Clinical Significance of Telomerase Reverse-Transcriptase Promoter Mutations in Hepatocellular Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Tissue Samples
2.2. DNA Extraction
2.3. TERTp Mutation Analysis by End-Point PCR and Sanger Sequencing
2.4. TERTp Mutation Analysis by ddPCR
2.5. Statistical Analysis
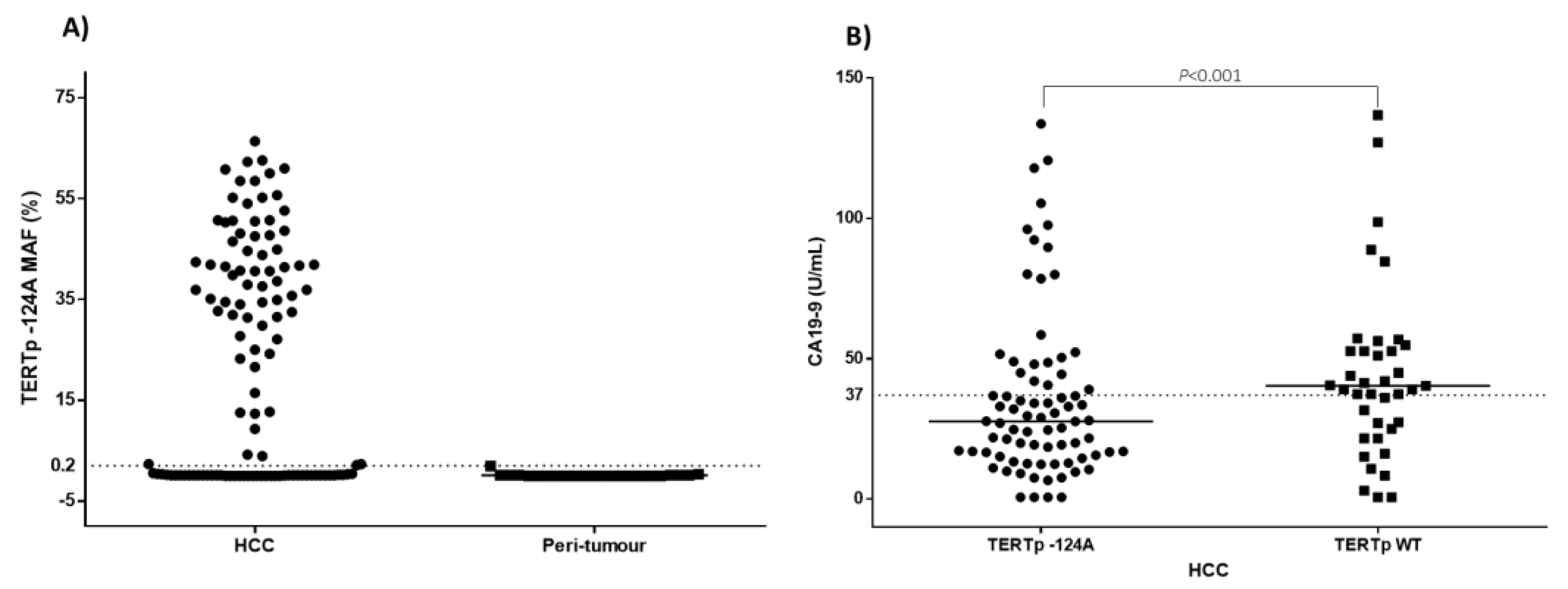
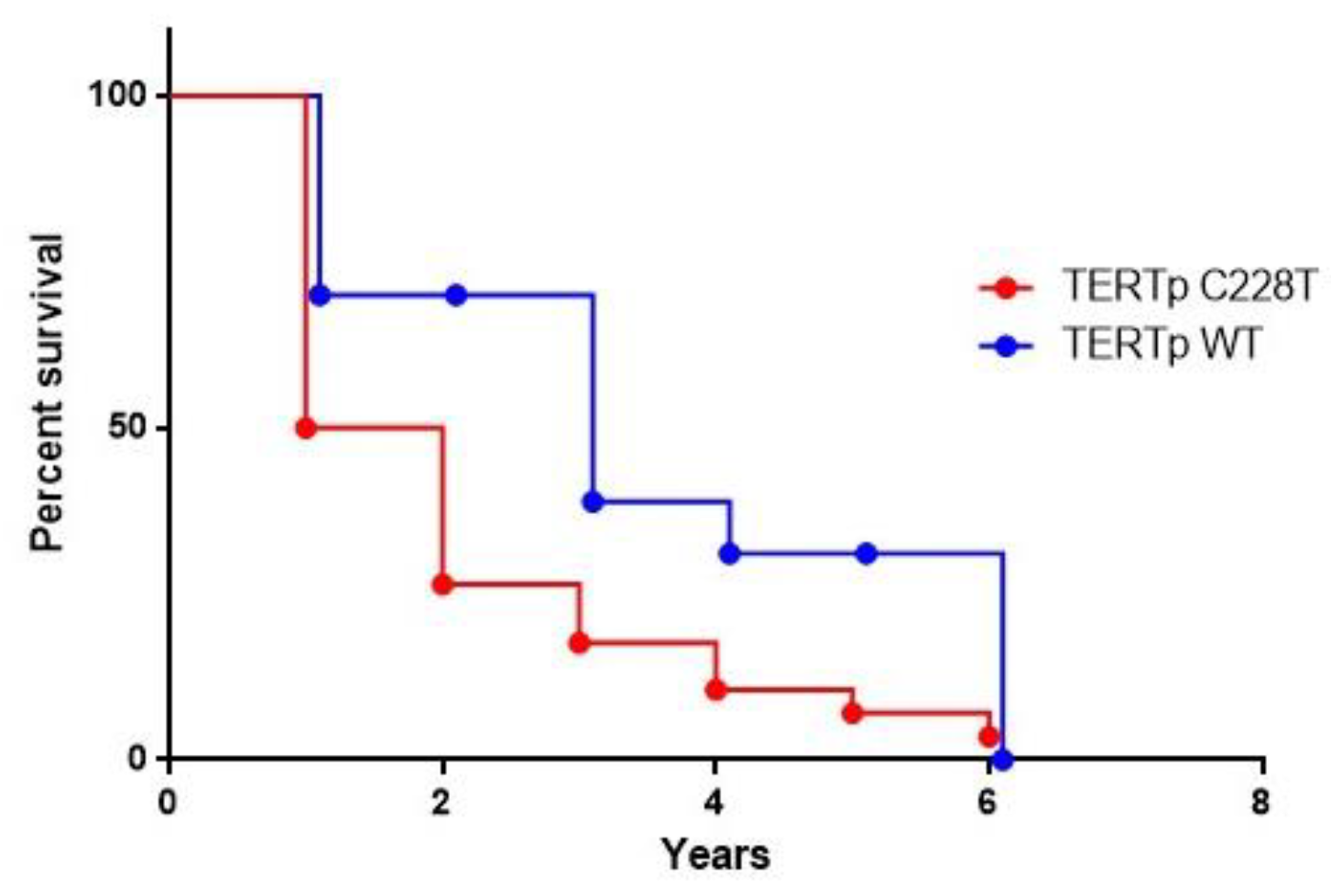
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Kulik, L.; El-Serag, H.B. Epidemiology and Management of Hepatocellular Carcinoma. Gastroenterology 2019, 156, 477–491. [Google Scholar] [CrossRef] [PubMed]
- McGlynn, K.A.; Petrick, J.L.; El-Serag, H.B. Epidemiology of Hepatocellular Carcinoma. Hepatology 2021, 73 (Suppl. 1), 4–13. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2021, 7, 6. [Google Scholar] [CrossRef]
- Balogh, J.; Victor, D., 3rd; Asham, E.H.; Burroughs, S.G.; Boktour, M.; Saharia, A.; Li, X.; Ghobrial, R.M.; Monsour, H.P., Jr. Hepatocellular carcinoma: A review. J. Hepatocell. Carcinoma 2016, 3, 41–53. [Google Scholar] [CrossRef] [Green Version]
- Banales, J.M.; Iñarrairaegui, M.; Arbelaiz, A.; Milkiewicz, P.; Muntané, J.; Muñoz-Bellvis, L.; La Casta, A.; Gonzalez, L.M.; Arretxe, E.; Alonso, C.; et al. Serum Metabolites as Diagnostic Biomarkers for Cholangiocarcinoma, Hepatocellular Carcinoma, and Primary Sclerosing Cholangitis. Hepatology 2019, 70, 547–562. [Google Scholar] [CrossRef] [Green Version]
- Mueller, C.; Waldburger, N.; Stampfl, U.; Kauczor, H.U.; Schirmacher, P.; Sommer, C.M.; Longerich, T. Non-invasive diagnosis of hepatocellular carcinoma revisited. Gut 2018, 67, 991–993. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2018, 69, 182–236. [Google Scholar] [CrossRef] [Green Version]
- Kudo, M.; Finn, R.S.; Qin, S.; Han, K.H.; Ikeda, K.; Piscaglia, F.; Baron, A.; Park, J.W.; Han, G.; Jassem, J.; et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: A randomised phase 3 non-inferiority trial. Lancet 2018, 391, 1163–1173. [Google Scholar] [CrossRef] [Green Version]
- Llovet, J.M.; Ricci, S.; Mazzaferro, V.; Hilgard, P.; Gane, E.; Blanc, J.F.; de Oliveira, A.C.; Santoro, A.; Raoul, J.L.; Forner, A.; et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008, 359, 378–390. [Google Scholar] [CrossRef]
