Accelerated T2-Weighted TSE Imaging of the Prostate Using Deep Learning Image Reconstruction: A Prospective Comparison with Standard T2-Weighted TSE Imaging
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Deep Learning Image Reconstruction Technique
2.3. Imaging Protocol
2.4. Image Analysis
2.5. Statistical Evaluation
3. Results
3.1. Patients’ Characteristics
3.2. Evaluation of Qualitative Imaging Parameters
3.3. PI-RADS Scoring and Lesion Conspicuity
4. Discussion
Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Weinreb, J.C.; Barentsz, J.O.; Choyke, P.L.; Cornud, F.; Haider, M.A.; Macura, K.J.; Margolis, D.; Schnall, M.D.; Shtern, F.; Verma, S.; et al. PI-RADS Prostate Imaging—Reporting and Data System: 2015, Version 2. Eur. Urol. 2016, 69, 16–40. [Google Scholar] [CrossRef]
- Giganti, F.; Rosenkrantz, A.B.; Villeirs, G.; Panebianco, V.; Stabile, A.; Emberton, M.; Moore, C.M. The Evolution of MRI of the Prostate: The Past, the Present, and the Future. AJR Am. J. Roentgenol. 2019, 213, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Cornud, F.; Delongchamps, N.B.; Mozer, P.; Beuvon, F.; Schull, A.; Muradyan, N.; Peyromaure, M. Value of multiparametric MRI in the work-up of prostate cancer. Curr. Urol. Rep. 2012, 13, 82–92. [Google Scholar] [CrossRef]
- Ahmed, H.U.; Bosaily, A.E.S.; Brown, L.C.; Gabe, R.; Kaplan, R.; Parmar, M.K.; PROMIS Study Group. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): A paired validating confirmatory study. Lancet 2017, 389, 815–822. [Google Scholar] [CrossRef] [Green Version]
