The Role of DND1 in Cancers
Abstract
Simple Summary
Abstract
1. Introduction
2. Molecular Mechanisms of DND1 Function
2.1. mRNA Stabilization
2.2. mRNA Repression or Degradation
2.3. Other Molecular Functions of DND1
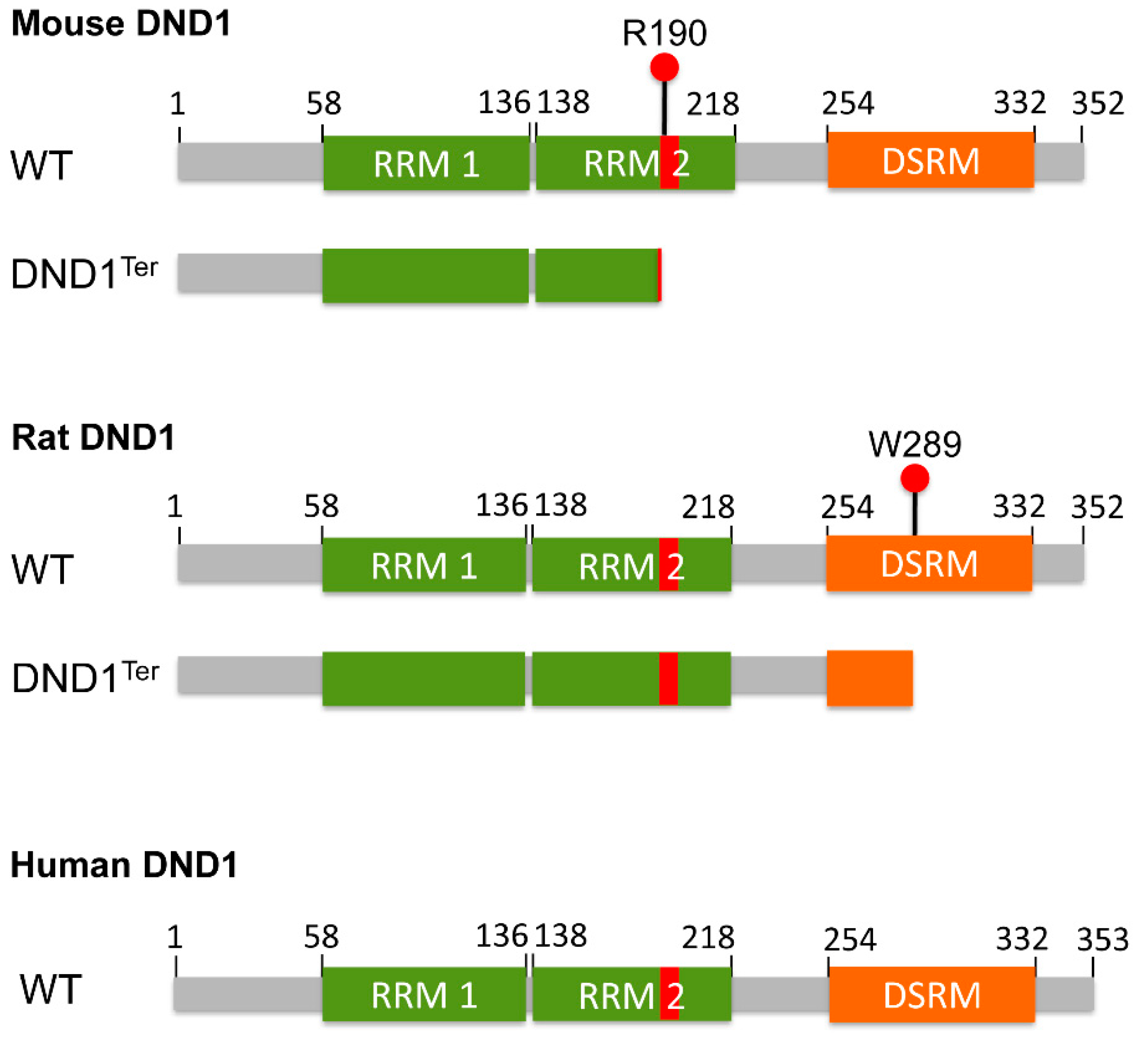
3. Dnd1Ter Mutation in Testicular and Ovarian Teratomas
4. The Emerging Role of DND1 in Somatic Cancers
5. Genetically Engineered Mouse Alleles of DND1
6. Conclusions and Future Perspective
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stevens, L.C. A new inbred subline of mice (129-terSv) with a high incidence of spontaneous congenital testicular teratomas. J. Natl. Cancer Inst. 1973, 50, 235–242. [Google Scholar] [CrossRef]
- Noguchi, T.; Noguchi, M. A recessive mutation (ter) causing germ cell deficiency and a high incidence of congenital testicular teratomas in 129/Sv-ter mice. J. Natl. Cancer Inst. 1985, 75, 385–392. [Google Scholar]
- Noguchi, T.; Stevens, L.C. Primordial germ cell proliferation in fetal testes in mouse strains with high and low incidences of congenital testicular teratomas. J. Natl. Cancer Inst. 1982, 69, 907–913. [Google Scholar] [CrossRef]
- Sakurai, T.; Katoh, H.; Moriwaki, K.; Noguchi, T.; Noguchi, M. The ter primordial germ cell deficiency mutation maps near Grl-1 on mouse chromosome 18. Mamm. Genome 1994, 5, 333–336. [Google Scholar] [CrossRef]
- Asada, Y.; Varnum, D.S.; Frankel, W.N.; Nadeau, J.H. A mutation in the Ter gene causing increased susceptibility to testicular teratomas maps to mouse chromosome 18. Nat. Genet. 1994, 6, 363–368. [Google Scholar] [CrossRef]
- Youngren, K.K.; Coveney, D.; Peng, X.; Bhattacharya, C.; Schmidt, L.S.; Nickerson, M.L.; Lamb, B.T.; Deng, J.M.; Behringer, R.R.; Capel, B.; et al. The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature 2005, 435, 360–364. [Google Scholar] [CrossRef]
- Kedde, M.; Strasser, M.J.; Boldajipour, B.; Vrielink, J.A.; Slanchev, K.; le Sage, C.; Nagel, R.; Voorhoeve, P.M.; van Duijse, J.; Orom, U.A.; et al. RNA-Binding Protein Dnd1 Inhibits MicroRNA Access to Target mRNA. Cell 2007, 131, 1273–1286. [Google Scholar] [CrossRef]
- Aguero, T.; Jin, Z.; Owens, D.; Malhotra, A.; Newman, K.; Yang, J.; King, M. Combined functions of two RRMs in Dead-end1 mimic helicase activity to promote nanos1 translation in the germline. Mol. Reprod. Dev. 2018, 85, 896–908. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Niimi, Y.; Shinmyozu, K.; Zhou, Z.; Kiso, M.; Saga, Y. Dead end1 is an essential partner of NANOS2 for selective binding of target RNAs in male germ cell development. EMBO Rep. 2016, 17, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, M.; Jishage, M.; Meyer, C.; Suryawanshi, H.; Der, E.; Yamaji, M.; Garzia, A.; Morozov, P.; Manickavel, S.; McFarland, H.L.; et al. DND1 maintains germline stem cells via recruitment of the CCR4-NOT complex to target mRNAs. Nature 2017, 543, 568–572. [Google Scholar] [CrossRef] [PubMed]
