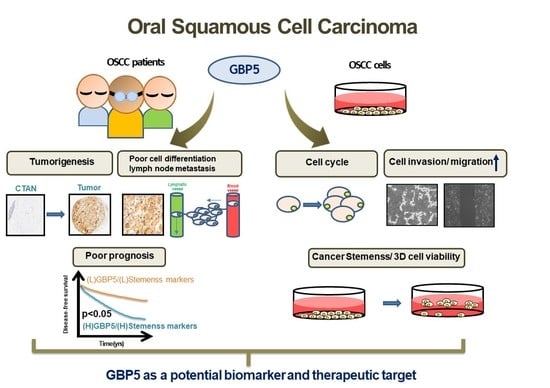
Clinical Significance and the Role of Guanylate-Binding Protein 5 in Oral Squamous Cell Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Tissue Specimens
2.2. Laser Capture Microdissection (LCM) and Next Generation Sequencing (NGS) of LCM-Captured Cells
2.3. Tissue Microarray (TMA) Construction and Immunohistochemistry (IHC)
2.4. Cell Culture
2.5. Transient and Stable Transfection
2.6. Real-Time PCR (RT-PCR)
2.7. Western Blotting
2.8. Clonogenic Assay
2.9. Tumor Sphere Formation
2.10. Cell Cycle Analysis
2.11. Cell Invasion and Migration
2.12. Statistical Analysis
3. Results
3.1. Expression Levels of GBP5 between Normal Tissues and Tumor Tissues in Oral Cancer Patients
3.2. The Association of GBP5 Expression with Prognosis in Oral Cancer Patients
3.3. Role of GBP5 in Cell Growth of OSCC Cells
3.4. The Role of GBP5 on Cell Cycle Control in OSCC Cells
3.5. The Role of GBP5 in the Migration and Invasion of OSCC Cells
3.6. The Role of GBP5 in the Cancer Stemness of OSCC Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhou, L.Q.; Hu, Y.; Xiao, H.J. The prognostic significance of survivin expression in patients with HNSCC: A systematic review and meta-analysis. BMC Cancer 2021, 21, 424. [Google Scholar] [CrossRef]
- Liu, L.; Chen, J.; Cai, X.; Yao, Z.; Huang, J. Progress in targeted therapeutic drugs for oral squamous cell carcinoma. Surg. Oncol. 2019, 31, 90–97. [Google Scholar] [CrossRef]
- Almangush, A.; Leivo, I.; Makitie, A.A. Biomarkers for Immunotherapy of Oral Squamous Cell Carcinoma: Current Status and Challenges. Front. Oncol. 2021, 11, 616629. [Google Scholar] [CrossRef] [PubMed]
- Spoerl, S.; Gerken, M.; Mamilos, A.; Fischer, R.; Wolf, S.; Nieberle, F.; Klingelhoffer, C.; Meier, J.K.; Spoerl, S.; Ettl, T.; et al. Lymph node ratio as a predictor for outcome in oral squamous cell carcinoma: A multicenter population-based cohort study. Clin. Oral Investig. 2021, 25, 1705–1713. [Google Scholar] [CrossRef] [PubMed]
- Vestal, D.J.; Jeyaratnam, J.A. The guanylate-binding proteins: Emerging insights into the biochemical properties and functions of this family of large interferon-induced guanosine triphosphatase. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 2011, 31, 89–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Z.H.; Cai, F.; Zhong, Y. Comprehensive Analysis of the Expression and Prognosis for GBPs in Head and neck squamous cell carcinoma. Sci. Rep. 2020, 10, 6085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braun, E.; Hotter, D.; Koepke, L.; Zech, F.; Gross, R.; Sparrer, K.M.J.; Muller, J.A.; Pfaller, C.K.; Heusinger, E.; Wombacher, R.; et al. Guanylate-Binding Proteins 2 and 5 Exert Broad Antiviral Activity by Inhibiting Furin-Mediated Processing of Viral Envelope Proteins. Cell Rep. 2019, 27, 2092–2104.e10. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Jiang, W.; Yu, Q.; Liu, W.; Zhou, P.; Li, J.; Xu, J.; Xu, B.; Wang, F.; Shao, F. Ubiquitination and degradation of GBPs by a Shigella effector to suppress host defence. Nature 2017, 551, 378–383. [Google Scholar] [CrossRef]