- Kasivisvanathan, V.; Rannikko, A.S.; Borghi, M.; Panebianco, V.; Mynderse, L.A.; Vaarala, M.H.; Moore, C.M. MRI-Targeted or Standard Biopsy for Prostate-Cancer Diagnosis. N. Engl. J. Med. 2018, 378, 1767–1777. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, L.; Ahmed, H.U.; Allen, C.; Barentsz, J.O.; Carey, B.; Futterer, J.J.; Emberton, M. Clinical applications of multiparametric MRI within the prostate cancer diagnostic pathway. Urol. Oncol. 2013, 31, 281–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McBee, M.P.; Awan, O.A.; Colucci, A.T.; Ghobadi, C.W.; Kadom, N.; Kansagra, A.P.; Auffermann, W.F. Deep Learning in Radiology. Acad. Radiol. 2018, 25, 1472–1480. [Google Scholar] [CrossRef] [Green Version]
- Walsh, S.L.F.; Calandriello, L.; Silva, M.; Sverzellati, N. Deep learning for classifying fibrotic lung disease on high-resolution computed tomography: A case-cohort study. Lancet Respir. Med. 2018, 6, 837–845. [Google Scholar] [CrossRef]
- Zhang, Y.D.; Wang, J.; Wu, C.J.; Bao, M.L.; Li, H.; Wang, X.N.; Shi, H.B. An imaging-based approach predicts clinical outcomes in prostate cancer through a novel support vector machine classification. Oncotarget 2016, 7, 78140–78151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, X.; Cao, R.; Shakeri, S.; Scalzo, F.; Lee, Y.; Enzmann, D.R.; Sung, K. Deep transfer learning-based prostate cancer classification using 3 Tesla multi-parametric MRI. Abdom. Radiol. 2019, 44, 2030–2039. [Google Scholar] [CrossRef]
- Padhani, A.R.; Turkbey, B. Detecting Prostate Cancer with Deep Learning for MRI: A Small Step Forward. Radiology 2019, 293, 618–619. [Google Scholar] [CrossRef]
- Kim, M.; Kim, H.S.; Kim, H.J.; Park, J.E.; Park, S.Y.; Kim, Y.H.; Lebel, M.R. Thin-Slice Pituitary MRI with Deep Learning-based Reconstruction: Diagnostic Performance in a Postoperative Setting. Radiology 2021, 298, 114–122. [Google Scholar] [CrossRef]