- Northrup, E.; Zschemisch, N.; Eisenblätter, R.; Glage, S.; Wedekind, D.; Cuppen, E.; Dorsch, M.; Hedrich, H. The ter mutation in the rat Dnd1 gene initiates gonadal teratomas and infertility in both genders. PLoS ONE. 2012, 7, e38001. [Google Scholar] [CrossRef]
- Naser, A.A.; Miyazaki, T.; Wang, J.; Takabayashi, S.; Pachoensuk, T.; Tokumoto, T. MC4R mutant mice develop ovarian teratomas. Sci. Rep. 2021, 11, 3483. [Google Scholar] [CrossRef]
- Liu, X.; Wang, A.; Heidbreder, C.E.; Jiang, L.; Yu, J.; Kolokythas, A.; Huang, L.; Dai, Y.; Zhou, X. MicroRNA-24 targeting RNA-binding protein DND1 in tongue squamous cell carcinoma. FEBS Lett. 2010, 584, 4115–4120. [Google Scholar] [CrossRef] [PubMed]
- Zechel, J.; Doerner, S.; Lager, A.; Tesar, P.; Heaney, J.; Nadeau, J. Contrasting effects of Deadend1 (Dnd1) gain and loss of function mutations on allelic inheritance, testicular cancer, and intestinal polyposis. BMC Genet. 2013, 14, 54. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, A.; Gordon, W.; Dizon, D.; Hopkin, A.S.; Gordon, E.; Yu, Z.; Andersen, B. The Grainyhead transcription factor Grhl3/Get1 suppresses miR-21 expression and tumorigenesis in skin: Modulation of the miR-21 target MSH2 by RNA-binding protein DND1. Oncogene 2013, 32, 1497–1507. [Google Scholar] [CrossRef] [PubMed]
- Wampfler, J.; Federzoni, E.A.; Torbett, B.E.; Fey, M.F.; Tschan, M.P. The RNA binding proteins RBM38 and DND1 are repressed in AML and have a novel function in APL differentiation. Leuk. Res. 2016, 41, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Pan, Y.; Lu, Y.M.; Zhu, L.; Chen, S. RNA-Binding Protein Dnd1 Promotes Breast Cancer Apoptosis by Stabilizing the Bim mRNA in a miR-221 Binding Site. Biomed. Res. Int. 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Gong, F.; Zhang, T.; Chi, B.; Wang, J. RNA-binding protein Dnd1 inhibits epithelial–mesenchymal transition and cancer stem cell-related traits on hepatocellular carcinoma cells. Biotechnol. Lett. 2017, 39, 1359–1367. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, W.; Liu, G.H.; Tang, W. MicroRNA-24 regulates the growth and chemosensitivity of the human colorectal cancer cells by targeting RNA-binding protein DND1. J. BUON 2019, 24, 1476–1481. [Google Scholar]
- Lloyd, R.; Erickson, L.; Jin, L.; Kulig, E.; Qian, X.; Cheville, J.; Scheithauer, B. p27kip1: A multifunctional cyclin-dependent kinase inhibitor with prognostic significance in human cancers. Am. J. Pathol. 1999, 154, 313–323. [Google Scholar] [CrossRef]
- Chen, S.; Zeng, M.; Sun, H.; Deng, W.; Lu, Y.; Tao, D.; Liu, Y.; Zhang, S.; Ma, Y. Zebrafish Dnd protein binds to 3’UTR of geminin mRNA and regulates its expression. BMB Rep. 2010, 43, 438–444. [Google Scholar] [CrossRef]
- Gu, W.; Mochizuki, K.; Otsuka, K.; Hamada, R.; Takehara, A.; Matsui, Y. Dnd1-mediated epigenetic control of teratoma formation in mouse. Biol. Open 2018, 7, bio032318. [Google Scholar] [CrossRef]
- Mei, W.; Jin, Z.; Lai, F.; Schwend, T.; Houston, D.; King, M.; Yang, J. Maternal Dead-End1 is required for vegetal cortical microtubule assembly during Xenopus axis specification. Development 2013, 140, 2334–2344. [Google Scholar] [CrossRef]