- Servetto, A.; Napolitano, F.; De Angelis, C.; De Placido, P.; Giuliano, M.; Arpino, G.; De Placido, S.; Bianco, R.; Formisano, L. A review of the use of next generation sequencing methodologies to identify biomarkers of resistance to CDK4/6 inhibitors in ER+/HER2- breast cancer. Crit. Rev. Oncol. Hematol. 2021, 157, 103191. [Google Scholar] [CrossRef]
- Haley, B.; Roudnicky, F. Functional Genomics for Cancer Drug Target Discovery. Cancer Cell 2020, 38, 31–43. [Google Scholar] [CrossRef]
- Liu, P.F.; Chen, H.C.; Shu, C.W.; Sie, H.C.; Lee, C.H.; Liou, H.H.; Cheng, J.T.; Tsai, K.W.; Ger, L.P. Guanylate-binding protein 6 is a novel biomarker for tumorigenesis and prognosis in tongue squamous cell carcinoma. Clin. Oral Investig. 2020, 24, 2673–2682. [Google Scholar] [CrossRef]
- Chen, H.C.; Tseng, Y.K.; Shu, C.W.; Fu, T.Y.; Liou, H.H.; Huang, C.H.; Chen, C.C.; Wang, J.S.; Wu, P.C.; Ger, L.P.; et al. Prognostic role of RECK in pathological outcome-dependent buccal mucosa squamous cell carcinoma. Oral Dis. 2020, 26, 62–71. [Google Scholar] [CrossRef]
- Chang, H.Y.; Tseng, Y.K.; Chen, Y.C.; Shu, C.W.; Lin, M.I.; Liou, H.H.; Fu, T.Y.; Lin, Y.C.; Ger, L.P.; Yeh, M.H.; et al. High snail expression predicts a poor prognosis in breast invasive ductal carcinoma patients with HER2/EGFR-positive subtypes. Surg. Oncol. 2018, 27, 314–320. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.S.; Tsai, W.L.; Liu, P.F.; Goan, Y.G.; Lin, C.W.; Tseng, H.H.; Lee, C.H.; Shu, C.W. The MAP3K7-mTOR Axis Promotes the Proliferation and Malignancy of Hepatocellu-lar Carcinoma Cells. Front. Oncol. 2019, 9, 474. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.F.; Tsai, K.L.; Hsu, C.J.; Tsai, W.L.; Cheng, J.S.; Chang, H.W.; Shiau, C.W.; Goan, Y.G.; Tseng, H.H.; Wu, C.H.; et al. Drug Repurposing Screening Identifies Tioconazole as an ATG4 Inhibitor that Suppresses Autophagy and Sensitizes Cancer Cells to Chemotherapy. Theranostics 2018, 8, 830–845. [Google Scholar] [CrossRef]
- Liang, W.Z.; Liu, P.F.; Fu, E.; Chung, H.S.; Jan, C.R.; Wu, C.H.; Shu, C.W.; Hsieh, Y.D. Selective cytotoxic effects of low-power laser irradiation on human oral cancer cells. Lasers Surg. Med. 2015, 47, 756–764. [Google Scholar] [CrossRef] [PubMed]
- Tzeng, Y.T.; Liu, P.F.; Li, J.Y.; Liu, L.F.; Kuo, S.Y.; Hsieh, C.W.; Lee, C.H.; Wu, C.H.; Hsiao, M.; Chang, H.T.; et al. Kinome-Wide siRNA Screening Identifies Src-Enhanced Resistance of Chemotherapeutic Drugs in Triple-Negative Breast Cancer Cells. Front. Pharmacol. 2018, 9, 1285. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.F.; Chang, H.W.; Cheng, J.S.; Lee, H.P.; Yen, C.Y.; Tsai, W.L.; Cheng, J.T.; Li, Y.J.; Huang, W.C.; Lee, C.H.; et al. Map1lc3b and Sqstm1 Modulated Autophagy for Tumorigenesis and Prognosis in Certain Subsites of Oral Squamous Cell Carcinoma. J. Clin. Med. 2018, 7, 478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, P.F.; Leung, C.M.; Chang, Y.H.; Cheng, J.S.; Chen, J.J.; Weng, C.J.; Tsai, K.W.; Hsu, C.J.; Liu, Y.C.; Hsu, P.C.; et al. ATG4B promotes colorectal cancer growth independent of autophagic flux. Autophagy 2014, 10, 1454–1465. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.F.; Chen, C.F.; Shu, C.W.; Chang, H.M.; Lee, C.H.; Liou, H.H.; Ger, L.P.; Chen, C.L.; Kang, B.H. UBE2C is a Potential Biomarker for Tumorigenesis and Prognosis in Tongue Squamous Cell Carcinoma. Diagnostics 2020, 10, 674. [Google Scholar] [CrossRef]
- Mabe, N.W.; Fox, D.B.; Lupo, R.; Decker, A.E.; Phelps, S.N.; Thompson, J.W.; Alvarez, J.V. Epigenetic silencing of tumor suppressor Par-4 promotes chemoresistance in recurrent breast cancer. J. Clin. Investig. 2018, 128, 4413–4428. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.M.; Zhang, J.G.; Zhang, X.; Li, Q. Targeting cancer stem cells for reversing therapy resistance: Mechanism, signaling, and prospective agents. Signal Transduct. Target. Ther. 2021, 6, 62. [Google Scholar] [CrossRef]
- Phi, L.T.H.; Sari, I.N.; Yang, Y.G.; Lee, S.H.; Jun, N.; Kim, K.S.; Lee, Y.K.; Kwon, H.Y. Cancer Stem Cells (CSCs) in Drug Resistance and their Therapeutic Implications in Cancer Treatment. Stem Cells Int. 2018, 2018, 5416923. [Google Scholar] [CrossRef] [Green Version]
- Santos, J.C.; Broz, P. Sensing of invading pathogens by GBPs: At the crossroads between cell-autonomous and innate immunity. J. Leukoc. Biol. 2018, 104, 729–735. [Google Scholar] [CrossRef]
- Li, L.; Ma, G.; Jing, C.; Liu, Z. Guanylate-binding protein 1 (GBP1) promotes lymph node metastasis in human esophageal squamous cell carcinoma. Discov. Med. 2015, 20, 369–378. [Google Scholar]
- Yu, C.J.; Chang, K.P.; Chang, Y.J.; Hsu, C.W.; Liang, Y.; Yu, J.S.; Chi, L.M.; Chang, Y.S.; Wu, C.C. Identification of guanylate-binding protein 1 as a potential oral cancer marker involved in cell invasion using omics-based analysis. J. Proteome Res. 2011, 10, 3778–3788. [Google Scholar] [CrossRef]
- Tipton, A.R.; Nyabuto, G.O.; Trendel, J.A.; Mazur, T.M.; Wilson, J.P.; Wadi, S.; Justinger, J.S.; Moore, G.L.; Nguyen, P.T.; Vestal, D.J. Guanylate-Binding Protein-1 protects ovarian cancer cell lines but not breast cancer cell lines from killing by paclitaxel. Biochem. Biophys. Res. Commun. 2016, 478, 1617–1623. [Google Scholar] [CrossRef] [Green Version]
- Carbotti, G.; Petretto, A.; Naschberger, E.; Sturzl, M.; Martini, S.; Mingari, M.C.; Filaci, G.; Ferrini, S.; Fabbi, M. Cytokine-Induced Guanylate Binding Protein 1 (GBP1) Release from Human Ovarian Cancer Cells. Cancers 2020, 12, 488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Li, X.; Liu, L.; Cao, J.; Goscinski, M.A.; Fan, H.; Li, H.; Suo, Z. Oncogenic Role of Guanylate Binding Protein 1 in Human Prostate Cancer. Front. Oncol. 2019, 9, 1494. [Google Scholar] [CrossRef] [PubMed]
