Deep Learning for Automatic Subclassification of Gastric Carcinoma Using Whole-Slide Histopathology Images
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Cohort
2.2. Pathologic Diagnosis
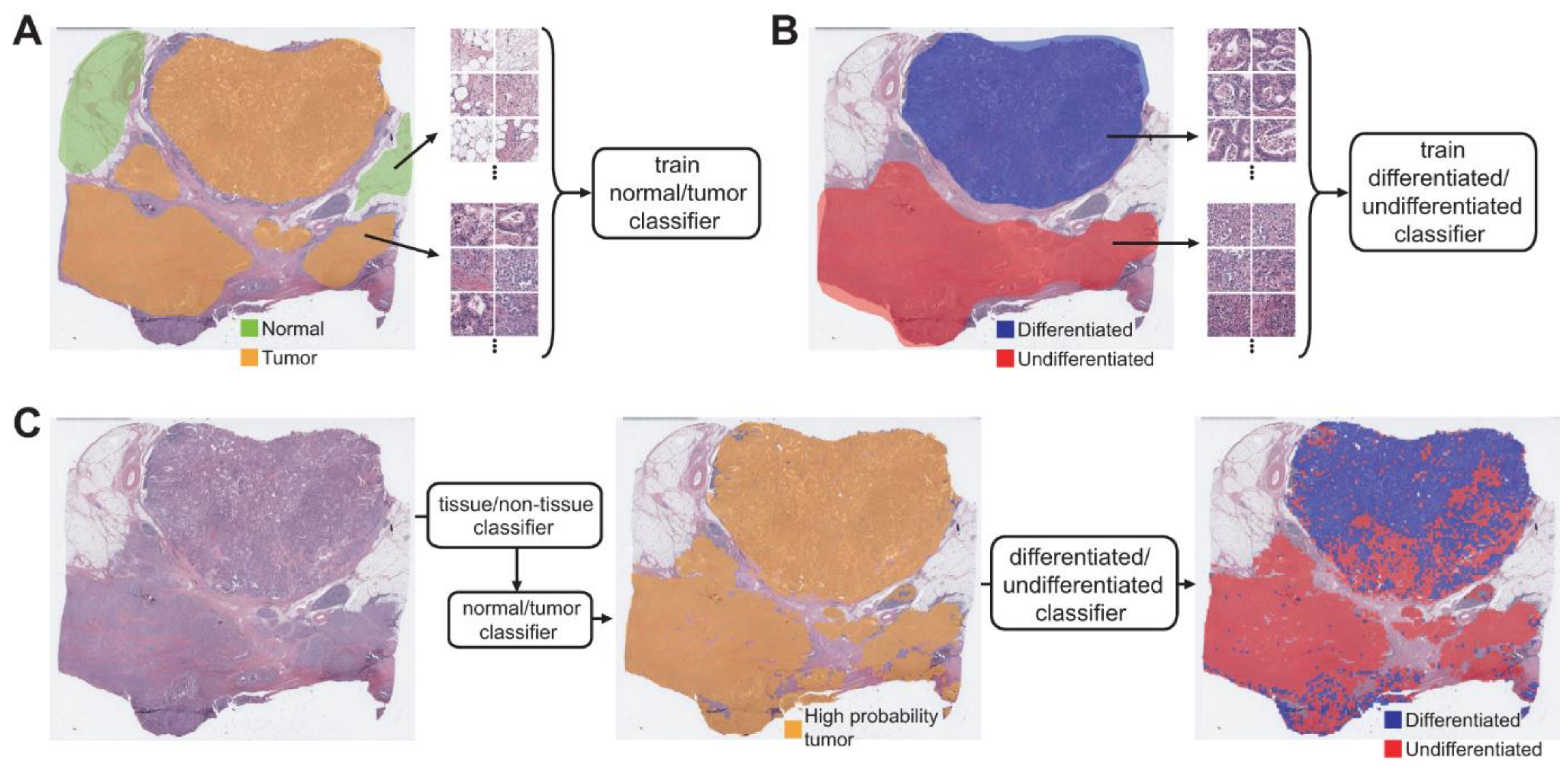
2.3. Deep Learning Model
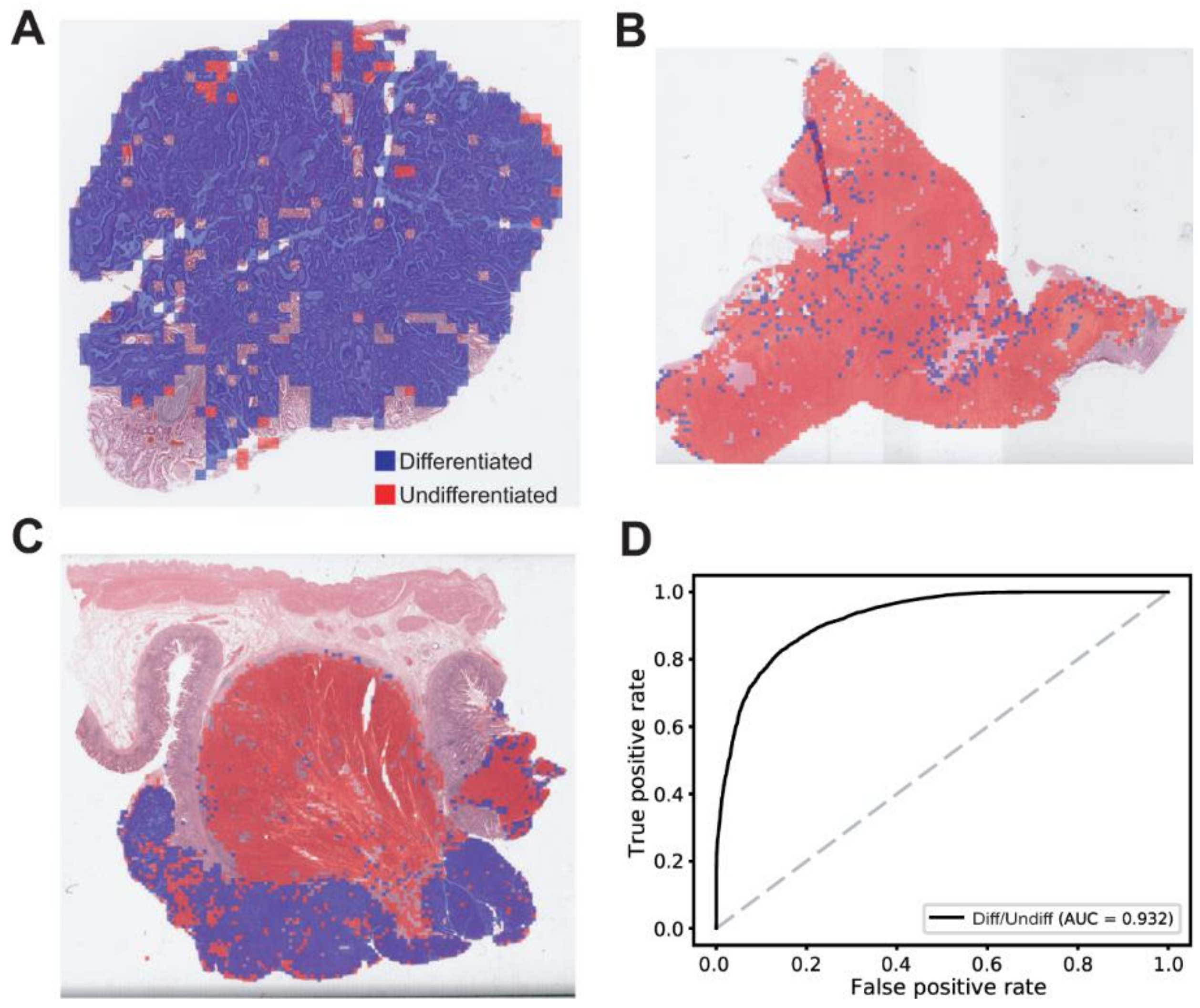
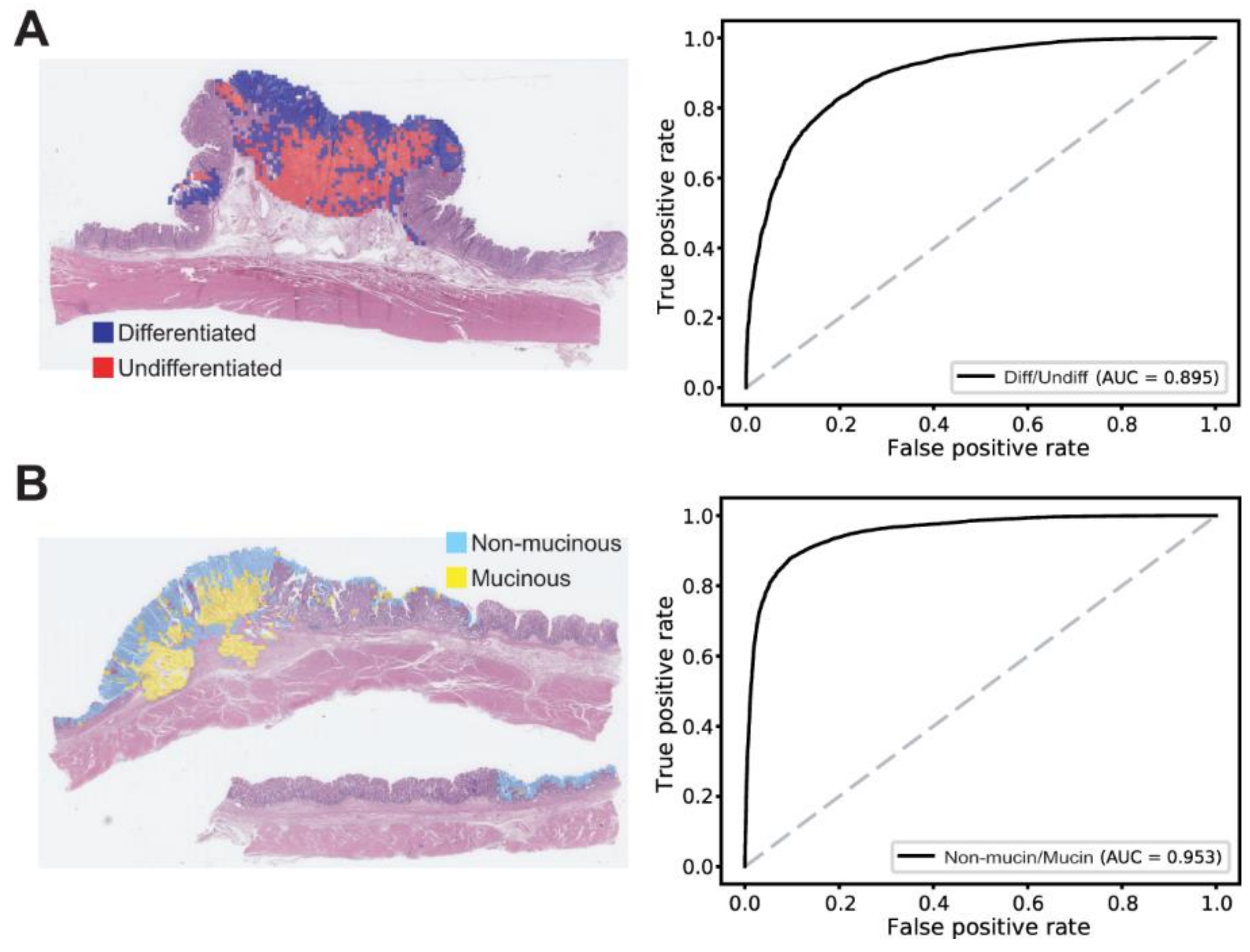
2.4. Statistics
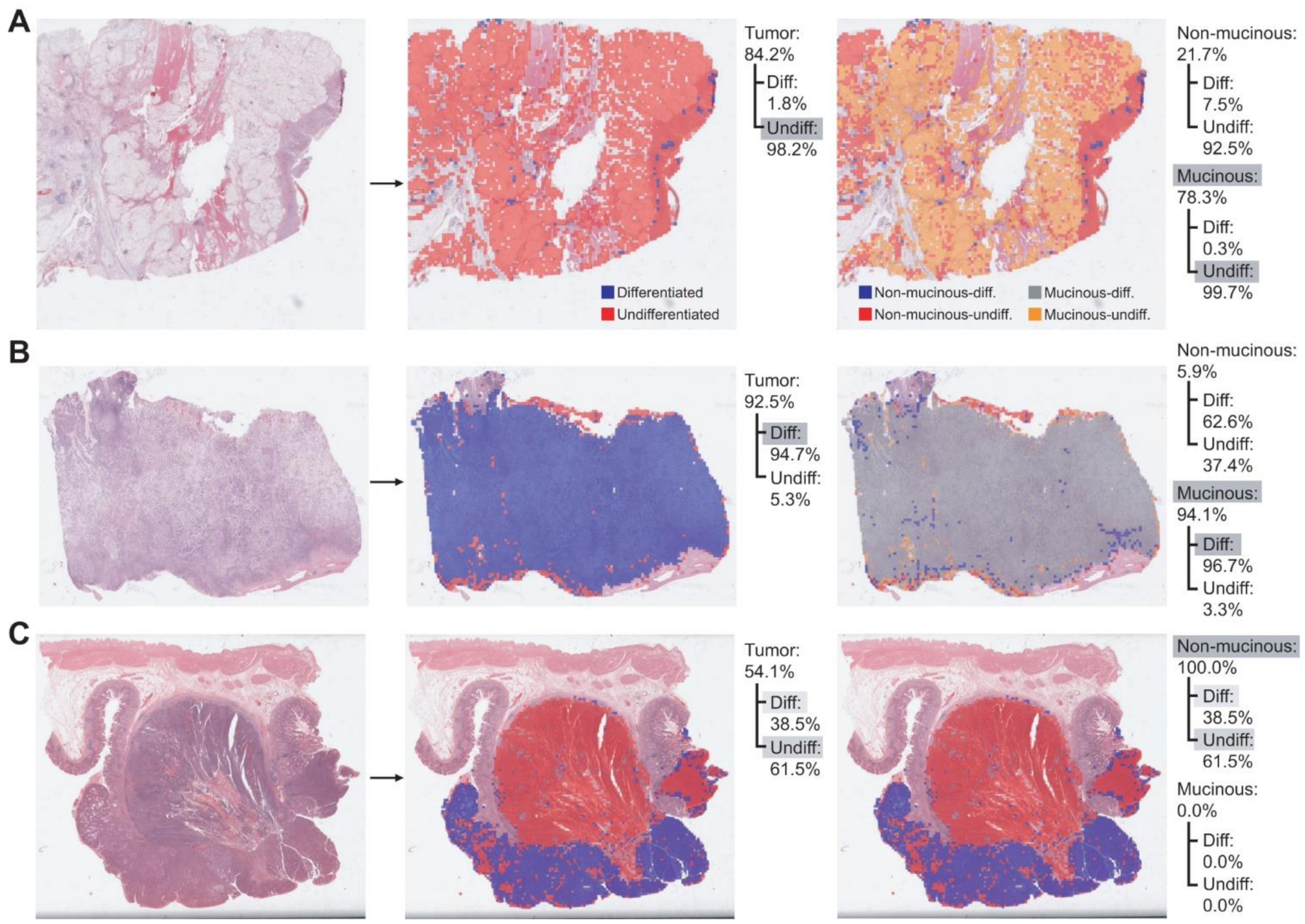
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Japanese Gastric Cancer Association. Japanese gastric cancer treatment guidelines 2018 (5th edition). Gastric Cancer 2021, 24, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Laurén, P. The Two Histological Main Types of Gastric Carcinoma: Diffuse and So-Called Intestinal-Type Carcinoma. An Attempt at a Histo-Clinical Classification. Acta Pathol. Microbiol. Scand. 1965, 64, 31–49. [Google Scholar] [CrossRef]
- Ming, S.C. Gastric carcinoma. A pathobiological classification. Cancer 1977, 39, 2475–2485. [Google Scholar] [CrossRef]
- Japanese Gastric Cancer Association. Japanese classification of gastric carcinoma: 3rd English edition. Gastric Cancer 2011, 14, 101–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunisaki, C.; Akiyama, H.; Nomura, M.; Matsuda, G.; Otsuka, Y.; Ono, H.A.; Nagahori, Y.; Takahashi, M.; Kito, F.; Shimada, H. Clinicopathological properties of poorly-differentiated adenocarcinoma of the stomach: Comparison of solid- and non-solid-types. Anticancer. Res. 2006, 26, 639–646. [Google Scholar] [PubMed]
