M.S. T cell activation. Annu Rev Immunol. 2009, 27, 591–619. [Google Scholar] [CrossRef]
- Cooper, M.A.; Fehniger, T.A.; Caligiuri, M.A. The biology of human natural killer-cell subsets. Trends Immunol. 2001, 22, 633–640. [Google Scholar] [CrossRef]
- Michel, T.; Poli, A.; Cuapio, A.; Briquemont, B.; Iserentant, G.; Ollert, M.; Zimmer, J. Human CD56bright NK Cells: An Update. J. Immunol. 2016, 196, 2923–2931. [Google Scholar] [CrossRef] [Green Version]
- Ochoa, M.C.; Minute, L.; Rodriguez, I.; Garasa, S.; Ruiz, E.P.; Inogés, S.; Melero, I.; Berraondo, P. Antibody-dependent cell cytotoxicity: Immunotherapy strategies enhancing effector NK cells. Immunol. Cell Biol. 2017, 95, 347–355. [Google Scholar] [CrossRef]
- Poggi, A.; Biassoni, R.; Pella, N.; Paolieri, F.; Bellomo, R.; Bertolini, A.; Moretta, L.; Mingari, M.C. In vitro expansion of CD3/TCR- human thymocyte populations that selectively lack CD3 delta gene expression: A phenotypic and functional analysis. J. Exp. Med. 1990, 172, 1409–1418. [Google Scholar] [CrossRef] [Green Version]
- Mingari, M.C.; Poggi, A.; Biassoni, R.; Bellomo, R.; Ciccone, E.; Pella, N.; Morelli, L.; Verdiani, S.; Moretta, A.; Moretta, L. In vitro proliferation and cloning of CD3- CD16+ cells from human thymocyte precursors. J. Exp. Med. 1991, 174, 21–26. [Google Scholar] [CrossRef] [Green Version]
- Spits, H.; Bernink, J.H.; Lanier, L. NK cells and type 1 innate lymphoid cells: Partners in host defense. Nat. Immunol. 2016, 17, 758–764. [Google Scholar] [CrossRef]
- Zhou, J.; Tian, Z.; Peng, H. Tissue-resident NK cells and other innate lymphoid cells. Adv. Immunol. 2020, 145, 37–53. [Google Scholar] [PubMed]
- Geiger, T.L.; Sun, J.C. Development and maturation of natural killer cells. Curr. Opin. Immunol. 2016, 39, 82–89. [Google Scholar] [CrossRef] [Green Version]
- Cherrier, D.E.; Serafini, N.; Di Santo, J.P. Innate Lymphoid Cell Development: A T Cell Perspective. Immunity 2018, 48, 1091–1103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simoni, Y.; Newell, E.W. Dissecting human ILC heterogeneity: More than just three subsets. Immunology 2018, 153, 297–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poggi, A.; Zocchi, M.R. NK Cell Autoreactivity and Autoimmune Diseases. Front. Immunol. 2014, 5, 27. [Google Scholar] [CrossRef]
- Long, E.O.; Kim, H.S.; Liu, D.; Peterson, M.E.; Rajagopalan, S. Controlling Natural Killer Cell Responses: Integration of Signals for Activation and Inhibition. Annu. Rev. Immunol. 2013, 31, 227–258. [Google Scholar] [CrossRef] [Green Version]
- Morvan, M.G.; Lanier, L.L. NK cells and cancer: You can teach innate cells new tricks. Nat. Rev. Cancer 2016, 16, 7–19. [Google Scholar] [CrossRef]
- Moretta, L.; Bottino, C.; Pende, D.; Vitale, M.; Mingari, M.; Moretta, A. Different checkpoints in human NK-cell activation. Trends Immunol. 2004, 25, 670–676. [Google Scholar] [CrossRef]
- Seillet, C.; Brossay, L.; Vivier, E. Natural killers or ILC1s? That is the question. Curr Opin Immunol. 2021, 68, 48–53. [Google Scholar] [CrossRef]
- Krabbendam, L.; Bernink, J.H.; Spits, H. Innate lymphoid cells: From helper to killer. Curr. Opin. Immunol. 2021, 68, 28–33. [Google Scholar] [CrossRef]
- Arima, N. Dual effects of natural killer cells in transplantation for leukemia. Crit. Rev. Oncol. Hematol. 2021, 158, 103206. [Google Scholar] [CrossRef] [PubMed]
- Asl, K.; Velaei, K.; Rafat, A.; Nasrabadi, H.T.; Movassaghpour, A.A.; Mahdavi, M.; Charoudeh, H.N. The role of KIR positive NK cells in diseases and its importance in clinical intervention. Int. Immunopharmacol. 2021, 92, 107361. [Google Scholar] [CrossRef]
- Kucuksezer, U.C.; Cetin, E.A.; Esen, F.; Tahrali, I.; Akdeniz, N.; Gelmez, M.Y.; Deniz, G. The Role of Natural Killer Cells in Autoimmune Diseases. Front. Immunol. 2021, 12. [Google Scholar] [CrossRef]
- Bödder, J.; Zahan, T.; van Slooten, R.; Schreibelt, G.; de Vries, I.J.M.; Flórez-Grau, G. Harnessing the cDC1-NK Cross-Talk in the Tumor Microenvironment to Battle Cancer. Front. Immunol. 2021, 11. [Google Scholar] [CrossRef]
- Rosenstock, P.; Kaufmann, T. Sialic Acids and Their Influence on Human NK Cell Function. Cells 2021, 10, 263. [Google Scholar] [CrossRef] [PubMed]
- Batista, I.A.; Quintas, S.T.; Melo, S.A. The Interplay of Exosomes and NK Cells in Cancer Biology. Cancers 2021, 13, 473. [Google Scholar] [CrossRef]
- Dixon, K.J.; Wu, J.; Walcheck, B. Engineering Anti-Tumor Monoclonal Antibodies and Fc Receptors to Enhance ADCC by Human NK Cells. Cancers 2021, 13, 312. [Google Scholar] [CrossRef]
- Spaggiari, G.M.; Carosio, R.; Pende, D.; Marcenaro, S.; Rivera, P.; Zocchi, M.R.; Moretta, L.; Poggi, A. NK cell-mediated lysis of autologous antigen-presenting cells is triggered by the engagement of the phosphatidylinositol 3-kinase upon ligation of the natural cytotoxicity receptors NKp30 and NKp46. Eur. J. Immunol. 2001, 31, 1656–1665. [Google Scholar] [CrossRef]