- Guimaraes, D.P.; Oliveira, I.M.; de Moraes, E.; Paiva, G.R.; Souza, D.M.; Barnas, C.; Olmedo, D.B.; Pinto, C.E.; Faria, P.A.; De Moura Gallo, C.V.; et al. Interferon-inducible guanylate binding protein (GBP)-2: A novel p53-regulated tumor marker in esophageal squamous cell carcinomas. Int. J. Cancer. 2009, 124, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Britzen-Laurent, N.; Lipnik, K.; Ocker, M.; Naschberger, E.; Schellerer, V.S.; Croner, R.S.; Vieth, M.; Waldner, M.; Steinberg, P.; Hohenadl, C.; et al. GBP-1 acts as a tumor suppressor in colorectal cancer cells. Carcinogenesis 2013, 34, 153–162. [Google Scholar] [CrossRef] [Green Version]
- Godoy, P.; Cadenas, C.; Hellwig, B.; Marchan, R.; Stewart, J.; Reif, R.; Lohr, M.; Gehrmann, M.; Rahnenfuhrer, J.; Schmidt, M.; et al. Interferon-inducible guanylate binding protein (GBP2) is associated with better prognosis in breast cancer and indicates an efficient T cell response. Breast Cancer 2014, 21, 491–499. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, X.; Liang, Q.; Wang, S.; Xiwen, L.; Pan, F.; Chen, H.; Li, D. Distinct prognostic value of mRNA expression of guanylate-binding protein genes in skin cutaneous melanoma. Oncol. Lett. 2018, 15, 7914–7922. [Google Scholar] [CrossRef]
- Fellenberg, F.; Hartmann, T.B.; Dummer, R.; Usener, D.; Schadendorf, D.; Eichmuller, S. GBP-5 splicing variants: New guanylate-binding proteins with tumor-associated expression and antigenicity. J. Invest. Derm. 2004, 122, 1510–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patil, P.A.; Blakely, A.M.; Lombardo, K.A.; Machan, J.T.; Miner, T.J.; Wang, L.J.; Marwaha, A.S.; Matoso, A. Expression of PD-L1, indoleamine 2,3-dioxygenase and the immune microenvironment in gastric adenocarcinoma. Histopathology 2018, 73, 124–136. [Google Scholar] [CrossRef]
- Jorissen, R.N.; Walker, F.; Pouliot, N.; Garrett, T.P.; Ward, C.W.; Burgess, A.W. Epidermal growth factor receptor: Mechanisms of activation and signalling. Exp. Cell Res. 2003, 284, 31–53. [Google Scholar] [CrossRef]
- Li, M.; Mukasa, A.; Inda, M.M.; Zhang, J.; Chin, L.; Cavenee, W.; Furnari, F. Guanylate binding protein 1 is a novel effector of EGFR-driven invasion in glioblastoma. J. Exp. Med. 2011, 208, 2657–2673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, X.; Zhu, H.; Dai, X.; Xi, Y.; Sheng, Y.; Gao, C.; Liu, H.; Xue, Y.; Liu, J.; Shi, J.; et al. Overexpression of GBP1 predicts poor prognosis and promotes tumor growth in human glioblastoma multiforme. Cancer Biomark. 2019, 25, 275–290. [Google Scholar] [CrossRef]
- Mustafa, D.A.M.; Pedrosa, R.; Smid, M.; van der Weiden, M.; de Weerd, V.; Nigg, A.L.; Berrevoets, C.; Zeneyedpour, L.; Priego, N.; Valiente, M.; et al. T lymphocytes facilitate brain metastasis of breast cancer by inducing Guanylate-Binding Protein 1 expression. Acta Neuropathol. 2018, 135, 581–599. [Google Scholar] [CrossRef] [Green Version]
- Januchowski, R.; Wojtowicz, K.; Zabel, M. The role of aldehyde dehydrogenase (ALDH) in cancer drug resistance. Biomed. Pharm. 2013, 67, 669–680. [Google Scholar] [CrossRef]