- Recht, M.P.; Zbontar, J.; Sodickson, D.K.; Knoll, F.; Yakubova, N.; Sriram, A.; Zitnick, C.L. Using Deep Learning to Accelerate Knee MRI at 3T: Results of an Interchangeability Study. AJR Am. J. Roentgenol. 2020. [Google Scholar] [CrossRef]
- Herrmann, J.; Gassenmaier, S.; Nickel, D.; Arberet, S.; Afat, S.; Lingg, A.; Othman, A.E. Diagnostic Confidence and Feasibility of a Deep Learning Accelerated HASTE Sequence of the Abdomen in a Single Breath-Hold. Investig. Radiol. 2020. [Google Scholar] [CrossRef]
- Herrmann, J.; Nickel, D.; Mugler, J.P., III; Arberet, S.; Gassenmaier, S.; Afat, S.; Othman, A.E. Development and Evaluation of Deep Learning-Accelerated Single-Breath-Hold Abdominal HASTE at 3 T Using Variable Refocusing Flip Angles. Investig. Radiol. 2021. [Google Scholar] [CrossRef]
- Hammernik, K.; Klatzer, T.; Kobler, E.; Recht, M.P.; Sodickson, D.K.; Pock, T.; Knoll, F. Learning a variational network for reconstruction of accelerated MRI data. Magn. Reson. Med. 2018, 79, 3055–3071. [Google Scholar] [CrossRef] [PubMed]
- Gassenmaier, S.; Afat, S.; Nickel, D.; Mostapha, M.; Herrmann, J.; Othman, A.E. Deep learning-accelerated T2-weighted imaging of the prostate: Reduction of acquisition time and improvement of image quality. Eur. J. Radiol. 2021, 137, 109600. [Google Scholar] [CrossRef]
- Becker, A.S.; Marcon, M.; Ghafoor, S.; Wurnig, M.C.; Frauenfelder, T.; Boss, A. Deep Learning in Mammography: Diagnostic Accuracy of a Multipurpose Image Analysis Software in the Detection of Breast Cancer. Investig. Radiol. 2017, 52, 434–440. [Google Scholar] [CrossRef] [PubMed]