- Kelley, R.K.; Mollon, P.; Blanc, J.F.; Daniele, B.; Yau, T.; Cheng, A.L.; Valcheva, V.; Marteau, F.; Guerra, I.; Abou-Alfa, G.K. Comparative Efficacy of Cabozantinib and Regorafenib for Advanced Hepatocellular Carcinoma. Adv. Ther. 2020, 37, 2678–2695. [Google Scholar] [CrossRef]
- Finn, R.S.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.Y.; Kudo, M.; Breder, V.; Merle, P.; Kaseb, A.O.; et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N. Engl. J. Med. 2020, 382, 1894–1905. [Google Scholar] [CrossRef]
- El-Khoueiry, A.B.; Sangro, B.; Yau, T.; Crocenzi, T.S.; Kudo, M.; Hsu, C.; Kim, T.Y.; Choo, S.P.; Trojan, J.; Welling, T.H.R.; et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): An open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet 2017, 389, 2492–2502. [Google Scholar] [CrossRef]
- Zhang, Y.; Shen, Y.; Cao, B.; Yan, A.; Ji, H. Elevated expression levels of androgen receptors and matrix metalloproteinase-2 and -9 in 30 cases of hepatocellular carcinoma compared with adjacent tissues as predictors of cancer invasion and staging. Exp. Ther. Med. 2015, 9, 905–908. [Google Scholar] [CrossRef] [Green Version]
- Takeda, H.; Takai, A.; Inuzuka, T.; Marusawa, H. Genetic basis of hepatitis virus-associated hepatocellular carcinoma: Linkage between infection, inflammation, and tumorigenesis. J. Gastroenterol. 2017, 52, 26–38. [Google Scholar] [CrossRef] [Green Version]
- De Re, V.; Tornesello, M.L.; De Zorzi, M.; Caggiari, L.; Pezzuto, F.; Leone, P.; Racanelli, V.; Lauletta, G.; Zanussi, S.; Repetto, O.; et al. PDCD1 and IFNL4 genetic variants and risk of developing hepatitis C virus-related diseases. Liver Int. 2021, 41, 133–149. [Google Scholar] [CrossRef]
- Schulze, K.; Imbeaud, S.; Letouzé, E.; Alexandrov, L.B.; Calderaro, J.; Rebouissou, S.; Couchy, G.; Meiller, C.; Shinde, J.; Soysouvanh, F.; et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet. 2015, 47, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Tornesello, M.L.; Buonaguro, L.; Tatangelo, F.; Botti, G.; Izzo, F.; Buonaguro, F.M. Mutations in TP53, CTNNB1 and PIK3CA genes in hepatocellular carcinoma associated with hepatitis B and hepatitis C virus infections. Genomics 2013, 102, 74–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shibata, T. Genomic landscape of hepatocarcinogenesis. J. Hum. Genet. 2021. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Sun, M.; Wang, M.; Feng, M.; Yang, F.; Li, L.; Zhao, J.; Chang, C.; Dong, H.; Xie, T.; et al. Dysregulation of Wnt/β-catenin signaling by protein kinases in hepatocellular carcinoma and its therapeutic application. Cancer Sci. 2021, 112, 1695–1706. [Google Scholar] [CrossRef] [PubMed]