- Koebernick, K.; Loeber, J.; Arthur, P.K.; Tarbashevich, K.; Pieler, T. Elr-type proteins protect Xenopus Dead end mRNA from miR-18-mediated clearance in the soma. Proc. Natl. Acad. Sci. USA 2010, 107, 16148–16153. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, C.; Aggarwal, S.; Kumar, M.; Ali, A.; Matin, A. Mouse apolipoprotein B editing complex 3 (APOBEC3) is expressed in germ cells and interacts with dead-end (DND1). PLoS ONE 2008, 3, e2315. [Google Scholar] [CrossRef]
- Ali, S.; Karki, N.; Bhattacharya, C.; Zhu, R.; MacDuff, D.; Stenglein, M.; Schumacher, A.; Demorest, Z.; Harris, R.; Matin, A.; et al. APOBEC3 inhibits DEAD-END function to regulate microRNA activity. BMC Mol. Biol. 2013, 14, 16. [Google Scholar] [CrossRef]
- Liu, W.; Collodi, P. Zebrafish dead end possesses ATPase activity that is required for primordial germ cell development. Faseb. J. 2010, 24, 2641–2650. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Li, Y.; Yang, S.; Huang, S.; Yan, M.; Ding, Y.; Tang, W.; Lou, X.; Yin, Q.; Sun, Z.; et al. CRISPR-Cas9-mediated base-editing screening in mice identifies DND1 amino acids that are critical for primordial germ cell development. Nat. Cell Biol. 2018, 11, 1315–1325. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Tani-Matsuhana, S.; Ohkawa, Y.; Sakamoto, H.; Inoue, K. DND protein functions as a translation repressor during zebrafish embryogenesis. Biochem. Biophys. Res. Commun. 2017, 484, 235–240. [Google Scholar] [CrossRef]
- Niimi, Y.; Imai, A.; Nishimura, H.; Yui, K.; Kikuchi, A.; Koike, H.; Saga, Y.; Suzuki, A. Essential role of mouse Dead end1 in the maintenance of spermatogonia. Dev. Biol. 2019, 445, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Imai, A.; Hagiwara, Y.; Niimi, Y.; Tokumoto, T.; Saga, Y.; Suzuki, A. Mouse dead end1 acts with Nanos2 and Nanos3 to regulate testicular teratoma incidence. PLoS ONE 2020, 15, e0232047. [Google Scholar] [CrossRef]
- Cook, M.S.; Munger, S.C.; Nadeau, J.H.; Capel, B. Regulation of male germ cell cycle arrest and differentiation by DND1 is modulated by genetic background. Development 2011, 138, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.; Iacovino, M.; Mahen, E.; Kyba, M.; Matin, A. Transcripts that associate with the RNA binding protein, DEAD-END (DND1), in embryonic stem (ES) cells. BMC Mol. Biol. 2011, 12, 37. [Google Scholar] [CrossRef] [PubMed]
- Ruthig, V.A.; Friedersdorf, M.B.; Garness, J.A.; Munger, S.C.; Bunce, C.; Keene, J.D.; Capel, B. The RNA-binding protein DND1 acts Sequentially as a negative regulator of pluripotency and a positive regulator of epigenetic modifiers required for germ cell reprogramming. Development 2019. [Google Scholar] [CrossRef]
- Aguero, T.; Jin, Z.; Chorghade, S.; Kalsotra, A.; King, M.; Yang, J. Maternal Dead-end 1 promotes translation of nanos1 by binding the eIF3 complex. Development 2017, 144, 3755–3765. [Google Scholar] [PubMed]