- Fayad, L.M.; Parekh, V.S.; de Castro Luna, R.; Ko, C.C.; Tank, D.; Fritz, J.; Jacobs, M.A. A Deep Learning System for Synthetic Knee Magnetic Resonance Imaging: Is Artificial Intelligence-Based Fat-Suppressed Imaging Feasible? Investig. Radiol. 2021, 56, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Kart, T.; Fischer, M.; Küstner, T.; Hepp, T.; Bamberg, F.; Winzeck, S.; Gatidis, S. Deep Learning-Based Automated Abdominal Organ Segmentation in the UK Biobank and German National Cohort Magnetic Resonance Imaging Studies. Investig. Radiol. 2021, 56, 401–408. [Google Scholar] [CrossRef]
- Almansour, H.; Gassenmaier, S.; Nickel, D.; Kannengiesser, S.; Afat, S.; Weiss, J.; Othman, A.E. Deep Learning-Based Superresolution Reconstruction for Upper Abdominal Magnetic Resonance Imaging: An Analysis of Image Quality, Diagnostic Confidence, and Lesion Conspicuity. Investig. Radiol. 2021. [Google Scholar] [CrossRef]
- Chartrand, G.; Cheng, P.M.; Vorontsov, E.; Drozdzal, M.; Turcotte, S.; Pal, C.J.; Tang, A. Deep Learning: A Primer for Radiologists. Radiographics 2017, 37, 2113–2131. [Google Scholar] [CrossRef] [Green Version]
- Schelb, P.; Kohl, S.; Radtke, J.P.; Wiesenfarth, M.; Kickingereder, P.; Bickelhaupt, S.; Bonekamp, D. Classification of Cancer at Prostate MRI: Deep Learning versus Clinical PI-RADS Assessment. Radiology 2019, 293, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Shao, W.; Banh, L.; Kunder, C.A.; Fan, R.E.; Soerensen, S.J.; Wang, J.B.; Rusu, M. ProsRegNet: A deep learning framework for registration of MRI and histopathology images of the prostate. Med. Image Anal. 2021, 68, 101919. [Google Scholar] [CrossRef]