- Zhang, Z.; Wu, N.; Lu, Y.; Davidson, D.; Colonna, M.; Veillette, A. DNAM-1 controls NK cell activation via an ITT-like motif. J. Exp. Med. 2015, 212, 2165–2182. [Google Scholar] [CrossRef]
- Kwon, H.J.; Choi, G.E.; Ryu, S.; Kwon, S.J.; Kim, S.C.; Booth, C.; Nichols, K.E.; Kim, H.S. Stepwise phosphorylation of p65 promotes NF-kappaB activation and NK cell responses during target cell recognition. Nat Commun. 2016, 7, 11686. [Google Scholar] [CrossRef]
- Zocchi, M.R.; Contini, P.; Alfano, M.; Poggi, A. Pertussis toxin (PTX) B subunit and the nontoxic PTX mutant PT9K/129G inhibit Tat-induced TGF-beta production by NK cells and TGF-beta-mediated NK cell apoptosis. J. Immunol. 2005, 174, 6054–6061. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.; Yin, T.; Zhou, F.; Cui, P.; Gou, S.; Wang, C. Valproic acid sensitizes pancreatic cancer cells to natural killer cell-mediated lysis by upregulating MICA and MICB via the PI3K/Akt signaling pathway. BMC Cancer 2014, 14, 370. [Google Scholar] [CrossRef] [Green Version]
- Poggi, A.; Musso, A.; Dapino, I.; Zocchi, M.R. Mechanisms of tumor escape from immune system: Role of mesenchymal stromal cells. Immunol. Lett. 2014, 159, 55–72. [Google Scholar] [CrossRef]
- Ganesh, K.; Massagué, J. Targeting metastatic cancer. Nat. Med. 2021, 27, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Marcucci, F.; Rumio, C. The tumor-promoting effects of the adaptive immune system: A cause of hyperprogressive disease in cancer? Cell. Mol. Life Sci. 2021, 78, 853–865. [Google Scholar] [CrossRef] [PubMed]
- Hussaini, S.; Chehade, R.; Boldt, R.G.; Raphael, J.; Blanchette, P.; Vareki, S.M.; Fernandes, R. Association between immune-related side effects and efficacy and benefit of immune checkpoint inhibitors—A systematic review and meta-analysis. Cancer Treat. Rev. 2021, 92, 102134. [Google Scholar] [CrossRef] [PubMed]
- Spaggiari, G.M.; Contini, P.; Dondero, A.; Carosio, R.; Puppo, F.; Indiveri, F.; Zocchi, M.R.; Poggi, A. Soluble HLA class I induces NK cell apoptosis upon the engagement of killer-activating HLA class I receptors through FasL-Fas interaction. Blood 2002, 100, 4098–4107. [Google Scholar] [CrossRef]
- Caligiuri, M.A. Human natural killer cells. Blood 2008, 112, 461–469. [Google Scholar] [CrossRef]
- Mace, E.M.; Monkley, S.J.; Critchley, D.R.; Takei, F. A Dual Role for Talin in NK Cell Cytotoxicity: Activation of LFA-1-Mediated Cell Adhesion and Polarization of NK Cells. J. Immunol. 2009, 182, 948–956. [Google Scholar] [CrossRef] [Green Version]
- Gross, C.; Brzostowski, J.A.; Liu, D.; Long, E.O. Tethering of Intercellular Adhesion Molecule on Target Cells Is Required for LFA-1–Dependent NK Cell Adhesion and Granule Polarization. J. Immunol. 2010, 185, 2918–2926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez-Madrid, F.; Simon, P.; Thompson, S.; Springer, T.A. Mapping of antigenic and functional epitopes on the alpha- and beta-subunits of two related mouse glycoproteins involved in cell interactions, LFA-1 and Mac-1. J. Exp. Med. 1983, 158, 586–602. [Google Scholar] [CrossRef] [PubMed]
- Poggi, A.; Spada, F.; Costa, P.; Tomasello, E.; Revello, V.; Pella, N.; Zocchi, M.R.; Moretta, L. Dissection of lymphocyte function-associated antigen 1-dependent adhesion and signal transduction in human natural killer cells shown by the use of cholera or pertussis toxin. Eur. J. Immunol. 1996, 26, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Donskov, F.; Basse, P.H.; Hokland, M. Expression and function of LFA-1 on A-NK and T-LAK cells: Role in tumor target killing and migration into tumor tissue. Nat. Immun. 1996, 15, 9162263. [Google Scholar]
- Castriconi, R.; Dondero, A.; Cantoni, C.; Della Chiesa, M.; Prato, C.; Nanni, M.; Fiorini, M.; Notarangelo, L.; Parolini, S.; Moretta, L.; et al. Functional characterization of natural killer cells in type I leukocyte adhesion deficiency. Blood 2007, 109, 4873–4881. [Google Scholar] [CrossRef] [PubMed]
- Bottino, C.; Castriconi, R.; Pende, D.; Rivera, P.; Nanni, M.; Carnemolla, B.; Cantoni, C.; Grassi, J.; Marcenaro, S.; Reymond, N.; et al. Identification of PVR (CD155) and Nectin-2 (CD112) as Cell Surface Ligands for the Human DNAM-1 (CD226) Activating Molecule. J. Exp. Med. 2003, 198, 557–567. [Google Scholar] [CrossRef] [Green Version]
- Salih, H.R.; Holdenrieder, S.; Steinle, A. Soluble NKG2D ligands: Prevalence, release, and functional impact. Front. Biosci. 2008, 13, 3448–3456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groh, V.; Wu, J.; Yee, C.; Spies, T. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature 2002, 419, 734–738. [Google Scholar] [CrossRef]
- González, S.; Groh, V.; Spies, T. Immunobiology of Human NKG2D and Its Ligands. Curr. Top. Microbiol. Immunol. 2006, 298, 121–138. [Google Scholar] [CrossRef]
- Groh, V.; Smythe, K.; Dai, Z.; Spies, T. Fas ligand–mediated paracrine T cell regulation by the receptor NKG2D in tumor immunity. Nat. Immunol. 2006, 7, 755–762. [Google Scholar] [CrossRef]