- Horn, S.; Figl, A.; Rachakonda, P.S.; Fischer, C.; Sucker, A.; Gast, A.; Kadel, S.; Moll, I.; Nagore, E.; Hemminki, K.; et al. TERT promoter mutations in familial and sporadic melanoma. Science 2013, 339, 959–961. [Google Scholar] [CrossRef] [Green Version]
- Huang, F.W.; Hodis, E.; Xu, M.J.; Kryukov, G.V.; Chin, L.; Garraway, L.A. Highly recurrent TERT promoter mutations in human melanoma. Science 2013, 339, 957–959. [Google Scholar] [CrossRef] [Green Version]
- Killela, P.J.; Reitman, Z.J.; Jiao, Y.; Bettegowda, C.; Agrawal, N.; Diaz, L.A., Jr.; Friedman, A.H.; Friedman, H.; Gallia, G.L.; Giovanella, B.C.; et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl. Acad. Sci. USA 2013, 110, 6021–6026. [Google Scholar] [CrossRef] [Green Version]
- Nault, J.C.; Ningarhari, M.; Rebouissou, S.; Zucman-Rossi, J. The role of telomeres and telomerase in cirrhosis and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 544–558. [Google Scholar] [CrossRef]
- In der Stroth, L.; Tharehalli, U.; Gunes, C.; Lechel, A. Telomeres and Telomerase in the Development of Liver Cancer. Cancers 2020, 12, 2048. [Google Scholar] [CrossRef]
- Lin, S.; Nascimento, E.M.; Gajera, C.R.; Chen, L.; Neuhöfer, P.; Garbuzov, A.; Wang, S.; Artandi, S.E. Distributed hepatocytes expressing telomerase repopulate the liver in homeostasis and injury. Nature 2018, 556, 244–248. [Google Scholar] [CrossRef]
- Heidenreich, B.; Kumar, R. TERT promoter mutations in telomere biology. Mutat. Res. 2017, 771, 15–31. [Google Scholar] [CrossRef]
- Müller, M.; Bird, T.G.; Nault, J.C. The landscape of gene mutations in cirrhosis and hepatocellular carcinoma. J. Hepatol. 2020, 72, 990–1002. [Google Scholar] [CrossRef] [Green Version]
- Nault, J.C.; Mallet, M.; Pilati, C.; Calderaro, J.; Bioulac-Sage, P.; Laurent, C.; Laurent, A.; Cherqui, D.; Balabaud, C.; Zucman-Rossi, J. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat. Commun. 2013, 4, 2218. [Google Scholar] [CrossRef] [Green Version]
- Totoki, Y.; Tatsuno, K.; Covington, K.R.; Ueda, H.; Creighton, C.J.; Kato, M.; Tsuji, S.; Donehower, L.A.; Slagle, B.L.; Nakamura, H.; et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat. Genet. 2014, 46, 1267–1273. [Google Scholar] [CrossRef]