- Wang, X.; Ma, J.; Bhosale, P.; Rovira, J.J.I.; Qayyum, A.; Sun, J.; Szklaruk, J. Novel deep learning-based noise reduction technique for prostate magnetic resonance imaging. Abdom. Radiol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Benkert, T.; Block, K.T.; Sodickson, D.K.; Otazo, R.; Chandarana, H. Compressed sensing for body MRI. J. Magn. Reson. Imaging 2017, 45, 966–987. [Google Scholar] [CrossRef] [PubMed]
- Griswold, M.A.; Jakob, P.M.; Heidemann, R.M.; Nittka, M.; Jellus, V.; Wang, J.; Haase, A. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn. Reson. Med. 2002, 47, 1202–1210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lustig, M.; Donoho, D.; Pauly, J.M. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn. Reson. Med. 2007, 58, 1182–1195. [Google Scholar] [CrossRef]
- Yanasak, N.E.; Kelly, M.J. MR imaging artifacts and parallel imaging techniques with calibration scanning: A new twist on old problems. Radiographics 2014, 34, 532–548. [Google Scholar] [CrossRef] [Green Version]
- Yang, R.K.; Roth, C.G.; Ward, R.J.; deJesus, J.O.; Mitchell, D.G. Optimizing abdominal MR imaging: Approaches to common problems. Radiographics 2010, 30, 185–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Rooij, M.; Hamoen, E.H.; Futterer, J.J.; Barentsz, J.O.; Rovers, M.M. Accuracy of multiparametric MRI for prostate cancer detection: A meta-analysis. AJR Am. J. Roentgenol. 2014, 202, 343–351. [Google Scholar] [CrossRef]
- Giganti, F.; Stabile, A.; Stavrinides, V.; Osinibi, E.; Retter, A.; Orczyk, C.; Moore, C.M. Natural history of prostate cancer on active surveillance: Stratification by MRI using the PRECISE recommendations in a UK cohort. Eur. Radiol. 2021, 31, 1644–1655. [Google Scholar] [CrossRef] [PubMed]