- Zingoni, A.; Vulpis, E.; LoConte, L.; Santoni, A. NKG2D Ligand Shedding in Response to Stress: Role of ADAM. Front. Immunol. 2020, 11, 447. [Google Scholar] [CrossRef]
- Melaiu, O.; Lucarini, V.; Cifaldi, L.; Fruci, D. Influence of the Tumor Microenvironment on NK Cell Function in Solid Tumors. Front. Immunol. 2020, 10. [Google Scholar] [CrossRef]
- Vujanovic, N.L. Role of TNF Family Ligands in Antitumor Activity of Natural Killer Cells. Int. Rev. Immunol. 2001, 20, 415–437. [Google Scholar] [CrossRef] [PubMed]
- Prager, I.; Watzl, C. Mechanisms of natural killer cell-mediated cellular cytotoxicity. J. Leukoc. Biol. 2019, 105, 1319–1329. [Google Scholar] [CrossRef] [PubMed]
- Meyaard, L. LAIR and collagens in immune regulation. Immunol. Lett. 2010, 128, 26–28. [Google Scholar] [CrossRef]
- Meyaard, L.; Adema, G.J.; Chang, C.; Woollatt, E.; Sutherland, G.R.; Lanier, L.L.; Phillips, J.H. LAIR-1, a Novel Inhibitory Receptor Expressed on Human Mononuclear Leukocytes. Immunity 1997, 7, 283–290. [Google Scholar] [CrossRef]
- Poggi, A.; Pella, N.; Morelli, L.; Spada, F.; Revello, V.; Sivori, S.; Augugliaro, R.; Moretta, L.; Moretta, A. p40, a novel surface molecule involved in the regulation of the non-major histocompatibility complex-restricted cytolytic activity in humans. Eur. J. Immunol. 1995, 25, 369–376. [Google Scholar] [CrossRef]
- Khan, M.; Arooj, S.; Wang, H. NK Cell-Based Immune Checkpoint Inhibition. Front. Immunol. 2020, 11, 167. [Google Scholar] [CrossRef] [PubMed]
- Jandus, C.; Boligan, K.F.; Chijioke, O.; Liu, H.; Dahlhaus, M.; Démoulins, T.; Schneider, C.; Wehrli, M.; Hunger, R.E.; Baerlocher, G.M.; et al. Interactions between Siglec-7/9 receptors and ligands influence NK cell–dependent tumor immunosurveillance. J. Clin. Investig. 2014, 124, 1810–1820. [Google Scholar] [CrossRef] [Green Version]
- Blake, S.J.; Dougall, W.C.; Miles, J.J.; Teng, M.W.; Smyth, M.J. Molecular Pathways: Targeting CD96 and TIGIT for Cancer Immunotherapy. Clin. Cancer Res. 2016, 22, 5183–5188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanietsky, N.; Simic, H.; Arapovic, J.; Toporik, A.; Levy, O.; Novik, A.; Levine, Z.; Beiman, M.; Dassa, L.; Achdout, H.; et al. The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. Proc. Natl. Acad. Sci. USA 2009, 106, 17858–17863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valhondo, I.; Hassouneh, F.; Lopez-Sejas, N.; Pera, A.; Sanchez-Correa, B.; Guerrero, B.; Bergua, J.M.; Arcos, M.J.; Bañas, H.; Casas-Avilés, I.; et al. Characterization of the DNAM-1, TIGIT and TACTILE Axis on Circulating NK, NKT-Like and T Cell Subsets in Patients with Acute Myeloid Leukemia. Cancers 2020, 12, 2171. [Google Scholar] [CrossRef]
- Fuchs, A.; Cella, M.; Giurisato, E.; Shaw, A.S.; Colonna, M. Cutting Edge: CD96 (Tactile) Promotes NK Cell-Target Cell Adhesion by Interacting with the Poliovirus Receptor (CD155). J. Immunol. 2004, 172, 3994–3998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.L.; O’Farrell, S.; Clayberger, C.; Krensky, A.M. Identification and molecular cloning of tactile. A novel human T cell activation antigen that is a member of the Ig gene superfamily. J. Immunol. 1992, 148, 2600–2608. [Google Scholar] [PubMed]
- Vari, F.; Arpon, D.; Keane, C.; Hertzberg, M.S.; Talaulikar, D.; Jain, S.; Cui, Q.; Han, E.; Tobin, J.; Bird, R.; et al. Immune evasion via PD-1/PD-L1 on NK cells and monocyte/macrophages is more prominent in Hodgkin lymphoma than DLBCL. Blood 2018, 131, 1809–1819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pesce, S.; Greppi, M.; Grossi, F.; Del Zotto, G.; Moretta, L.; Sivori, S.; Genova, C.; Marcenaro, E. PD/1-PD-Ls Checkpoint: Insight on the Potential Role of NK Cells. Front. Immunol. 2019, 10, 1242. [Google Scholar] [CrossRef]
- Vacca, P.; Pesce, S.; Greppi, M.; Fulcheri, E.; Munari, E.; Olive, D.; Mingari, M.C.; Moretta, A.; Moretta, L.; Marcenaro, E. PD-1 is expressed by and regulates human group 3 innate lymphoid cells in human decidua. Mucosal Immunol. 2019, 12, 624–631. [Google Scholar] [CrossRef]
- Tan, S.; Xu, Y.; Wang, Z.; Wang, T.; Du, X.; Song, X.; Guo, X.; Peng, J.; Zhang, J.; Liang, Y.; et al. Tim-3 hampers tumor surveillance of liver resident and conventional NK cells by disrupting PI3K signaling. Cancer Res. 2020, 80, 1130–1142. [Google Scholar] [CrossRef] [Green Version]
- Gleason, M.K.; Lenvik, T.R.; McCullar, V.; Felices, M.; O’Brien, M.S.; Cooley, S.A.; Verneris, M.R.; Cichocki, F.; Holman, C.J.; Panoskaltsis-Mortari, A.; et al. Tim-3 is an inducible human natural killer cell receptor that enhances interferon gamma production in response to galectin-9. Blood 2012, 119, 3064–3072. [Google Scholar] [CrossRef]
- Dao, T.N.; Utturkar, S.; Atallah Lanman, N.; Matosevic, S. TIM-3 Expression Is Downregulated on Human NK Cells in Response to Cancer Targets in Synergy with Activation. Cancers 2020, 12, 2417. [Google Scholar] [CrossRef]
- D’Atri, S.; Tentori, L.; Fuggetta, M.P.; Marini, S.; Bonmassar, E. A miniaturized cell-mediated cytotoxicity assay with human effector mononuclear cells. Int. J. Tissue React. 1986, 8, 383–390. [Google Scholar]