- Takizawa, K.; Ono, H.; Kakushima, N.; Tanaka, M.; Hasuike, N.; Matsubayashi, H.; Yamagichi, Y.; Bando, E.; Terashima, M.; Kusafuka, K.; et al. Risk of lymph node metastases from intramucosal gastric cancer in relation to histological types: How to manage the mixed histological type for endoscopic submucosal dissection. Gastric Cancer 2012, 16, 531–536. [Google Scholar] [CrossRef] [PubMed]
- Gotoda, T.; Yanagisawa, A.; Sasako, M.; Ono, H.; Nakanishi, Y.; Shimoda, T.; Kato, Y. Incidence of lymph node metastasis from early gastric cancer: Estimation with a large number of cases at two large centers. Gastric Cancer 2000, 3, 219–225. [Google Scholar] [CrossRef] [Green Version]
- Mikami, K.; Hirano, Y.; Futami, K.; Maekawa, T. Expansion of lymph node metastasis in mixed-type submucosal invasive gastric cancer. Asian J. Surg. 2018, 41, 462–466. [Google Scholar] [CrossRef] [PubMed]
- Kook, M.-C. Risk Factors for Lymph Node Metastasis in Undifferentiated-Type Gastric Carcinoma. Clin. Endosc. 2019, 52, 15–20. [Google Scholar] [CrossRef]
- Lee, I.S.; Lee, S.; Park, Y.S.; Gong, C.S.; Yook, J.H.; Kim, B.S. Applicability of endoscopic submucosal dissection for undifferentiated early gastric cancer: Mixed histology of poorly differentiated adenocarcinoma and signet ring cell carcinoma is a worse predictive factor of nodal metastasis. Surg. Oncol. 2017, 26, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.S.; Lee, G.E.; Kang, M.G.; Han, K.H.; Jung, E.S.; Song, K.Y. Mixed Histology Is a Risk Factor for Lymph Node Metastasis in Early Gastric Cancer. J. Surg. Res. 2019, 236, 271–277. [Google Scholar] [CrossRef]