- Kawasoe, M.; Yamamoto, Y.; Okawa, K.; Funato, T.; Takeda, M.; Hara, T.; Tsurumi, H.; Moriwaki, H.; Arioka, Y.; Takemura, M.; et al. Acquired resistance of leukemic cells to AraC is associated with the upregulation of aldehyde dehydrogenase 1 family member A2. Exp. Hematol. 2013, 41, 597–603.e2. [Google Scholar] [CrossRef] [Green Version]
- Hoofd, C.; Wang, X.; Lam, S.; Jenkins, C.; Wood, B.; Giambra, V.; Weng, A.P. CD44 promotes chemoresistance in T-ALL by increased drug efflux. Exp. Hematol. 2016, 44, 166–171.e17. [Google Scholar] [CrossRef] [PubMed]
- An, Y.; Ongkeko, W.M. ABCG2: The key to chemoresistance in cancer stem cells? An, Y. and Ongkeko, W.M., 2009. ABCG2: The key to chemoresistance in cancer stem cells? Expert Opin. Drug Metab. Toxicol. 2009, 5, 1529–1542. [Google Scholar] [CrossRef]
- Wan, Q.; Qu, J.; Li, L.; Gao, F. Guanylate-binding protein 1 correlates with advanced tumor features, and serves as a prognostic biomarker for worse survival in lung adenocarcinoma patients. J. Clin. Lab. Anal. 2021, 35, e23610. [Google Scholar] [CrossRef]
- Cheng, L.; Gou, L.; Wei, T.; Zhang, J. GBP1 promotes erlotinib resistance via PGK1activated EMT signaling in nonsmall cell lung cancer. Int. J. Oncol. 2020, 57, 858–870. [Google Scholar] [CrossRef] [PubMed]
- Wadi, S.; Tipton, A.R.; Trendel, J.A.; Khuder, S.A.; Vestal, D.J. hGBP-1 Expression Predicts Shorter Progression-Free Survival in Ovarian Cancers, While Contributing to Paclitaxel Resistance. J. Cancer Ther. 2016, 7, 994–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreoli, M.; Persico, M.; Kumar, A.; Orteca, N.; Kumar, V.; Pepe, A.; Mahalingam, S.; Alegria, A.E.; Petrella, L.; Sevciunaite, L.; et al. Identification of the first inhibitor of the GBP1:PIM1 interaction. Implications for the development of a new class of anticancer agents against paclitaxel resistant cancer cells. J. Med. Chem. 2014, 57, 7916–7932. [Google Scholar] [CrossRef] [Green Version]





| Variables | Normal Tissue | Tumor Adjacent Normal | Tumor | χ2 | p-Value * | |||
|---|---|---|---|---|---|---|---|---|
| Mean ± SD | Median | Mean ± SD | Median | Mean ± SD | Median | |||
| BMSCC | (n = 30) | (n = 117) | (n = 182) | |||||
| 2.33 ± 2.09 | 2.00 | 2.50 ± 1.95 | 3.00 | 3.73 ± 1.97 | 4.00 | 28.233 | <0.001 | |
| TSCC | (n = 31) | (n = 187) | (n = 245) | |||||
| 1.00 ± 1.41 | 0.00 | 1.07 ± 1.51 | 0.00 | 3.21 ± 1.90 | 3.00 | 138.654 | <0.001 | |
| LSCC | (n = 15) | (n = 49) | (n = 72) | |||||
| 2.33 ± 1.54 | 2.00 | 1.53 ± 1.85 | 0.00 | 3.68 ± 1.91 | 4.00 | 32.544 | <0.001 | |
| OSCC | (n = 76) | (n = 353) | (n = 499) | |||||
| 1.79 ± 1.84 | 2.00 | 1.61 ± 1.83 | 2.00 | 3.47 ± 1.94 | 3.00 | 177.689 | <0.001 | |
| Variable | No. (%) | CHR (95% CI) | p-Value * | AHR (95% CI) | p-Value † | |
|---|---|---|---|---|---|---|
| Sex | ||||||
| Female | Low | 39 (47.6) | 1.00 | 1.00 | ||
| High | 43 (52.4) | 1.00 (0.51–1.94) | 0.994 | 1.25 (0.63–2.48) | 0.520 a | |
| Male | Low | 110 (60.4) | 1.00 | 1.00 | ||
| High | 72 (39.6) | 1.39 (0.89–2.16) | 0.147 | 1.49 (0.95–2.33) | 0.080 a | |
| Age, years | ||||||
| ≤60 | Low | 73 (65.8) | 1.00 | 1.00 | ||
| High | 38 (34.2) | 1.46 (0.81–2.63) | 0.206 | 1.60 (0.89–2.90) | 0.119 a | |