- Pizao, P.E.; Lyaruu, D.M.; Peters, G.J.; Van Ark-Otte, J.; Winograd, B.; Giaccone, G.; Pinedo, H.M. Growth, morphology and chemosensitivity studies on postconfluent cells cultured in ’V’-bottomed microtiter plates. Br. J. Cancer 1992, 66, 660–665. [Google Scholar] [CrossRef] [Green Version]
- Kawasaki, A.; Shinkai, Y.; Yagita, H.; Okumura, K. Expression of perforin in murine natural killer cells and cytotoxic T lymphocytes in vivo. Eur. J. Immunol. 1992, 22, 1215–1219. [Google Scholar] [CrossRef]
- Bonci, F.; Zabogli, E.; Conti, F.; Merico, A.; Freer, G.; Bendinelli, M.; Pistello, M. A novel method for producing target cells and assessing cytotoxic T lymphocyte activity in outbred hosts. BMC Biotechnol. 2009, 9, 18. [Google Scholar] [CrossRef] [Green Version]
- Elsner, L.; Dressel, R. 51Cr-release to monitor NK cell cytotoxicity. Methods Enzymol. 2020, 631, 497–512. [Google Scholar]
- Goldberg, J.E.; Sherwood, S.W.; Clayberger, C. A novel method for measuring CTL and NK cell-mediated cytotoxicity using annexin V and two-color flow cytometry. J. Immunol. Methods 1999, 224, 1–9. [Google Scholar] [CrossRef]
- Kang, S.M.; Kim, D.; Lee, J.H.; Takayama, S.; Park, J.Y. Engineered Microsystems for Spheroid and Organoid Studies. Adv. Healthc. Mater. 2021, 10, e2001284. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Chen, S.; Win Naing, M. A review of manufacturing capabilities of cell spheroid generation technologies and future development. Biotechnol. Bioeng. 2021, 118, 542–554. [Google Scholar] [CrossRef] [PubMed]
- Courau, T.; Bonnereau, J.; Chicoteau, J.; Bottois, H.; Remark, R.; Assante Miranda, L.; Toubert, A.; Blery, M.; Aparicio, T.; Allez, M.; et al. Cocultures of human colorectal tumor spheroids with immune cells reveal the therapeutic potential of MICA/B and NKG2A targeting for cancer treatment. J. Immunother. Cancer 2019, 7, 74. [Google Scholar] [CrossRef] [Green Version]
- Lanuza, P.M.; Vigueras, A.; Olivan, S.; Prats, A.-C.; Costas, S.; Llamazares, G.; Sanchez-Martinez, D.; Ayuso, J.M.; Fernandez, L.; Ochoa, I.; et al. Activated human primary NK cells efficiently kill colorectal cancer cells in 3D spheroid cultures irrespectively of the level of PD-L1 expression. OncoImmunology 2018, 7, e1395123. [Google Scholar] [CrossRef] [PubMed]
- Maas, R.J.; Evert, J.S.H.-V.; Van der Meer, J.M.; Mekers, V.; Rezaeifard, S.; Korman, A.J.; de Jonge, P.K.; Cany, J.; Woestenenk, R.; Schaap, N.P.; et al. TIGIT blockade enhances functionality of peritoneal NK cells with altered expression of DNAM-1/TIGIT/CD96 checkpoint molecules in ovarian cancer. OncoImmunology 2020, 9, 1843247. [Google Scholar] [CrossRef] [PubMed]
- Herter, S.; Morra, L.; Schlenker, R.; Sulcova, J.; Fahrni, L.; Waldhauer, I.; Lehmann, S.; Reisländer, T.; Agarkova, I.; Kelm, J.M.; et al. A novel three-dimensional heterotypic spheroid model for the assessment of the activity of cancer immunotherapy agents. Cancer Immunol. Immunother. CII 2017, 66, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Giannattasio, A.; Weil, S.; Kloess, S.; Ansari, N.; Stelzer, E.H.K.; Cerwenka, A.; Steinle, A.; Koehl, U.; Koch, J. Cytotoxicity and infiltration of human NK cells in in vivo-like tumor spheroids. BMC Cancer 2015, 15, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stojanovic, A.; Cerwenka, A. Natural Killer Cells and Solid Tumors. J. Innate Immun. 2011, 3, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Yang, Z.; Xu, S.; Li, X.; Yang, X.; Jin, P.; Liu, Y.; Zhou, X.; Zhang, T.; Gong, C.; et al. Heterotypic CAF-tumor spheroids promote early peritoneal metastasis of ovarian cancer. J. Exp. Med. 2019, 216, 688–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sargenti, A.; Musmeci, F.; Bacchi, F.; Delprete, C.; Cristaldi, D.A.; Cannas, F.; Bonetti, S.; Pasqua, S.; Gazzola, D.; Costa, D.; et al. Physical Characterization of Colorectal Cancer Spheroids and Evaluation of NK Cell Infiltration Through a Flow-Based Analysis. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef]
- Di Mascolo, D.; Varesano, S.; Benelli, R.; Mollica, H.; Salis, A.; Zocchi, M.R.; Decuzzi, P.; Poggi, A. Cancers Nanoformulated Zoledronic Acid Boosts the Vδ2 T Cell Immunotherapeutic Potential in Colorectal Cancer. Cancer 2019, 12, 104. [Google Scholar] [CrossRef] [Green Version]
- Varesano, S.; Zocchi, M.R.; Poggi, A. Zoledronate Triggers Vδ2 T Cells to Destroy and Kill Spheroids of Colon Carcinoma: Quantitative Image Analysis of Three-Dimensional Cultures. Front. Immunol. 2018, 9, 998. [Google Scholar] [CrossRef]
- Misun, P.M.; Birchler, A.K.; Lang, M.; Hierlemann, A.; Frey, O. Fabrication and Operation of Microfluidic Hanging-Drop Networks. Adv. Struct. Saf. Stud. 2018, 1771, 183–202. [Google Scholar] [CrossRef]
- Leung, B.M.; Lesher-Perez, S.C.; Matsuoka, T.; Moraes, C.; Takayama, S. Media additives to promote spheroid circularity and compactness in hanging drop platform. Biomater. Sci. 2014, 3, 336–344. [Google Scholar] [CrossRef] [Green Version]