- Hindson, C.M.; Chevillet, J.R.; Briggs, H.A.; Gallichotte, E.N.; Ruf, I.K.; Hindson, B.J.; Vessella, R.L.; Tewari, M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 2013, 10, 1003–1005. [Google Scholar] [CrossRef]
- Sykes, P.J.; Neoh, S.H.; Brisco, M.J.; Hughes, E.; Condon, J.; Morley, A.A. Quantitation of targets for PCR by use of limiting dilution. Biotechniques 1992, 13, 444–449. [Google Scholar]
- Pinheiro, L.B.; Coleman, V.A.; Hindson, C.M.; Herrmann, J.; Hindson, B.J.; Bhat, S.; Emslie, K.R. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal. Chem. 2012, 84, 1003–1011. [Google Scholar] [CrossRef]
- Denis, J.A.; Nectoux, J.; Lamy, P.J.; Rouillac Le Sciellour, C.; Guermouche, H.; Alary, A.S.; Kosmider, O.; Sarafan-Vasseur, N.; Jovelet, C.; Busser, B.; et al. Development of digital PCR molecular tests for clinical practice: Principles, practical implementation and recommendations. Ann. Biol. Clin. 2018, 76, 505–523. [Google Scholar] [CrossRef]
- Taylor, S.C.; Laperriere, G.; Germain, H. Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: From variable nonsense to publication quality data. Sci. Rep. 2017, 7, 2409. [Google Scholar] [CrossRef] [Green Version]
- Pezzuto, F.; Izzo, F.; Buonaguro, L.; Annunziata, C.; Tatangelo, F.; Botti, G.; Buonaguro, F.M.; Tornesello, M.L. Tumor specific mutations in TERT promoter and CTNNB1 gene in hepatitis B and hepatitis C related hepatocellular carcinoma. Oncotarget 2016, 7, 54253–54262. [Google Scholar] [CrossRef] [Green Version]
- Edmondson, H.A.; Steiner, P.E. Primary carcinoma of the liver: A study of 100 cases among 48,900 necropsies. Cancer 1954, 7, 462–503. [Google Scholar] [CrossRef]
- Huggett, J.F. The Digital MIQE Guidelines Update: Minimum Information for Publication of Quantitative Digital PCR Experiments for 2020. Clin. Chem. 2020, 66, 1012–1029. [Google Scholar] [CrossRef]
- Dong, R.; Zheng, S.; Dong, K. TERT promoter mutation during development of hepatoblastoma to hepatocellular carcinoma. J. Hepatol. 2015, 62, 497. [Google Scholar] [CrossRef] [Green Version]
- Nault, J.C.; Zucman-Rossi, J. TERT promoter mutations in primary liver tumors. Clin. Res. Hepatol. Gastroenterol. 2016, 40, 9–14. [Google Scholar] [CrossRef]