| >60 | Low | 76 (49.7) | 1.00 | 1.00 | ||
| High | 77 (50.3) | 1.06(0.66–1.71) | 0.809 | 1.19 (0.73–1.93) | 0.480 a | |
| Cell differentiation | ||||||
| Well | Low | 19 (50.0) | >1.00 | 1.00 | ||
| High | 19 (50.0) | 1.00 (0.32–3.13) | 1.000 | 0.74 (0.22–2.51) | 0.624 b | |
| Moderate, poor | >Low | 130 (57.5) | n">1.00 | 1.00 | ||
| High | 96 (42.5) | 1.29 (0.87–1.90) | 0.201 | 1.49 (1.01–2.21) | 0.047 b | |
| AJCC pathological stage | ||||||
| I, II | >Low | >26 (45.6) | 1.00 | 1.00 | ||
| High | 31 (54.4) | 2.82 (0.72–11.01) | 0.136 | 2.80 (0.71–11.05) | 0.141 c | |
| III, IV | Low | 123 (59.4) | 1.00 | 1.00 | ||
| High | 84 (40.6) | 1.23 (0.84–1.82) | 0.293 | 1.31 (0.88–1.94) | 0.179 c | |
| T classification | ||||||
| T1, T2 | Low | 57 (54.8) | 1.00 | 1.00 | ||
| High | 47 (45.2) | 1.43 (0.65–3.14) | 0.372 | 1.67 (0.75–3.74) | 0.212 d | |
| T3, T4 | Low | 92 (57.5) | 1.00 | 1.00 | ||
| High | 68 (42.5) | 1.27 (0.83–1.94) | 0.266 | 1.31 (0.86–2.01) | 0.209 d | |
| N classification | ||||||
| N0 | Low | 63 (53.4) | 1.00 | 1.00 | ||
| High | 55 (46.6) | 1.29 (0.68–2.43) | 0.442 | 1.39 (0.73–2.65) | 0.314 e | |
| N1, N2, N3 | Low | 86 (58.9) | 1.00 | 1.00 | ||
| High | 60 (41.1) | 1.31 (0.83–2.06) | 0.241 | 1.33 (0.84–2.10) | 0.225 e | |
| Postoperative RT | ||||||
| No | Low | 54 (54.5) | 1.00 | 1.00 | ||
| High | 45 (45.5) | 1.24 (0.70–2.22) | 0.464 | 1.61 (0.89–2.92) | 0.118 a | |
| Yes | Low | 78 (56.1) | 1.00 | 1.00 | ||
| High | 61 (43.9) | 1.25 (0.72–2.19) | 0.430 | 1.35 (0.77–2.37) | 0.293 a |
| Variable | No. (%) | CHR (95% CI) | p-Value * | AHR (95% CI) | p-Value † | |
|---|---|---|---|---|---|---|
| Sex | ||||||
| Female | Low | 31 (42.5) | 1.00 | 1.00 | ||
| High | 42 (57.5) | 1.62 (0.54–4.85) | 0.390 | 1.75 (0.57–5.36) | 0.328 a | |
| Male | Low | 88 (57.5) | 1.00 | 1.00 | ||
| High | 65 (42.5) | 1.73 (0.94–3.20) | 0.081 | 1.83 (0.99–3.40) | 0.056 a | |
| Age, years | ||||||
| ≤60 | Low | 57 (60.6) | 1.00 | 1.00 | ||
| High | 37 (39.4) | 1.65 (0.69–3.98) | 0.261 | 1.83 (0.76–4.41) | 0.177 a | |
| >60 | Low | 62 (47.0) | 1.00 | 1.00 | ||
| High | 70 (53.0) | 1.54(0.78–3.04) | 0.214 | 1.64 (0.82–3.26) | 0.159 a | |
| Cell differentiation | ||||||
| Well | Low | 17 (48.6) | 1.00 | 1.00 | ||
| High | 18 (51.4) | 1.69 (0.28–10.13) | 0.567 | 1.08 (0.18–6.57) | 0.931 b | |
| Moderate, poor | Low | 102 (53.4) | 1.00 | 1.00 | ||
| High | 89 (46.6) | 1.67 (0.95–2.93) | 0.073 | 1.81 (1.03–3.19) | 0.040 b | |
| AJCC pathological stage | ||||||
| I, II | Low | 23 (44.2) | 1.00 | 1.00 | ||
| High | 29 (55.8) | 59.84 (0.20–17,884.73) | 0.159 | 48.43 (0.19–12,494.06) | 0.171 c | |
| III, IV | Low | 96 (55.2) | 1.00 | 1.00 | ||
| High | 78 (44.8) | 1.35 (0.76–2.40) | 0.307 | 1.40 (0.78–2.50) | 0.256 c | |
| T classification | ||||||
| T1, T2 | Low | 49 (51.0) | 1.00 | 1.00 | ||
| High | 47 (49.0) | 2.28 (0.85–6.07) | 0.101 | 2.50 (0.92–6.78) | 0.072 d | |
| T3, T4 | Low | 70 (53.8) | 1.00 | 1.00 | ||
| High | 60 (46.2) | 1.48 (0.77–2.85) | 0.240 | 1.50 (0.77–2.89) | 0.231 d | |