- Isobe, T.; Hashimoto, K.; Kizaki, J.; Matono, S.; Murakami, N.; Kinugasa, T.; Aoyagi, K.; Akagi, Y. Characteristics and prognosis of mucinous gastric carcinoma. Mol. Clin. Oncol. 2015, 3, 44–50. [Google Scholar] [CrossRef] [Green Version]
- Sharma, H.; Zerbe, N.; Klempert, I.; Hellwich, O.; Hufnagl, P. Deep convolutional neural networks for automatic classification of gastric carcinoma using whole slide images in digital histopathology. Comput. Med. Imaging Graph. 2017, 61, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, Q.-C.; Xu, M.-D.; Zhang, Z.; Cheng, J.; Zhong, Y.-S.; Zhang, Y.-Q.; Chen, W.-F.; Yao, L.-Q.; Zhou, P.-H.; et al. Application of convolutional neural network in the diagnosis of the invasion depth of gastric cancer based on conventional endoscopy. Gastrointest. Endosc. 2019, 89, 806–815.e1. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.-O.; Lee, S.H.; Jang, H.-J. Feasibility of fully automated classification of whole slide images based on deep learning. Korean J. Physiol. Pharmacol. 2020, 24, 89–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Nam, S.; Chong, Y.; Jung, C.K.; Kwak, T.-Y.; Lee, J.Y.; Park, J.; Rho, M.J.; Go, H. Introduction to digital pathology and computer-aided pathology. J. Pathol. Transl. Med. 2020, 54, 125–134. [Google Scholar] [CrossRef] [Green Version]
- Jang, H.-J.; Lee, A.; Kang, J.; Song, I.H.; Lee, S.H. Prediction of clinically actionable genetic alterations from colorectal cancer histopathology images using deep learning. World J. Gastroenterol. 2020, 26, 6207–6223. [Google Scholar] [CrossRef]