- Pezzuto, F.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. Frequency and geographic distribution of TERT promoter mutations in primary hepatocellular carcinoma. Infect. Agent Cancer 2017, 12, 27. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Wang, R.L.; Liu, J.J.; Zhou, J.; Li, X.; Hu, W.W.; Jiang, W.J.; Hao, N.B. The prognostic significance of hTERT overexpression in cancers: A systematic review and meta-analysis. Medicine 2018, 97, e11794. [Google Scholar] [CrossRef]
- Ningarhari, M.; Caruso, S.; Hirsch, T.Z.; Bayard, Q.; Franconi, A.; Vedie, A.L.; Noblet, B.; Blanc, J.F.; Amaddeo, G.; Ganne, N.; et al. Telomere length is key to hepatocellular carcinoma diversity and telomerase addiction is an actionable therapeutic target. J. Hepatol. 2021, 74, 1155–1166. [Google Scholar] [CrossRef]
- Lee, T.; Teng, T.Z.J.; Shelat, V.G. Carbohydrate antigen 19-9—Tumor marker: Past, present, and future. World J. Gastrointest. Surg. 2020, 12, 468–490. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.C.; Goyal, A.; Iuga, A.; Krishnamoorthy, S.; Lee, V.; Verna, E.C.; Wang, S.; Chen, F.N.; Rodriguez, R.; Emond, J.; et al. Elevated CA19-9 Is Associated With Increased Mortality In A Prospective Cohort Of Hepatocellular Carcinoma Patients. Clin. Transl. Gastroenterol. 2015, 6, e74. [Google Scholar] [CrossRef]
- Liu, R.; Zhang, T.; Zhu, G.; Xing, M. Regulation of mutant TERT by BRAF V600E/MAP kinase pathway through FOS/GABP in human cancer. Nat. Commun. 2018, 9, 579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, J.; Liu, R.; Zhu, G.; Umbricht, C.B.; Xing, M. TERT promoter mutation determines apoptotic and therapeutic responses of BRAF-mutant cancers to BRAF and MEK inhibitors: Achilles Heel. Proc. Natl. Acad. Sci. USA 2020, 117, 15846–15851. [Google Scholar] [CrossRef] [PubMed]
- Litchfield, K.; Stanislaw, S.; Spain, L.; Gallegos, L.L.; Rowan, A.; Schnidrig, D.; Rosenbaum, H.; Harle, A.; Au, L.; Hill, S.M.; et al. Representative Sequencing: Unbiased Sampling of Solid Tumor Tissue. Cell Rep. 2020, 31, 107550. [Google Scholar] [CrossRef]

| Sample ID | Sanger TERTp | ddPCR TERTp | MAF (%) | Number of Alleles Screened | Sample ID | Sanger TERTp | ddPCR TERTp | MAF (%) | Number of Alleles Screened |
|---|---|---|---|---|---|---|---|---|---|
| 57 | WT | WT | - | 3084 | 307 | −124A | −124A | 41.71 | 4057 |
| 60 | WT | WT | - | 1934 | 314 | WT | WT | - | 4620 |
| 66 | −124A | −124A | 47.48 | 7736 | 315 | −124A | −124A | 41.42 | 10285 |