| N classification | ||||||
| N0 | Low | 55 (51.4) | 1.00 | 1.00 | ||
| High | 52 (48.6) | 1.26 (0.55–2.86) | 0.585 | 1.36 (0.60–3.12) | 0.462 e | |
| N1, N2, N3 | Low | 64 (53.8) | 1.00 | 1.00 | ||
| High | 55 (46.2) | 2.00 (0.99–4.06) | 0.054 | 2.21 (1.09–4.50) | 0.028 e | |
| Postoperative RT | ||||||
| No | Low | 47 (53.4) | 1.00 | 1.00 | ||
| High | 41 (46.6) | 2.04 (0.74–5.62) | 0.167 | 2.54 (0.90–7.15) | 0.078 a | |
| Yes | Low | 67 (52.3) | 1.00 | 1.00 | ||
| High | 61 (47.7) | 1.52 (0.79–2.92) | 0.216 | 1.57 (0.81–3.04) | 0.180 a |
| Variable | No. (%) | CHR (95% CI) | p-Value | AHR (95% CI) | p-Value | |
|---|---|---|---|---|---|---|
| GBP5 | Low | 149 (56.4) | 1.00 | 1.00 | ||
| High | 115 (43.6) | 1.23 (0.85–1.77) | 0.280 a | 1.39 (0.96–2.02) | 0.084 b | |
| ALDH1A1 | Low | 227 (86.0) | 1.00 | 1.00 | ||
| High | 37 (14.0) | 1.61 (1.03–2.53) | 0.038 a | 1.33 (0.84–2.10) | 0.225 b | |
| ALDH1A2 | Low | 55 (20.8) | 1.00 | 1.00 | ||
| High | 209 (79.2) | 2.06 (1.15–3.66) | 0.015 a | 2.04 (1.14–3.64) | 0.016 b | |
| CD166 | Low | 222 (84.1) | 1.00 | 1.00 | ||
| High | 42 (15.9) | 1.89 (1.23–2.90) | 0.004 a | 1.89 (1.23–2.90) | 0.004 b | |
| ABCG2 | Low | 187 (70.8) | 1.00 | 1.00 | ||
| High | 77 (29.2) | 1.60 (1.09–2.35) | 0.016 a | 1.38 (0.93–2.04) | 0.106 b | |
| GBP5 (L), ALDH1A1 (L) | 120 (45.5) | 1.00 | 1.00 | |||
| GBP5 (H), ALDH1A1 (L) | 107 (40.5) | 1.10 (0.76–1.59) | 0.624 a | 1.32 (0.87–2.00) | 0.190 c | |
| GBP5 (L), ALDH1A1 (H) | 29 (11.0) | 1.42 (0.85–2.34) | 0.178 a | 1.69 (0.97–2.93) | 0.065 c | |
| GBP5 (H), ALDH1A1(H) | 8 (3.0) | 2.17 (0.95–4.95) | 0.066 a | 2.63 (1.11–6.19) | 0.027 c | |
| GBP5 (L), ALDH1A2 (L) | 30 (11.4) | 1.00 | 1.00 | |||
| GBP5 (H), ALDH1A2 (L) | 25 (9.5) | 0.53 (0.24–1.22) | 0.139 a | 1.06 (0.35–3.15) | 0.922 c | |
| GBP5 (L), ALDH1A2 (H) | 119 (45.1) | 1.02 (0.70–1.47) | 0.934 a | 1.90 (0.86–4.17) | 0.112 c | |
| GBP5 (H), ALDH1A2(H) | 90 (34.1) | 1.48 (1.02–2.15) | 0.040 a | 2.40 (1.09–5.32) | 0.030 c | |
| GBP5 (L), CD166 (L) | 120 (45.5) | 1.00 | 1.00 | |||
| GBP5 (H), CD166 (L) | 102 (38.6) | 1.09 (0.75–1.59) | 0.643 a | 1.42 (0.93–2.17) | 0.108 c | |
| GBP5 (L), CD166 (H) | 29 (11.0) | 1.80 (1.11–2.92) | 0.018 a | 2.23 (1.30–3.82) | 0.004 c | |
| GBP5 (H), CD166 (H) | 13 (4.9) | 1.76 (0.86–3.62) | 0.125 a | 2.30 (1.07–4.92) | 0.032 c | |
| GBP5 (L), ABCG2 (L) | 104 (39.4) | 1.00 | 1.00 | |||
| GBP5 (H), ABCG2 (L) | 83 (31.4) | 0.93 (0.62–1.38) | 0.700 a | 1.19 (0.75–1.90) | 0.456 c | |
| GBP5 (L), ABCG2 (H) | 45 (17.0) | 1.25 (0.79–1.98) | 0.335 a | 1.52 (0.90–2.56) | 0.117 c | |
| GBP5 (H), ABCG2(H) | 32 (12.1) | 1.82 (1.11–2.98) | 0.019 a | 2.12 (1.21–3.70) | 0.008 c |
| Variable | No. (%) | CHR (95% CI) | p-Value | AHR (95% CI) | p-Value | |
|---|---|---|---|---|---|---|
| GBP5 | Low | 119 (52.7) | 1.00 | 1.00 | ||
| High | 107 (47.3) | 1.63 (0.95–2.78) | 0.074 a | 1.78 (1.04–3.05) | 0.037 b | |
| ALDH1A1 | Low | 212 (93.8) | 1.00 | 1.00 | ||
| High | 14 (6.2) | 1.95 (0.83–4.56) | 0.123 a | 1.74 (0.74–4.07) | 0.205 b | |