- Turkbey, B.; Rosenkrantz, A.B.; Haider, M.A.; Padhani, A.R.; Villeirs, G.; Macura, K.J.; Weinreb, J.C. Prostate Imaging Reporting and Data System Version 2.1: 2019 Update of Prostate Imaging Reporting and Data System Version 2. Eur. Urol. 2019, 76, 340–351. [Google Scholar] [CrossRef] [PubMed]

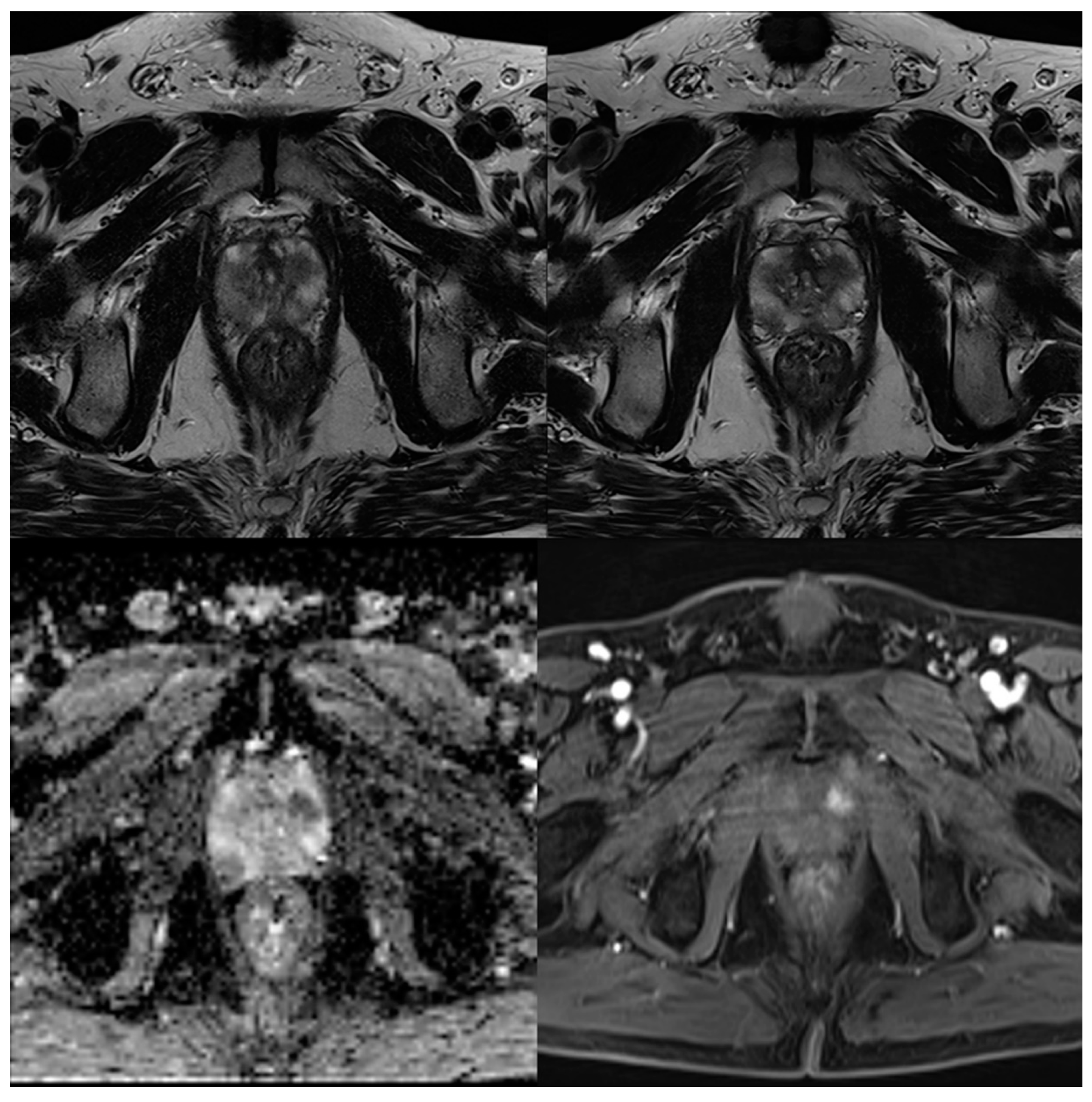
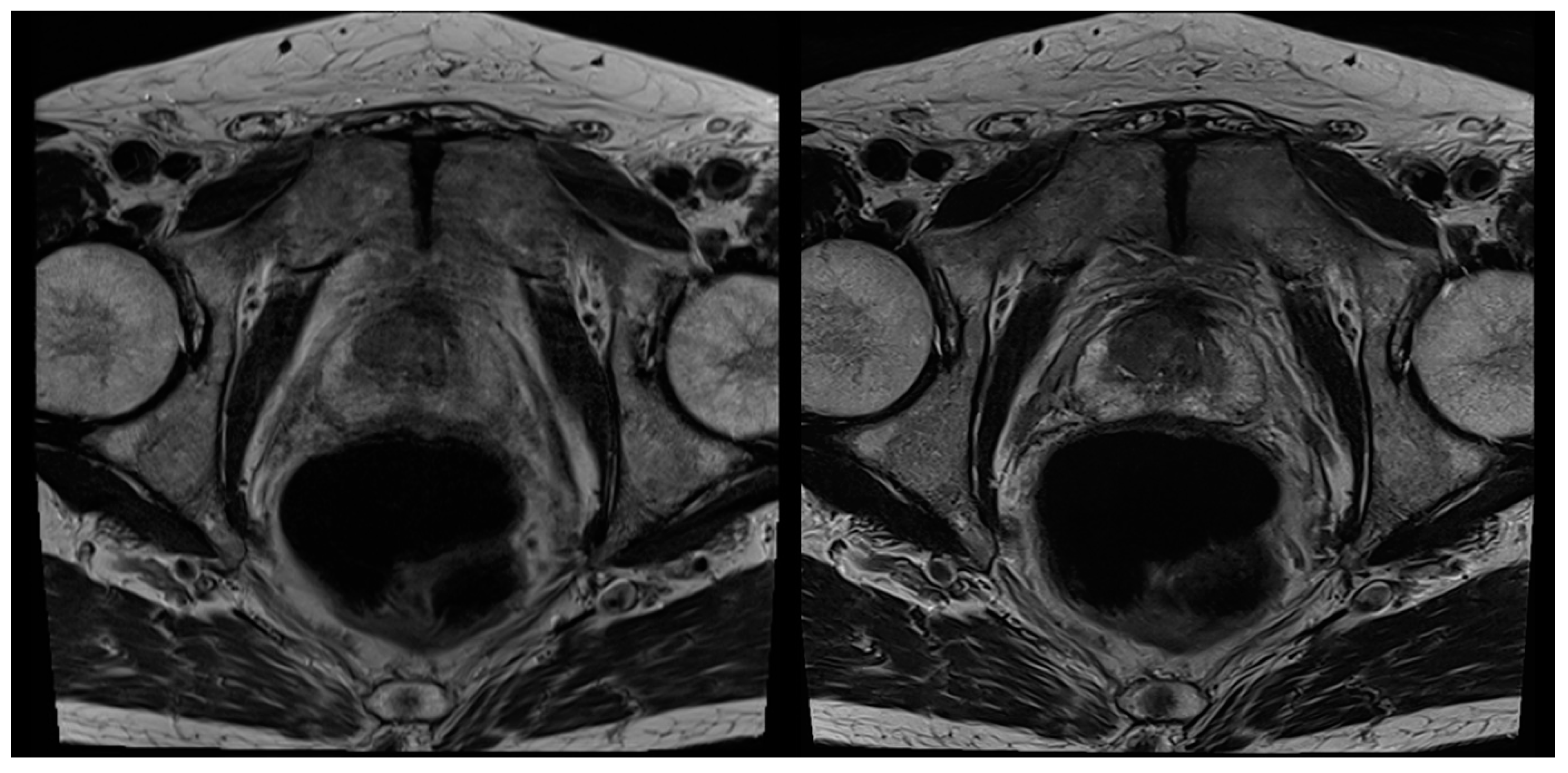
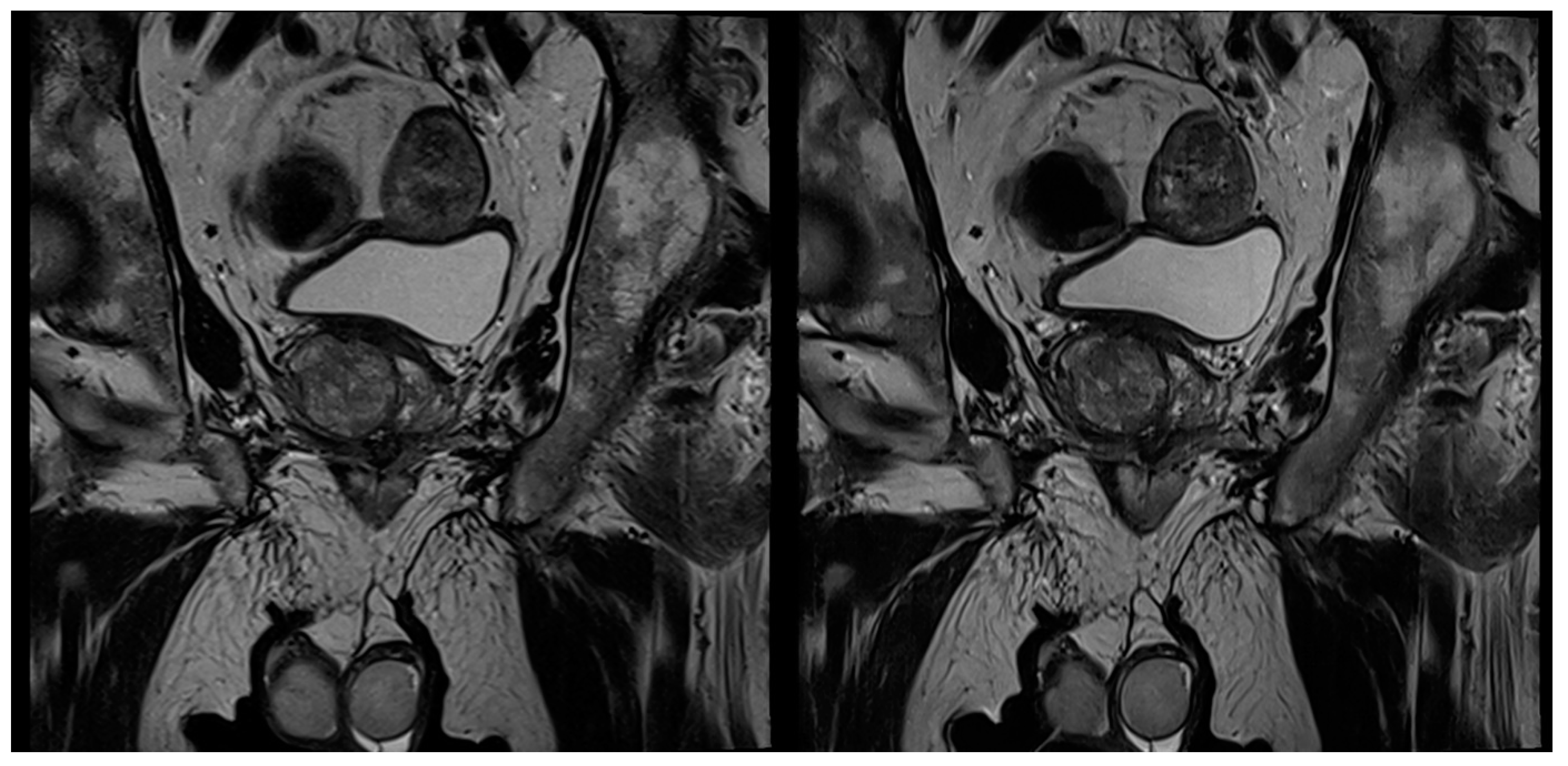
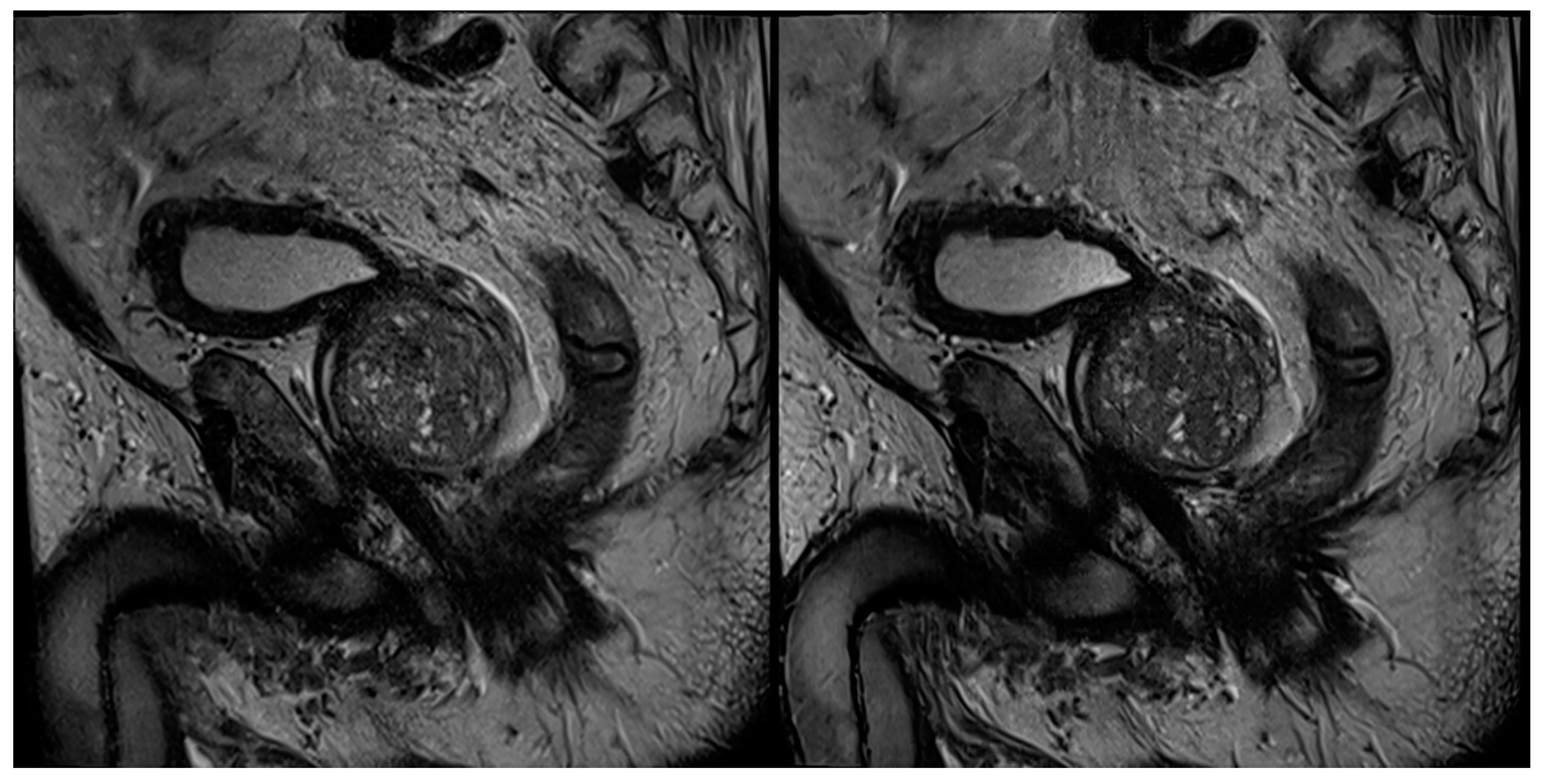
| Characteristics | Values |
|---|---|
| Number of patients | n = 60 |
| Age, mean ± standard deviation | 69 ± 9 years |
| Sex | 100% male |
| PSA, median (interquartile range) | 7.2 ng/mL (5.9–9.7 ng/mL) |
| Axial | Coronal | Sagittal | ||||
|---|---|---|---|---|---|---|
| T2S | T2DLR | T2S | T2DLR | T2S | T2DLR | |
| TR (ms) | 4470 | 4470 | 7480 | 7760 | 7480 | 6900 |
| TE (ms) | 104 | 104 | 101 | 101 | 101 | 101 |
| Averages | 3 | 1 | 3 | 1 | 2 | 1 |
| Voxel size (mm) | 0.3 × 0.3 × 3.0 | 0.3 × 0.3 × 3.0 | 0.3 × 0.3 × 3.0 | 0.3 × 0.3 × 3.0 | 0.3 × 0.3 × 3.0 | 0.3 × 0.3 × 3.0 |
| Field of view (mm) | 200 | 200 | 200 | 200 | 200 | 200 |
| Slice thickness (mm) | 3 | 3 | 3 | 3 | 3 | 3 |
| Parallel imaging factor | 3 | 3 | 3 | 3 | 2 | 3 |
| Acquisition time (min:sec) | 4:37 | 1:38 | 3:07 | 1:10 | 2:37 | 1:02 |
| Characteristics | Reader 1 | Reader 2 | ||||
|---|---|---|---|---|---|---|
| T2S | T2DLR | p-Value | T2S | T2DLR | p-Value | |
| Image noise axial | 3 (3–3) | 4 (4–4) | <0.001 | 3 (3–3) | 4 (4–4) | <0.001 |
| Image noise coronal | 3 (3–3) | 4 (4–4) | <0.001 | 3 (3–4) | 4 (4–4) | <0.001 |