- Eder, T.; Eder, I.E. 3D Hanging Drop Culture to Establish Prostate Cancer Organoids. Methods Mol. Biol. 2017, 1612, 167–175. [Google Scholar]
- Panek, M.; Grabacka, M.; Pierzchalska, M. The formation of intestinal organoids in a hanging drop culture. Cytotechnology 2018, 70, 1085–1095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.; Kim, H.; Park, J.K. Microfluidic channel-integrated hanging drop array chip operated by pushbuttons for spheroid culture and analysis. Analyst 2020, 145, 6974–6980. [Google Scholar] [CrossRef] [PubMed]
- Bert, B.; Dörendahl, A.; Leich, N.; Vietze, J.; Steinfath, M.; Chmielewska, J.; Hensel, A.; Grune, B.; Schönfelder, G. Rethinking 3R strategies: Digging deeper into AnimalTestInfo promotes transparency in in vivo biomedical research. PLoS Biol. 2017, 15, e2003217. [Google Scholar] [CrossRef] [Green Version]
- Törnqvist, E.; Annas, A.; Granath, B.; Jalkesten, E.; Cotgreave, I.; Öberg, M. Strategic Focus on 3R Principles Reveals Major Reductions in the Use of Animals in Pharmaceutical Toxicity Testing. PLoS ONE 2014, 9, e101638. [Google Scholar] [CrossRef] [Green Version]
- Shaheen, S.; Ahmed, M.; Lorenzi, F.; Nateri, A.S. Spheroid-Formation (Colonosphere) Assay for in Vitro Assessment and Expansion of Stem Cells in Colon Cancer. Stem Cell. Rev. Rep. 2016, 12, 492–499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourn, M.D.; Batchelor, D.V.B.; Ingram, N.; McLaughlan, J.R.; Coletta, P.L.; Evans, S.D.; Peyman, S.A. High-throughput microfluidics for evaluating microbubble enhanced delivery of cancer therapeutics in spheroid cultures. J. Control. Release 2020, 326, 13–24. [Google Scholar] [CrossRef]
- Reidy, E.; Leonard, N.A.; Treacy, O.; Ryan, A.E. A 3D View of Colorectal Cancer Models in Predicting Therapeutic Responses and Resistance. Cancers 2021, 13, 227. [Google Scholar] [CrossRef]
- Jung, G.; Hernández-Illán, E.; Moreira, L.; Balaguer, F.; Goel, A. Epigenetics of colorectal cancer: Biomarker and therapeutic potential. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 111–130. [Google Scholar] [CrossRef]
- Sveen, A.; Kopetz, S.; Lothe, R.A. Biomarker-guided therapy for colorectal cancer: Strength in complexity. Nat. Rev. Clin. Oncol. 2020, 17, 11–32. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, L.; Wang, Q.; Zhang, M.; Wang, B.; Jiang, K.; Ye, Y.; Wang, S.; Shen, Z. Molecular Characterization and Clinical Relevance of RNA Binding Proteins in Colorectal Cancer. Front. Genet. 2020, 11, 580149. [Google Scholar] [CrossRef]
- Keum, N.; Giovannucci, E. Global burden of colorectal cancer: Emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 713–732. [Google Scholar] [CrossRef]
- Morse, M.A.; Hochster, H.; Benson, A. Perspectives on Treatment of Metastatic Colorectal Cancer with Immune Checkpoint Inhibitor Therapy. Oncologist 2019, 25, 33–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Gastroenterol. Rev. 2019, 14, 89–103. [Google Scholar] [CrossRef]
- Müller, M.F.; Ibrahim, A.E.K.; Arends, M.J. Molecular pathological classification of colorectal cancer. Virchows Arch. 2016, 469, 125–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bürtin, F.; Mullins, C.S.; Linnebacher, M. Mouse models of colorectal cancer: Past, present and future perspectives. World J. Gastroenterol. 2020, 26, 1394–1426. [Google Scholar] [CrossRef]
- O’Rourke, K.P.; Loizou, E.; Livshits, G.; Schatoff, E.M.; Baslan, T.; Manchado, E.; Simon, J.; Romesser, P.B.; Leach, B.; Han, T.; et al. Transplantation of engineered organoids enables rapid generation of metastatic mouse models of colorectal cancer. Nat. Biotechnol. 2017, 35, 577–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blomme, A.; Van Simaeys, G.; Doumont, G.; Costanza, B.; Bellier, J.; Otaka, Y.; Sherer, F.; Lovinfosse, P.; Boutry, S.; Palacios, A.P.; et al. Murine stroma adopts a human-like metabolic phenotype in the PDX model of colorectal cancer and liver metastases. Oncogene 2018, 37, 1237–1250. [Google Scholar] [CrossRef]
- Liu, C.; Lewin Mejia, D.; Chiang, B.; Luker, K.E.; Luker, G.D. Hybrid collagen alginate hydrogel as a platform for 3D tumor spheroid invasion. Acta Biomater. 2018, 75, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, M.V.; Gaspar, V.M.; Ferreira, L.; Mano, J.F. Hydrogel 3D in vitro tumor models for screening cell aggregation mediated drug response. Biomater. Sci. 2020, 8, 1855–1864. [Google Scholar] [CrossRef]
- Zhao, Y.; Yao, R.; Ouyang, L.; Ding, H.; Zhang, T.; Zhang, K.; Cheng, S.; Sun, W. Three-dimensional printing of Hela cells for cervical tumor model in vitro. Biofabrication 2014, 6, 035001. [Google Scholar] [CrossRef]
- Antunes, J.; Gaspar, V.M.; Ferreira, L.; Monteiro, M.; Henrique, R.; Jeronimo, C.; Mano, J.F. In-air production of 3D co-culture tumor spheroid hydrogels for expedited drug screening. Acta Biomater. 2019, 94, 392–409. [Google Scholar] [CrossRef] [PubMed]