- Zhang, Y.; Su, Y.L.; Li, L.S.; Yang, Z.; Chen, S.; Xiong, J.; Fu, X.H.; Peng, X.N. Mouse dead end 1-β interacts with c-Jun and stimulates activator protein 1 transactivation. Mol. Med. Rep. 2015, 11, 1701–1707. [Google Scholar] [CrossRef][Green Version]
- Slanchev, K.; Stebler, J.; Goudarzi, M.; Cojocaru, V.; Weidinger, G.; Raz, E. Control of dead end localization and activity-implications for the function of the protein in antagonizing miRNA function. Mech. Dev. 2009, 126, 270–277. [Google Scholar] [CrossRef]
- Taguchi, A.; Watanabe, K.; Orii, H. Intracellular localizations of the Dead End protein in Xenopus primordial germ cells. Int. J. Dev. Biol. 2014, 58, 793–798. [Google Scholar] [CrossRef]
- Bhattacharya, C.; Aggarwal, S.; Zhu, R.; Kumar, M.; Zhao, M.; Meistrich, M.L.; Matin, A. The mouse dead-end gene isoform alpha is necessary for germ cell and embryonic viability. Biochem. Biophys. Res. Commun. 2007, 355, 194–199. [Google Scholar] [CrossRef]
- Kumari, P.; Bhavesh, N. Human DND1-RRM2 forms a non-canonical domain swapped dimer. Protein Sci. 2021, 30, 1184–1195. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Thul, P.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.; et al. A subcellular map of the human proteome. Science 2017, 356, eaal3321. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef] [PubMed]
- An, Y.; Sekinaka, T.; Tando, Y.; Okamura, D.; Tanaka, K.; Ito-Matsuoka, Y.; Takehara, A.; Yaegashi, N.; Matsui, Y. Derivation of pluripotent stem cells from nascent undifferentiated teratoma. Dev. Biol. 2019, 446, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, T.; Ikeda, Y.; Kubo, I.; Suganuma, S.; Fujita, N.; Itakura, M.; Hayashi, T.; Takabayashi, S.; Katoh, H.; Ohira, Y.; et al. Identification of genomic locus responsible for experimentally induced testicular teratoma 1 (ett1) on mouse Chr 18. Mamm. Genome 2014, 25, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, T.; Fukui, M.; Inagaki, E.; Miki, K.; Takabayashi, S.; Katoh, H.; Ohira, Y.; Noguchi, M.; Tokumoto, T. Identification of Two Additional Genomic Loci Responsible for experimentally induced testicular teratoma 2 and 3 (ett2 and ett3). Zoolog. Sci. 2018, 35, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-X.; Wright, E.; Ross, J.W. Expression of RNA-binding protein DND1 and FXR1 in the porcine ovary, and during oocyte maturation and early embryo development. Mol. Reprod. Dev. 2012, 79, 541–552. [Google Scholar] [CrossRef]
- Hwang, H.; Jin, Z.; Krishnamurthy, V.; Saha, A.; Klein, P.; Garcia, B.; Mei, W.; King, M.; Zhang, K.; Yang, J. Novel functions of the ubiquitin-independent proteasome system in regulating Xenopus germline development. Development 2019, 146, dev172700. [Google Scholar] [CrossRef] [PubMed]
- Linger, R.; Dudakia, D.; Huddart, R.; Tucker, K.; Friedlander, M.; Phillips, K.A.; Hogg, D.; Jewett, M.A.; Lohynska, R.; Daugaard, G.; et al. Analysis of the DND1 gene in men with sporadic and familial testicular germ cell tumors. Genes Chromosomes Cancer 2008, 47, 247–252. [Google Scholar] [CrossRef]