| Image noise sagittal | 3 (3–3) | 4 (3–4) | <0.001 | 4 (3–4) | 4 (3–4) | 0.005 |
| Artifacts axial | 3 (3–4) | 4 (4–4) | 0.003 | 4 (3–4) | 4 (4–4) | 0.003 |
| Artifacts coronal | 3 (3–4) | 4 (4–4) | 0.002 | 3 (3–4) | 4 (4–4) | 0.014 |
| Artifacts sagittal | 3 (3–4) | 4 (3–4) | 0.002 | 3 (3–4) | 4 (3–4) | 0.011 |
| Natural appearance axial | 4 (4–4) | 4 (4–4) | 1 | 4 (4–4) | 4 (4–4) | 0.630 |
| Natural appearance coronal | 4 (4–4) | 4 (4–4) | 1 | 4 (4–4) | 4 (4–4) | 1 |
| Natural appearance sagittal | 4 (3–4) | 4 (3–4) | 1 | 4 (3–4) | 4 (3–4) | 1 |
| Overall image quality axial | 3 (3–4) | 4 (4–4) | <0.001 | 3 (3–4) | 4 (4–4) | <0.001 |
| Overall image quality coronal | 3 (3–4) | 4 (4–4) | 0.002 | 3 (3–4) | 4 (4–4) | <0.001 |
| Overall image quality sagittal | 3 (3–4) | 4 (3–4) | 0.002 | 3 (3–4) | 4 (3–4) | 0.005 |
| Diagnostic confidence | 4 (3–4) | 4 (4–4) | 0.03 | 4 (3–4) | 4 (4–4) | 0.06 |
| T2 and PI-RADS Scoring | Reader 1 | Reader 2 | ||
|---|---|---|---|---|
| T2S | T2DLR | T2S | T2DLR | |
| T2 score | ||||
| 1 | 0 | 0 | 0 | 0 |
| 2 | 21 | 21 | 21 | 21 |
| 3 | 14 | 13 | 13 | 14 |
| 4 | 15 | 16 | 16 | 15 |
| 5 | 10 | 10 | 10 | 10 |
| PI-RADS score | ||||
| 1 | 0 | 0 | 0 | 0 |
| 2 | 21 | 21 | 21 | 21 |
| 3 | 4 | 5 | 5 | 5 |
| 4 | 25 | 24 | 24 | 24 |
| 5 | 10 | 10 | 10 | 10 |
| Intra-Reader Agreement | |
| Reader 1 T2 score | 0.931 |
| Reader 2 T2 score | 0.905 |
| Reader 1 PI-RADS score | 0.960 |
| Reader 2 PI-RADS score | 0.946 |
| Inter-Reader Agreement | |
| T2 score T2S | 0.823 |
| T2 score T2DLR | 0.932 |
| PI-RADS T2S | 0.905 |
| PI-RADS T2DLR | 0.946 |
| Characteristics | Reader 1 | Reader 2 | ||||
|---|---|---|---|---|---|---|