| 78 | WT | WT | - | 8560 | 324 | −124A | −124A | 43.76 | 4991 |
| 86 | −124A | −124A | 39.82 | 1871 | 334 | WT | −124A | 24.98 | 8406 |
| 88 | WT | −124A | 0.36 | 4876 | 338 | −124A | WT | - | 5279 |
| 117 | −124A | −124A | 54.05 | 8145 | 339 | −124A | −124A | 38.62 | 6007 |
| 126 | WT | −124A | 0.23 | 2645 | 340 | −124A | −124A | 27.70 | 6112 |
| 128 | WT | −124A | 2.32 | 1813 | 342 | −124A | −124A | 35.67 | 7580 |
| 129 | −124A | −124A | 44.57 | 3812 | 353 | WT | WT | - | 4207 |
| 132 | −124A | −124A | 12.45 | 1935 | 354 | −124A | −124A | 47.74 | 5754 |
| 134 | WT | −124A | 2.26 | 5493 | 355 | WT | WT | - | 6078 |
| 137 | WT | −124A | 0.35 | 1733 | 361 | WT | −124A | 52.61 | 6441 |
| 138 | WT | WT | - | 263 | 365 | −124A | −124A | 29.85 | 4798 |
| 144 | WT | −124A | 4.20 | 940 | 366 | −124A | −124A | 31.47 | 4894 |
| 146 | −124A | −124A | 12.70 | 4024 | 369 | WT | WT | - | 5635 |
| 152 | −124A | WT | - | 1220 | 370 | −124A | −124A | 37.65 | 8123 |
| 153 | WT | WT | - | 5077 | 372 | −124A | −124A | 31.92 | 4533 |
| 157 | WT | −124A | 0.32 | 2479 | 373 | −124A | −124A | 58.54 | 10,925 |
| 158 | WT | −124A | 12.28 | 477 | 374 | −124A | −124A | 35.13 | 4535 |
| 164 | WT | −124A | 9.26 | 1614 | 375 | −124A | −124A | 60.81 | 5586 |
| 167 | −124A | −124A | 44.94 | 6662 | 377 | WT | WT | - | 4255 |
| 169 | WT | −124A | 0.54 | 2508 | 382 | −124A | −124A | 50.30 | 6402 |
| 170 | WT | WT | - | 1857 | 386 | −124A | −124A | 34.90 | 4433 |
| 172 | −124A | −124A | 16.43 | 2258 | 387 | WT | WT | - | 6664 |
| 174 | WT | WT | - | 773 | 391 | WT | WT | - | 3067 |
| 175 | WT | −124A | 36.88 | 1214 | 392 | −124A | −124A | 36.87 | 3699 |
| 177 | WT | WT | - | 2357 | 393 | −124A | −124A | 50.49 | 4710 |
| 179 | −124A | −124A | 21.58 | 5584 | 394 | WT | WT | - | 7026 |
| 180 | −124A | −124A | 42.45 | 940 | 396 | −124A | −124A | 62.57 | 4133 |
| 181 | −124A | −124A | 41.86 | 6366 | 398 | WT | WT | - | 5286 |
| 186 | −124A | −124A | 0.25 | 3257 | 399 | −124A | −124A | 35.37 | 4241 |
| 187 | −124A | −124A | 40.60 | 298 | 402 | −124A | −124A | 50.72 | 4237 |
| 189 | −124A | −124A | 60.02 | 4650 | 403 | −124A | −124A | 48.65 | 10,681 |
| 190 | −124A | −124A | 62.28 | 920 | 404 | −124A | −124A | 52.82 | 4806 |
| 193 | −124A | −124A | 34.05 | 1536 | 406 | WT | WT | - | 3916 |
| 195 | −124A | −124A | 32.68 | 4932 | 407 | WT | −124A | 0.45 | 5839 |