- Sijmons, R.; Vos, Y.; Herkert, J.; Bos, K.; Holzik, M.; Hoekstra-Weebers, J.; Hofstra, R.; Hoekstra, H. Screening for germline DND1 mutations in testicular cancer patients. Fam. Cancer 2010, 9, 439–442. [Google Scholar] [CrossRef][Green Version]
- Milardi, D.; Grande, G.; Vincenzoni, F.; Pierconti, F.; Pontecorvi, A. Proteomics for the Identification of Biomarkers in Testicular Cancer-Review. Front. Endocrinol. 2019, 10, 462. [Google Scholar] [CrossRef] [PubMed]
- Nicholls, P.; Schorle, H.; Naqvi, S.; Hu, Y.; Fan, Y.; Carmell, M.; Dobrinski, I.; Watson, A.; Carlson, D.; Fahrenkrug, S.; et al. Mammalian germ cells are determined after PGC colonization of the nascent gonad. Proc. Natl. Acad. Sci. USA 2019, 116, 25677–25687. [Google Scholar] [CrossRef]
- Webster, N.; Maywald, R.; Benton, S.; Dawson, E.; Murillo, O.; LaPlante, E.; Milosavljevic, A.; Lanza, D.; Heaney, J. Testicular germ cell tumors arise in the absence of sex-specific differentiation. Development. 2021, 148, dev197111. [Google Scholar] [CrossRef] [PubMed]
- Voorhoeve, P.M.; le Sage, C.; Schrier, M.; Gillis, A.J.M.; Stoop, H.; Nagel, R.; Liu, Y.-P.; van Duijse, J.; Drost, J.; Griekspoor, A.; et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 2006, 124, 1169–1181. [Google Scholar] [CrossRef] [PubMed]
- Almstrup, K.; Lobo, J.; Mørup, N.; Belge, G.; Rajpert-De Meyts, E.; Looijenga, L.; Dieckmann, K. Application of miRNAs in the diagnosis and monitoring of testicular germ cell tumours. Nat. Rev. Urol. 2020, 17, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Kashem, M.; Sultana, N.; Balcar, V.J. Exposure of Rat Neural Stem Cells to Ethanol Affects Cell Numbers and Alters Expression of 28 Proteins. Neurochem. Res. 2018, 43, 1841–1854. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.; Sumer, S.; Aksoy, B.; Jacobsen, A.; Byrne, C.; Heuer, M.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Nunez, L.; Mokkapati, S.; Yu, C.; Deng, J.; Behringer, R.; Matin, A. Generation of a novel mouse strain with conditional, cell-type specific, expression of DND1. Genesis 2019, 57, e23335. [Google Scholar] [CrossRef]
- Ruthig, V.; Yokonishi, T.; Friedersdorf, M.; Batchvarova, S.; Hardy, J.; Garness, J.; Keene, J.; Capel, B. A transgenic DND1GFP fusion allele reports in vivo expression and RNA-binding targets in undifferentiated mouse germ cells. Biol. Reprod. 2021, 104, 861–874. [Google Scholar] [CrossRef]
- Bell, R.; Kimpel, M.; McClintick, J.; Strother, W.; Carr, L.; Liang, T.; Rodd, Z.; Mayfield, D.; Edenberg, H.; McBride, W. Gene expression changes in the nucleus accumbens of alcoholpreferring rats following chronic ethanol consumption. Pharmacol. Biochem. Behav. 2009, 94, 131–147. [Google Scholar] [CrossRef] [PubMed]

| Tumor Type | Endogenous DND1 Status in Human Cell or Tissue Samples | Phenotypes Caused by Experimental Alteration of DND1 | Mechanism of DND1 Function | Reference |
|---|---|---|---|---|
| Breast cancer | Lower expression of DND1 levels in breast cancer tissue compared to normal. | Knockdown of DND1 in MCF-7 cells decreased BIM expression and inhibited apoptosis. | DND1 increases expression of BIM by blocking miR-221 from BIM-3′UTR. | [17] |