- Lee, S.H.; Song, I.H.; Jang, H. Feasibility of deep learning-based fully automated classification of microsatellite instability in tissue slides of colorectal cancer. Int. J. Cancer 2021, 149, 728–740. [Google Scholar] [CrossRef]
- Jang, H.-J.; Song, I.H.; Lee, S.H. Generalizability of Deep Learning System for the Pathologic Diagnosis of Various Cancers. Appl. Sci. 2021, 11, 808. [Google Scholar] [CrossRef]
- Venkatraman, E.S. A Permutation Test to Compare Receiver Operating Characteristic Curves. Biometrics 2000, 56, 1134–1138. [Google Scholar] [CrossRef]
- Min, Y.W.; Lee, J.H. Endoscopic Resection for Early Gastric Cancer beyond Absolute Indication with Emphasis on Controversial Issues. J. Gastric Cancer 2014, 14, 7–14. [Google Scholar] [CrossRef] [Green Version]
- Sekiguchi, M.; Oda, I.; Taniguchi, H.; Suzuki, H.; Morita, S.; Fukagawa, T.; Sekine, S.; Kushima, R.; Katai, H. Risk stratification and predictive risk-scoring model for lymph node metastasis in early gastric cancer. J. Gastroenterol. 2016, 51, 961–970. [Google Scholar] [CrossRef]
- Kanesaka, T.; Nagahama, T.; Uedo, N.; Doyama, H.; Ueo, T.; Uchita, K.; Yoshida, N.; Takeda, Y.; Imamura, K.; Wada, K.; et al. Clinical predictors of histologic type of gastric cancer. Gastrointest. Endosc. 2018, 87, 1014–1022. [Google Scholar] [CrossRef]
- Maehara, Y.; Anai, H.; Kusumoto, H.; Sugimachi, K. Poorly differentiated human gastric carcinoma is more sensitive to antitumor drugs than is well differentiated carcinoma. Eur. J. Surg. Oncol. 1987, 13, 203–206. [Google Scholar]