| 197 | −124A | −124A | 55.21 | 3735 | 408 | −124A | −124A | 31.41 | 7501 |
| 199 | −124A | −124A | 55.70 | 4501 | 409 | −124A | −124A | 41.49 | 3155 |
| 203 | WT | WT | - | 1641 | 411 | WT | WT | - | 4043 |
| 206 | WT | WT | - | 3139 | 412 | WT | WT | - | 6960 |
| 208 | −124A | −124A | 50.61 | 4545 | 414 | −124A | WT | - | 5194 |
| 211 | −124A | −124A | 41.87 | 8866 | 415 | WT | WT | - | 3871 |
| 212 | WT | WT | - | 2403 | 416 | WT | WT | - | 7303 |
| 221 | −124A | −124A | 58.54 | 6917 | 417 | WT | WT | - | 4082 |
| 226 | WT | WT | - | 5609 | 418 | −124A | −124A | 55.15 | 7318 |
| 229 | WT | WT | - | 5934 | 421 | WT | WT | - | 5523 |
| 233 | WT | WT | - | 3502 | 422 | −124A | −124A | 37.95 | 5658 |
| 235 | −124A | −124A | 27.10 | 5022 | 423 | −124A | −124A | 66.37 | 11,773 |
| 236 | WT | WT | - | 5060 | 424 | WT | WT | - | 6444 |
| 241 | WT | WT | - | 4670 | 425 | −124A | −124A | 48.11 | 6117 |
| 242 | WT | WT | - | 1217 | 426 | WT | −124A | 40.73 | 5949 |
| 243 | −124A | −124A | 34.54 | 4100 | 427 | −124A | −124A | 40.57 | 9056 |
| 245 | WT | WT | - | 4708 | 428 | WT | WT | - | 6447 |
| 247 | −124A | −124A | 16.22 | 2226 | 429 | WT | −124A | 2.01 | 5240 |
| 248 | −124A | −124A | 0.41 | 2089 | 430 | WT | WT | - | 8393 |
| 259 | −124A | −124A | 3.37 | 4388 | 431 | WT | WT | - | 6915 |
| 273 | −124A | −124A | 47.21 | 5484 | 432 | WT | WT | - | 15,202 |
| 282 | −124A | −124A | 32.43 | 3170 | 433 | −124A | −124A | 50.68 | 7675 |
| 287 | WT | −124A | 2.95 | 4738 | 434 | WT | WT | - | 5818 |
| 297 | WT | WT | - | 3814 |
| Sanger vs. ddPCR | Sanger Sequencing | Performance | |||
|---|---|---|---|---|---|
| ddPCR | TERTp −124A | TERTp wildtype | Total | Sensitivity | 95.2% |
| TERTp −124A | 60 | 17 | 77 | Specificity | 70.7% |
| TERTp wildtype | 3 | 41 | 44 | Concordance | 83.5% |
| Total | 63 | 58 | 121 | ||
| Characteristics | HCC | TERTp WT (n = 35) | TERTp −124A (n = 79) | p Value * |
|---|---|---|---|---|
| (n = 114) | ||||
| Age (years), n (%) | 0.116 | |||
| ≤65 years | 37 (32.5) | 15 (42.9) | 22 (27.8) | |
| >65 years | 77 (67.5) | 20 (57.1) | 57 (72.2) | |
| Gender, n (%) | 0.259 | |||
| Male | 86 (75.4) | 24 (68.6) | 62 (78.5) | |
| Female | 28 (24.6) | 11 (31.4) | 17 (21.5) | |
| Etiology, n (%) | 0.387 | |||
| HBV+ | 10 (8.8) | 4 (11.4) | 6 (7.6) | |
| HCV+ | 99 (86.8) | 27 (77.1) | 72 (91.1) | |
| No virus | 5 (4.4) | 4 (11.4) | 1 (1.3) | |