- Edwards, S.J.; Carannante, V.; Kuhnigk, K.; Ring, H.; Tararuk, T.; Hallböök, F.; Blom, H.; Önfelt, B.; Brismar, H. High-Resolution Imaging of Tumor Spheroids and Organoids Enabled by Expansion Microscopy. Front. Mol. Biosci. 2020, 7, 208. [Google Scholar] [CrossRef]
- Lee, J.M.; Park, D.Y.; Yang, L.; Kim, E.-J.; Ahrberg, C.D.; Lee, K.-B.; Chung, B.G. Generation of uniform-sized multicellular tumor spheroids using hydrogel microwells for advanced drug screening. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Hachey, S.J.; Movsesyan, S.; Nguyen, Q.H.; Burton-Sojo, G.; Tankazyan, A.; Wu, J.; Hoang, T.; Zhao, D.; Wang, S.; Hatch, M.M.; et al. An in vitro vascularized micro-tumor model of human colorectal cancer recapitulates in vivo responses to standard-of-care therapy. Lab Chip 2021, 21, 1333–1351. [Google Scholar] [CrossRef]
- Diosdi, A.; Hirling, D.; Kovacs, M.; Toth, T.; Harmati, M.; Koos, K.; Buzas, K.; Piccinini, F.; Horvath, P. A quantitative metric for the comparative evaluation of optical clearing protocols for 3D multicellular spheroids. Comput. Struct. Biotechnol. J. 2021, 19, 1233–1243. [Google Scholar] [CrossRef] [PubMed]
- Bowers, H.J.; Fannin, E.E.; Thomas, A.; Weis, J.A. Characterization of multicellular breast tumor spheroids using image data-driven biophysical mathematical modeling. Sci. Rep. 2020, 10, 11583. [Google Scholar] [CrossRef] [PubMed]
- Balaji, P.; Murugadas, A.; Ramkumar, A.; Thirumurugan, R.; Shanmugaapriya, S.; Akbarsha, M.A. Characterization of Hen’s Egg White To Use It as a Novel Platform To Culture Three-Dimensional Multicellular Tumor Spheroids. ACS Omega 2020, 5, 19760–19770. [Google Scholar] [CrossRef]
- Van Zundert, I.; Fortuni, B.; Rocha, S. From 2D to 3D Cancer Cell Models-The Enigmas of Drug Delivery Research. Nanomaterials 2020, 10, 2236. [Google Scholar] [CrossRef] [PubMed]
- Lanuza, P.M.; Pesini, C.; Arias, M.A.; Calvo, C.; Ramirez-Labrada, A.; Pardo, J. Recalling the Biological Significance of Immune Checkpoints on NK Cells: A Chance to Overcome LAG3, PD1, and CTLA4 Inhibitory Pathways by Adoptive NK Cell Transfer? Front. Immunol. 2020, 10. [Google Scholar] [CrossRef] [Green Version]
- Nishi, K.; Ishikura, S.; Umebayashi, M.; Morisaki, T.; Inozume, T.; Kinugasa, T.; Aoki, M.; Nimura, S.; Swain, A.; Yoshida, Y.; et al. Mutant KRAS Promotes NKG2D+ T Cell Infiltration and CD155 Dependent Immune Evasion. Anticancer Res. 2020, 40, 4663–4674. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Martínez, D.; Azaceta, G.; Muntasell, A.; Aguiló, N.; Núñez, D.; Gálvez, E.M.; Naval, J.; Anel, A.; Palomera, L.; Vilches, C. Human NK cells activated by EBV lymphoblastoidcells overcome anti-apoptotic mechanisms of drug resistance in haematological cancer cells. Oncoimmunology 2015, 4, e991613. [Google Scholar] [CrossRef] [Green Version]
- Heinze, A.; Grebe, B.; Bremm, M.; Huenecke, S.; Munir, T.A.; Graafen, L.; Frueh, J.T.; Merker, M.; Rettinger, E.; Soerensen, J.; et al. The Synergistic Use of IL-15 and IL-21 for the Generation of NK Cells From CD3/CD19-Depleted Grafts Improves Their ex vivo Expansion and Cytotoxic Potential Against Neuroblastoma: Perspective for Optimized Immunotherapy Post Haploidentical Stem Cell Transplantation. Front. Immunol. 2019, 10, 2816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Bano, J.; Florès-Florès, R.; Josselin, E.; Goubard, A.; Ganier, L.; Castellano, R.; Chames, P.; Baty, D.; Kerfelec, B. A Bispecific Antibody-Based Approach for Targeting Mesothelin in Triple Negative Breast Cancer. Front. Immunol. 2019, 10, 1593. [Google Scholar] [CrossRef] [Green Version]
- Veneziani, I.; Infante, P.; Ferretti, E.; Melaiu, O.; Battistelli, C.; Lucarini, V.; Compagnone, M.; Nicoletti, C.; Castellano, A.; Petrini, S.; et al. Nutlin-3a Enhances Natural Killer Cell–Mediated Killing of Neuroblastoma by Restoring p53-Dependent Expression of Ligands for NKG2D and DNAM-1 Receptors. Cancer Immunol. Res. 2021, 9, 170–183. [Google Scholar] [CrossRef]
- Neal, J.; Li, X.; Zhu, J.; Giangarra, V.; Grzeskowiak, C.L.; Ju, J.; Liu, I.H.; Chiou, S.-H.; Salahudeen, A.A.; Smith, A.R.; et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell 2018, 175, 1972–1988.e16. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Ootani, A.; Kuo, C. An Air–Liquid Interface Culture System for 3D Organoid Culture of Diverse Primary Gastrointestinal Tissues. Methods Mol. Biol. 2016, 1422, 33–40. [Google Scholar] [PubMed]
- Drost, J.; Clevers, H. Organoids in cancer research. Nat. Rev. Cancer 2018, 18, 407–418. [Google Scholar] [CrossRef]
- Sakalem, M.E.; De Sibio, M.T.; da Costa, F.A.D.S.; de Oliveira, M. Historical evolution of spheroids and organoids, and possibilities of use in life sciences and medicine. Biotechnol. J. 2021, 16, e2000463. [Google Scholar] [CrossRef]
- Dijkstra, K.; Cattaneo, C.M.; Weeber, F.; Chalabi, M.; Van De Haar, J.; Fanchi, L.F.; Slagter, M.; Van Der Velden, D.L.; Kaing, S.; Kelderman, S.; et al. Generation of Tumor-Reactive T Cells by Co-culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell 2018, 174, 1586–1598.e12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dutta, D.; Heo, I.; Clevers, H. Disease Modeling in Stem Cell-Derived 3D Organoid Systems. Trends Mol. Med. 2017, 23, 393–410. [Google Scholar] [CrossRef] [PubMed]