- Adachi, Y.; Yasuda, K.; Inomata, M.; Sato, K.; Shiraishi, N.; Kitano, S. Pathology and prognosis of gastric carcinoma: Well versus poorly differentiated type. Cancer 2000, 89, 1418–1424. [Google Scholar] [CrossRef]
- Shim, C.N.; Chung, H.; Park, J.C.; Lee, H.; Shin, S.K.; Kil Lee, S.; Lee, Y.C. Early gastric cancer with mixed histology predominantly of differentiated type is a distinct subtype with different therapeutic outcomes of endoscopic resection. Surg. Endosc. 2014, 29, 1787–1794. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Ichikawa, D.; Miyamae, M.; Shimizu, H.; Konishi, H.; Shiozaki, A.; Fujiwara, H.; Okamoto, K.; Kishimoto, M.; Otsuji, E. Histological mixed-type as an independent prognostic factor in stage I gastric carcinoma. World J. Gastroenterol. 2015, 21, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Choi, I.J.; Han, H.S.; Kim, Y.-W.; Ryu, K.W.; Yoon, H.M.; Eom, B.W.; Kim, C.G.; Lee, J.Y.; Cho, S.-J.; et al. Risk of Lymph Node Metastasis in Differentiated Type Mucosal Early Gastric Cancer Mixed with Minor Undifferentiated Type Histology. Ann. Surg. Oncol. 2014, 22, 1813–1819. [Google Scholar] [CrossRef]
- Yuan, Y.; Chen, Z.; Chen, J.; Huang, W.; Peng, J.; Ye, J.; He, W.; Wang, L.; Xu, J.; Cai, S.; et al. Mucinous gastric carcinoma: An update of clinicopathologic features and prognostic value from a retrospective study of clinical series. Int. J. Clin. Exp. Pathol. 2018, 11, 813–821. [Google Scholar]
- Ho, S.W.T.; Tan, P. Dissection of gastric cancer heterogeneity for precision oncology. Cancer Sci. 2019, 110, 3405–3414. [Google Scholar] [CrossRef] [Green Version]