| Hepatocellular carcinoma (HCC) | DND1 mRNA and protein levels significantly decreased in HCC sphere cells. | DND1 overexpression inhibited spheroid formation; suppressed HCC cancer cell stemness; inhibited epithelial-mesenchymal transition; increased sensitivity of HCC cells to sorafenib. | DND1 binds to LATS2 3′-UTR, elevating LATS2 level and YAP phosphorylation and retention in the cytoplasm, therefore diminishing transcriptional activity of YAP. | [18] |
| Intestinal polyposis | N/A | Apc+/MinDnd1+/Ter mice had higher polyp numbers compared to Apc+/MinDnd1+/+ mice. | N/A | [14] |
| Colorectal cancer (CRC) | DND1 expression significantly up-regulated in CRC cell lines. | Silencing DND1 reduced SW48 cell line viability and overexpression of DND1 promoted cell proliferation. | DND1 is the potential target of miR-24 in SW48 cells and involved in miR-24 mediated inhibitory effects on cell proliferation. | [19] |
| Tongue squamous cell carcinoma (TSCC) | Reduced expression of DND1 in TSCC cells and tissues. | DND1 knockdown in TSCC cell lines enhanced cell proliferation and reduced apoptosis. Enhanced DND1 expression reduced cell proliferation and increased apoptosis. | DND1 is a direct target of miR-24. miR-24 suppressed DND1, leading to reduced CDKN1B. | [13] |
| Acute myeloid leukemia (AML) | Lower DND1 mRNA levels in AML blasts and CD34+ progenitor cells. | Inhibition of both RBM38 and DND1 mRNAs significantly attenuated NB4 differentiation and resulted in decreased p21(CIP1) mRNA. | Activity of RBM38 and DND1 during neutrophil differentiation antagonize the activity of oncomiRs to protect mRNAs, for example p21CIP1 that are important for myeloid differentiation. | [16] |
| Skin cancer | Reduced expression of DND1 mRNA in transformed HaCaT cells and loss of DND1 mRNA and protein in tumors from transformed HaCaT cells. | Expression of DND1 in transformed HaCaT cells interfered with miR-21-mediated repression of MSH2; Knockdown of DND1 reduced MSH2 RNA, an effect further enhanced by miR-21. | DND1, which decreases sensitivity of MSH2 to miR-21, is down-regulated during tumorigenesis therefore increasing the effectiveness of miR-21 in tumors. | [15] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Godavarthi, J.D.; Williams-Villalobo, A.; Polk, S.; Matin, A. The Role of DND1 in Cancers. Cancers 2021, 13, 3679. https://doi.org/10.3390/cancers13153679
Zhang Y, Godavarthi JD, Williams-Villalobo A, Polk S, Matin A. The Role of DND1 in Cancers. Cancers. 2021; 13(15):3679. https://doi.org/10.3390/cancers13153679
Chicago/Turabian StyleZhang, Yun, Jyotsna D. Godavarthi, Abie Williams-Villalobo, Shahrazad Polk, and Angabin Matin. 2021. "The Role of DND1 in Cancers" Cancers 13, no. 15: 3679. https://doi.org/10.3390/cancers13153679
APA StyleZhang, Y., Godavarthi, J. D., Williams-Villalobo, A., Polk, S., & Matin, A. (2021). The Role of DND1 in Cancers. Cancers, 13(15), 3679. https://doi.org/10.3390/cancers13153679