| T2S | T2DLR | p-Value | T2S | T2DLR | p-Value | |
| Lesion size (mm) | 13 (10–16) | 13 (11–16) | 1 | 12 (10–16) | 13 (10–17) | 0.840 |
| Lesion conspicuity | 3 (3–4) | 4 (4–4) | <0.001 | 3 (3–4) | 4 (4–4) | 0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gassenmaier, S.; Afat, S.; Nickel, M.D.; Mostapha, M.; Herrmann, J.; Almansour, H.; Nikolaou, K.; Othman, A.E. Accelerated T2-Weighted TSE Imaging of the Prostate Using Deep Learning Image Reconstruction: A Prospective Comparison with Standard T2-Weighted TSE Imaging. Cancers 2021, 13, 3593. https://doi.org/10.3390/cancers13143593
Gassenmaier S, Afat S, Nickel MD, Mostapha M, Herrmann J, Almansour H, Nikolaou K, Othman AE. Accelerated T2-Weighted TSE Imaging of the Prostate Using Deep Learning Image Reconstruction: A Prospective Comparison with Standard T2-Weighted TSE Imaging. Cancers. 2021; 13(14):3593. https://doi.org/10.3390/cancers13143593
Chicago/Turabian StyleGassenmaier, Sebastian, Saif Afat, Marcel Dominik Nickel, Mahmoud Mostapha, Judith Herrmann, Haidara Almansour, Konstantin Nikolaou, and Ahmed E. Othman. 2021. "Accelerated T2-Weighted TSE Imaging of the Prostate Using Deep Learning Image Reconstruction: A Prospective Comparison with Standard T2-Weighted TSE Imaging" Cancers 13, no. 14: 3593. https://doi.org/10.3390/cancers13143593
APA StyleGassenmaier, S., Afat, S., Nickel, M. D., Mostapha, M., Herrmann, J., Almansour, H., Nikolaou, K., & Othman, A. E. (2021). Accelerated T2-Weighted TSE Imaging of the Prostate Using Deep Learning Image Reconstruction: A Prospective Comparison with Standard T2-Weighted TSE Imaging. Cancers, 13(14), 3593. https://doi.org/10.3390/cancers13143593