- Gullo, I.; Carneiro, F.; Oliveira, C.; Almeida, G. Heterogeneity in Gastric Cancer: From Pure Morphology to Molecular Classifications. Pathobiology 2018, 85, 50–63. [Google Scholar] [CrossRef]
- Liu, M.; Liu, Y.; Di, J.; Su, Z.; Yang, H.; Jiang, B.; Wang, Z.; Zhuang, M.; Bai, F.; Su, X. Multi-region and single-cell sequencing reveal variable genomic heterogeneity in rectal cancer. BMC Cancer 2017, 17, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Cui, H.; Zhou, Y.; Yang, B.; Kong, P.; Zhang, Y.; Liu, Y.; Wang, B.; Cheng, Y.; Li, J.; et al. Multi-region sequencing unveils novel actionable targets and spatial heterogeneity in esophageal squamous cell carcinoma. Nat. Commun. 2019, 10, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bera, K.; Schalper, K.A.; Rimm, D.L.; Velcheti, V.; Madabhushi, A. Artificial intelligence in digital pathology—New tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 2019, 16, 703–715. [Google Scholar] [CrossRef]
- Mobadersany, P.; Yousefi, S.; Amgad, M.; Gutman, D.A.; Barnholtz-Sloan, J.; Vega, J.E.V.; Brat, D.J.; Cooper, L.A.D. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc. Natl. Acad. Sci. USA 2018, 115, E2970–E2979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colling, R.; Pitman, H.; Oien, K.; Rajpoot, N.; Macklin, P.S.; CM-Path AI in Histopathology Working Group; Snead, D.; Sackville, T.; Verrill, C. Artificial intelligence in digital pathology: A roadmap to routine use in clinical practice. J. Pathol. 2019, 249, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Rudin, C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat. Mach. Intell. 2019, 1, 206–215. [Google Scholar] [CrossRef] [Green Version]