| AFP (ng/mL), n (%) | 0.338 | |||
| ≤20 ng/mL | 50 (43.9) | 13 (37.1) | 37 (46.8) | |
| >20 ng/mL | 64 (56.1) | 22(62.9) | 42 (53.2) | |
| CA19-9 (U/)mL, n (%) | 0.0105 | |||
| ≤37 U/mL | 69 (60.5) | 15 (42.9) | 54 (68.3) | |
| >37 U/mL | 45 (39.5) | 20 (57.1) | 25 (31.7) | |
| CEA (ng/mL), n (%) | 0.124 | |||
| ≤3 ng/mL | 56 (49.1) | 21 (60.0) | 35 (44.3) | |
| >3 ng/mL | 58 (50.9) | 14 (40.0) | 44 (55.7) | |
| ALT (U/l), n (%) | 0.995 | |||
| ≤33 U/l | 13 (11.4) | 4 (11.4) | 9 (11.4) | |
| >33 U/l | 101 (88.6) | 31 (88.6) | 70 (88.6) | |
| AST (U/l), n (%) | 0.527 | |||
| ≤32 U/l | 16 (14.0) | 6 (17.4) | 10 (12.7) | |
| >32 U/l | 98 (86.0) | 29 (82.9) | 69 (87.3) | |
| GGT (U/l), n (%) | 0.928 | |||
| ≤40 U/l | 19 (16.7) | 6 (17.1) | 13 (16.5) | |
| >40 U/l | 95 (83.3) | 29 (82.9) | 66 (83.5) | |
| Tumour size, n (%) | 0.048 | |||
| ≤4 cm | 54 (47.4) | 12 (34.3) | 42 (53.2) | |
| >4 cm | 60 (52.6) | 23 (65.7) | 37 (46.8) | |
| Tumour nodules, n (%) | 0.045 | |||
| Single | 76 (66.7) | 28 (80.0) | 48 (60.8) | |
| Multiple | 38 (33.3) | 7 (20.0) | 31 (39.2) | |
| Tumour differentiation, n (%) | 0.815 | |||
| G1 | 1 (0.9) | 0 | 1 (1.3) | |
| G2 | 109 (95.6) | 34 (97.1) | 75 (94.9) | |
| G3 | 3 (2.6) | 1 (2.9) | 2 (2.5) | |
| G4 | 1 (0.9) | 0 | 1 (1.3) | |
| Child pugh, n(%) | 0.593 | |||
| A | 91 (79.8) | 29 (82.9) | 62 (78.5) | |
| B | 23 (20.2) | 6 (17.1) | 17 (21.5) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pezzuto, F.; Izzo, F.; De Luca, P.; Biffali, E.; Buonaguro, L.; Tatangelo, F.; Buonaguro, F.M.; Tornesello, M.L. Clinical Significance of Telomerase Reverse-Transcriptase Promoter Mutations in Hepatocellular Carcinoma. Cancers 2021, 13, 3771. https://doi.org/10.3390/cancers13153771
Pezzuto F, Izzo F, De Luca P, Biffali E, Buonaguro L, Tatangelo F, Buonaguro FM, Tornesello ML. Clinical Significance of Telomerase Reverse-Transcriptase Promoter Mutations in Hepatocellular Carcinoma. Cancers. 2021; 13(15):3771. https://doi.org/10.3390/cancers13153771
Chicago/Turabian StylePezzuto, Francesca, Francesco Izzo, Pasquale De Luca, Elio Biffali, Luigi Buonaguro, Fabiana Tatangelo, Franco Maria Buonaguro, and Maria Lina Tornesello. 2021. "Clinical Significance of Telomerase Reverse-Transcriptase Promoter Mutations in Hepatocellular Carcinoma" Cancers 13, no. 15: 3771. https://doi.org/10.3390/cancers13153771