- Abel, A.M.; Yang, C.; Thakar, M.S.; Malarkannan, S. Natural Killer Cells: Development, Maturation, and Clinical Utilization. Front. Immunol. 2018, 9, 1869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbas, A.K.; Trotta, E.; Simeonov, D.R.; Marson, A.; Bluestone, J.A. SciRevisiting IL-2: Biology and therapeutic prospects. Immunology 2018, 3, eaat1482. [Google Scholar] [CrossRef] [Green Version]
- Sim, G.C.; Radvanyi, L. The IL-2 cytokine family in cancer immunotherapy. Cytokine Growth Factor Rev. 2014, 25, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.; Lin, J.X.; Leonard, W.J. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity 2013, 38, 13–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meazza, R.; Azzarone, B.; Orengo, A.M.; Ferrini, S. Role of Common-Gamma Chain Cytokines in NK Cell Development and Function: Perspectives for Immunotherapy. J. Biomed. Biotechnol. 2011, 2011, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Braun, D.A.; Bakouny, Z.; Hirsch, L.; Flippot, R.; Van Allen, E.M.; Wu, C.J.; Choueiri, T.K. Beyond conventional immune-checkpoint inhibition—novel immunotherapies for renal cell carcinoma. Nat. Rev. Clin. Oncol. 2021, 18, 199–214. [Google Scholar] [CrossRef] [PubMed]
- Marcon, F.; Zuo, J.; Pearce, H.; Nicol, S.; Margielewska-Davies, S.; Farhat, M.; Mahon, B.; Middleton, G.; Brown, R.; Roberts, K.J.; et al. NK cells in pancreatic cancer demonstrate impaired cytotoxicity and a regulatory IL-10 phenotype. OncoImmunology 2020, 9, 1845424. [Google Scholar] [CrossRef] [PubMed]
- Schnalzger, T.E.; De Groot, M.H.; Zhang, C.; Mosa, M.H.; Michels, B.E.; Röder, J.; Darvishi, T.; Wels, W.S.; Farin, H.F. 3D model for CAR-mediated cytotoxicity using patient-derived colorectal cancer organoids. EMBO J. 2019, 38. [Google Scholar] [CrossRef]
- Gan, H.K.; Cvrljevic, A.N.; Johns, T.G. The epidermal growth factor receptor variant III (EGFRvIII): Where wild things are altered. FEBS J. 2013, 280, 5350–5370. [Google Scholar] [CrossRef]
- Phillips, A.C.; Boghaert, E.R.; Vaidya, K.S.; Falls, H.D.; Mitten, M.J.; Devries, P.J.; Benatuil, L.; Hsieh, C.-M.; Meulbroek, J.A.; Panchal, S.C.; et al. Characterization of ABBV-221, a Tumor-Selective EGFR-Targeting Antibody Drug Conjugate. Mol. Cancer Ther. 2018, 17, 795–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slaney, C.Y.; Kershaw, M.H.; Darcy, P.K. Trafficking of T Cells into Tumors. Cancer Res. 2014, 74, 7168–7174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertoni, A.; Alabiso, O.; Galetto, A.S.; Baldanzi, G. Integrins in T Cell Physiology. Int. J. Mol. Sci. 2018, 19, 485. [Google Scholar] [CrossRef] [Green Version]
- Maghazachi, A. Compartmentalization of human natural killer cells. Mol. Immunol. 2005, 42, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Korbecki, J.; Grochans, S.; Gutowska, I.; Barczak, K.; Baranowska-Bosiacka, I. CC Chemokines in a Tumor: A Review of Pro-Cancer and Anti-Cancer Properties of Receptors CCR5, CCR6, CCR7, CCR8, CCR9, and CCR10 Ligands. Int. J. Mol. Sci. 2020, 21, 7619. [Google Scholar] [CrossRef] [PubMed]
- Najafi, M.; Farhood, B.; Mortezaee, K. Extracellular matrix (ECM) stiffness and degradation as cancer drivers. J. Cell. Biochem. 2019, 120, 2782–2790. [Google Scholar] [CrossRef] [PubMed]
- Theocharis, A.D.; Manou, D.; Karamanos, N.K. The extracellular matrix as a multitasking player in disease. FEBS J. 2019, 286, 2830–2869. [Google Scholar] [CrossRef] [Green Version]
- Winkler, J.; Abisoye-Ogunniyan, A.; Metcalf, K.J.; Werb, Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 2020, 11, 5120. [Google Scholar] [CrossRef]
- Poggi, A.; Varesano, S.; Zocchi, M.R. How to Hit Mesenchymal Stromal Cells and Make the Tumor Microenvironment Immunostimulant Rather Than Immunosuppressive. Front. Immunol. 2018, 9, 262. [Google Scholar] [CrossRef] [Green Version]
- Galland, S.; Stamenkovic, I. Mesenchymal stromal cells in cancer: A review of their immunomodulatory functions and dual effects on tumor progression. J. Pathol. 2020, 250, 555–572. [Google Scholar] [CrossRef] [Green Version]
- Varderidou-Minasian, S.; Lorenowicz, M.J. Mesenchymal stromal/stem cell-derived extracellular vesicles in tissue repair: Challenges and opportunities. Theranostics 2020, 10, 5979–5997. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.J.; Solloa, E.H.; Shannon, M.J.; Mace, E.M. Generation of cell-derived matrices that support human NK cell migration and differentiation. J. Leukoc. Biol. 2020, 108, 1369–1378. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Matosevic, S. Chemokine networks modulating natural killer cell trafficking to solid tumors. Cytokine Growth Factor Rev. 2021, 59, 36–45. [Google Scholar] [CrossRef]