- Serag, A.; Ion-Margineanu, A.; Qureshi, H.; McMillan, R.; Saint Martin, M.J.; Diamond, J.; O’Reilly, P.; Hamilton, P. Translational AI and Deep Learning in Diagnostic Pathology. Front. Med. 2019, 6, 185. [Google Scholar] [CrossRef] [Green Version]
- Djuric, U.; Zadeh, G.; Aldape, K.; Diamandis, P. Precision histology: How deep learning is poised to revitalize histomorphology for personalized cancer care. NPJ Precis. Oncol. 2017, 1, 1–5. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jang, H.-J.; Song, I.-H.; Lee, S.-H. Deep Learning for Automatic Subclassification of Gastric Carcinoma Using Whole-Slide Histopathology Images. Cancers 2021, 13, 3811. https://doi.org/10.3390/cancers13153811
Jang H-J, Song I-H, Lee S-H. Deep Learning for Automatic Subclassification of Gastric Carcinoma Using Whole-Slide Histopathology Images. Cancers. 2021; 13(15):3811. https://doi.org/10.3390/cancers13153811
Chicago/Turabian StyleJang, Hyun-Jong, In-Hye Song, and Sung-Hak Lee. 2021. "Deep Learning for Automatic Subclassification of Gastric Carcinoma Using Whole-Slide Histopathology Images" Cancers 13, no. 15: 3811. https://doi.org/10.3390/cancers13153811
APA StyleJang, H.-J., Song, I.-H., & Lee, S.-H. (2021). Deep Learning for Automatic Subclassification of Gastric Carcinoma Using Whole-Slide Histopathology Images. Cancers, 13(15), 3811. https://doi.org/10.3390/cancers13153811