| ALDH1A2 | Low | 198 (87.6) | 1.00 | 1.00 | ||
| High | 28 (12.4) | 1.16 (0.55–2.46) | 0.702 a | 1.01 (0.47–2.15) | 0.989 b | |
| CD166 | Low | 148 (65.5) | 1.00 | 1.00 | ||
| High | 78 (34.5) | 1.45 (0.85–2.47) | 0.173 a | 1.27 (0.74–2.18) | 0.386 b | |
| ABCG2 | Low | 105 (46.5) | 1.00 | 1.00 | ||
| High | 121 (53.5) | 1.79 (1.03–3.13) | 0.040 a | 1.73 (0.99–3.02) | 0.054 b | |
| GBP5 (L), ALDH1A1 (L) | 108 (47.8) | 1.00 | 1.00 | |||
| GBP5 (H), ALDH1A1 (L) | 104 (46.0) | 1.58 (0.93–2.69) | 0.092 a | 1.87 (1.05–3.33) | 0.034 c | |
| GBP5 (L), ALDH1A1 (H) | 11 (4.9) | 1.97 (0.79–4.96) | 0.148 a | 2.79 (1.04–7.51) | 0.042 c | |
| GBP5 (H), ALDH1A1(H) | 3 (1.3) | 1.69 (0.23–12.25) | 0.604 a | 2.48 (0.33–18.62) | 0.376 c | |
| GBP5 (L), ALDH1A2 (L) | 106 (46.9) | 1.00 | 1.00 | |||
| GBP5 (H), ALDH1A2 (L) | 92 (40.7) | 1.36 (0.80–2.32) | 0.256 a | 1.47 (0.83–2.62) | 0.185 c | |
| GBP5 (L), ALDH1A2 (H) | 13 (5.8) | 0.54 (0.13–2.22) | 0.390 a | 0.68 (0.16–2.90) | 0.598 c | |
| GBP5 (H), ALDH1A2(H) | 15 (6.6) | 1.82 (0.78–4.26) | 0.166 a | 2.14 (0.87–5.28) | 0.100 c | |
| GBP5 (L), CD166 (L) | 76 (33.6) | 1.00 | 1.00 | |||
| GBP5 (H), CD166 (L) | 72 (31.9) | 1.32 (0.76–2.29) | 0.320 a | 2.11 (1.01–4.40) | 0.047 c | |
| GBP5 (L), CD166 (H) | 43 (19.0) | 1.19 (0.64–2.21) | 0.594 a | 1.99 (0.89–4.45) | 0.093 c | |
| GBP5 (H), CD166(H) | 35 (15.5) | 1.52 (0.79–2.96) | 0.212 a | 2.48 (1.08–5.74) | 0.033 c | |
| GBP5 (L), ABCG2 (L) | 55 (24.3) | 1.00 | 1.00 | |||
| GBP5 (H), ABCG2 (L) | 50 (22.1) | 0.68 (0.33–1.39) | 0.293 a | 1.07 (0.43–2.63) | 0.889 c | |
| GBP5 (L), ABCG2 (H) | 64 (28.3) | 0.82 (0.45–1.51) | 0.522 a | 1.26 (0.56–2.83) | 0.582 c | |
| GBP5 (H), ABCG2(H) | 57 (25.2) | 2.37 (1.38–4.08) | 0.002 a | 2.65 (1.25–5.61) | 0.011 c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, P.-F.; Shu, C.-W.; Lee, C.-H.; Sie, H.-C.; Liou, H.-H.; Cheng, J.-T.; Ger, L.-P.; Chen, C.-L.; Chen, C.-C.; Chen, C.-F. Clinical Significance and the Role of Guanylate-Binding Protein 5 in Oral Squamous Cell Carcinoma. Cancers 2021, 13, 4043. https://doi.org/10.3390/cancers13164043
Liu P-F, Shu C-W, Lee C-H, Sie H-C, Liou H-H, Cheng J-T, Ger L-P, Chen C-L, Chen C-C, Chen C-F. Clinical Significance and the Role of Guanylate-Binding Protein 5 in Oral Squamous Cell Carcinoma. Cancers. 2021; 13(16):4043. https://doi.org/10.3390/cancers13164043
Chicago/Turabian StyleLiu, Pei-Feng, Chih-Wen Shu, Cheng-Hsin Lee, Huei-Cin Sie, Huei-Han Liou, Jiin-Tsuey Cheng, Luo-Ping Ger, Chun-Lin Chen, Chien-Chou Chen, and Chun-Feng Chen. 2021. "Clinical Significance and the Role of Guanylate-Binding Protein 5 in Oral Squamous Cell Carcinoma" Cancers 13, no. 16: 4043. https://doi.org/10.3390/cancers13164043
APA StyleLiu, P.-F., Shu, C.-W., Lee, C.-H., Sie, H.-C., Liou, H.-H., Cheng, J.-T., Ger, L.-P., Chen, C.-L., Chen, C.-C., & Chen, C.-F. (2021). Clinical Significance and the Role of Guanylate-Binding Protein 5 in Oral Squamous Cell Carcinoma. Cancers, 13(16), 4043. https://doi.org/10.3390/cancers13164043