- Chouaib, S.; Kieda, C.; Benlalam, H.; Noman, M.Z.; Mami-Chouaib, F.; Rüegg, C. Endothelial cells as key determinants of the tumor microenvironment: Interaction with tumor cells, extracellular matrix and immune killer cells. Crit. Rev. Immunol. 2010, 30, 529–545. [Google Scholar] [CrossRef]
- Temples, M.N.; Adjei, I.M.; Nimocks, P.M.; Djeu, J.; Sharma, B. Engineered Three-Dimensional Tumor Models to Study Natural Killer Cell Suppression. ACS Biomater. Sci. Eng. 2020, 6, 4179–4199. [Google Scholar] [CrossRef] [PubMed]
- Hinshaw, D.C.; Shevde, L.A. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar] [CrossRef] [Green Version]
- Rossi, G.R.; Trindade, E.S.; Souza-Fonseca-Guimaraes, F. Tumor Microenvironment-Associated Extracellular Matrix Components Regulate NK Cell Function. Front. Immunol. 2020, 11, 73. [Google Scholar] [CrossRef]
- Barry, K.C.; Hsu, J.; Broz, M.L.; Cueto, F.J.; Binnewies, M.; Combes, A.J.; Nelson, A.E.; Loo, K.; Kumar, R.; Rosenblum, M.D.; et al. A natural killer–dendritic cell axis defines checkpoint therapy–responsive tumor microenvironments. Nat. Med. 2018, 24, 1178–1191. [Google Scholar] [CrossRef] [PubMed]
- Novikov, N.M.; Zolotaryova, S.Y.; Gautreau, A.M.; Denisov, E.V. Mutational drivers of cancer cell migration and invasion. Br. J. Cancer 2021, 124, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, M.; Naik, R.; Lingaraju, S.M.; Susheela, S.P.; Patil, S.; Srinivasachar, G.K.; Thungappa, S.C.; Murugan, K.; Jayappa, S.B.; Bhattacharjee, S.; et al. Landscape of clinically actionable mutations in breast cancer ‘A cohort study’. Transl. Oncol. 2021, 14, 100877. [Google Scholar] [CrossRef]
- Miles, B.; Tadi, P. Genetics, Somatic Mutation. 2020 Apr. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Persi, E.; Wolf, Y.I.; Horn, D.; Ruppin, E.; Demichelis, F.; Gatenby, R.A.; Gillies, R.J.; Koonin, E.V. Mutation–selection balance and compensatory mechanisms in tumour evolution. Nat. Rev. Genet. 2021, 22, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Strickler, J.H.; Hanks, B.A.; Khasraw, M. Tumor Mutational Burden as a Predictor of Immunotherapy Response: Is More Always Better? Clin. Cancer Res. 2021, 27, 1236–1241. [Google Scholar] [CrossRef] [PubMed]
- Luengo, A.; Gui, D.Y.; Heiden, M.G.V. Targeting Metabolism for Cancer Therapy. Cell Chem. Biol. 2017, 24, 1161–1180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koundouros, N.; Poulogiannis, G. Reprogramming of fatty acid metabolism in cancer. Br. J. Cancer 2020, 122, 4–22. [Google Scholar] [CrossRef] [Green Version]
- Park, J.H.; Pyun, W.Y.; Park, H.W. Cancer Metabolism: Phenotype, Signaling and Therapeutic Targets. Cells 2020, 9, 2308. [Google Scholar] [CrossRef]
- Bergers, G.; Fendt, S.-M. The metabolism of cancer cells during metastasis. Nat. Rev. Cancer 2021, 21, 162–180. [Google Scholar] [CrossRef]
- Hernández Vera, R.; O’Callaghan, P.; Fatsis-Kavalopoulos, N.; Kreuger, J. Modular microfluidic systems cast from 3D-printed molds for imaging leukocyte adherence to differentially treated endothelial cultures. Sci. Rep. 2019, 9, 11321. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Jang, J.H.; Wang, C.; He, B.; Zhang, K.; Zhang, P.; Vu, T.; Qin, L. Microfluidics Cell Loading-Dock System: Ordered Cellular Array for Dynamic Lymphocyte-Communication Study. Adv. Biosyst. 2017, 1. [Google Scholar] [CrossRef]
- Perozziello, G.; La Rocca, R.; Cojoc, G.; Liberale, C.; Malara, N.; Simone, G.; Candeloro, P.; Anichini, A.; Tirinato, L.; Gentile, F.; et al. Microfluidic Devices Modulate Tumor Cell Line Susceptibility to NK Cell Recognition. Small 2012, 8, 2886–2894. [Google Scholar] [CrossRef]
- Briones, J.C.; Espulgar, W.V.; Koyama, S.; Yoshikawa, H.; Park, J.; Naito, Y.; Kumanogoh, A.; Tamiya, E.; Takamatsu, H.; Saito, M. A Microfluidic Platform for Single Cell Fluorometric Granzyme B Profiling. Theranostics 2020, 10, 123–132. [Google Scholar] [CrossRef]
- Ayuso, J.M.; Truttschel, R.; Gong, M.M.; Humayun, M.; Virumbrales-Munoz, M.; Vitek, R.; Felder, M.; Gillies, S.D.; Sondel, P.; Wisinski, K.B.; et al. Evaluating natural killer cell cytotoxicity against solid tumors using a microfluidic model. OncoImmunology 2018, 8, 1553477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadovska, L.; Zandberga, E.; Sagini, K.; Jēkabsons, K.; Riekstiņa, U.; Kalniņa, Z.; Llorente, A.; Linē, A. A novel 3D heterotypic spheroid model for studying extracellular vesicle-mediated tumour and immune cell communication. Biochem. Biophys. Res. Commun. 2018, 495, 1930–1935. [Google Scholar] [CrossRef] [PubMed]
- Borten, M.A.; Bajikar, S.S.; Sasaki, N.; Clevers, H.; Janes, K.A. Automated brightfield morphometry of 3D organoid populations by OrganoSeg. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Piccinini, F.; Balassa, T.; Carbonaro, A.; Diosdi, A.; Toth, T.; Moshkov, N.; Tasnadi, E.A.; Horvath, P. Software tools for 3D nuclei segmentation and quantitative analysis in multicellular aggregates. Comput. Struct. Biotechnol. J. 2020, 18, 1287–1300. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).