Non-Coding RNAs in Pancreatic Cancer Diagnostics and Therapy: Focus on lncRNAs, circRNAs, and piRNAs
Abstract
:Simple Summary
Abstract
1. Introduction
2. Biogenesis and Functions of lncRNA, circRNA and piRNA
2.1. lncRNA
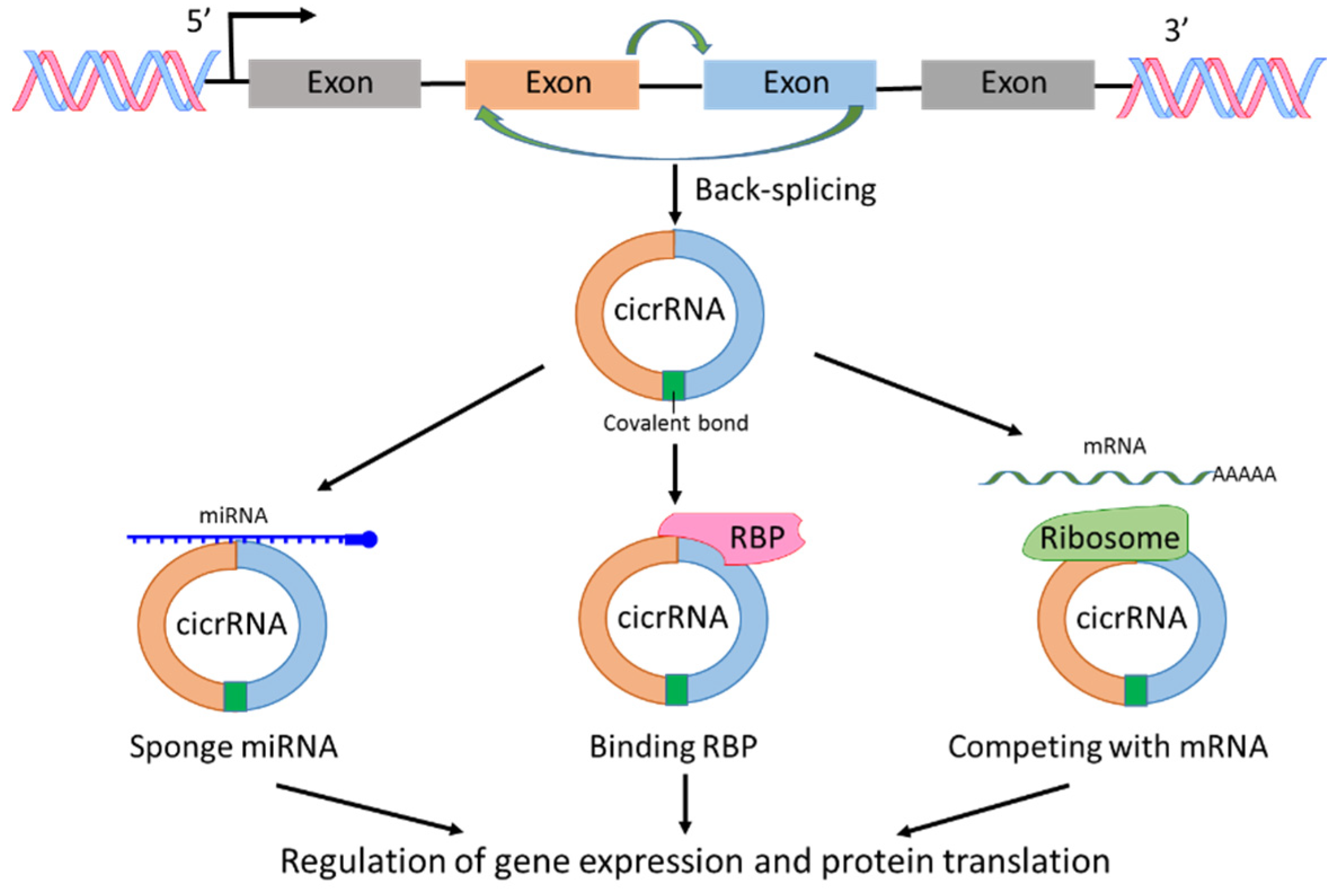
2.2. circRNA
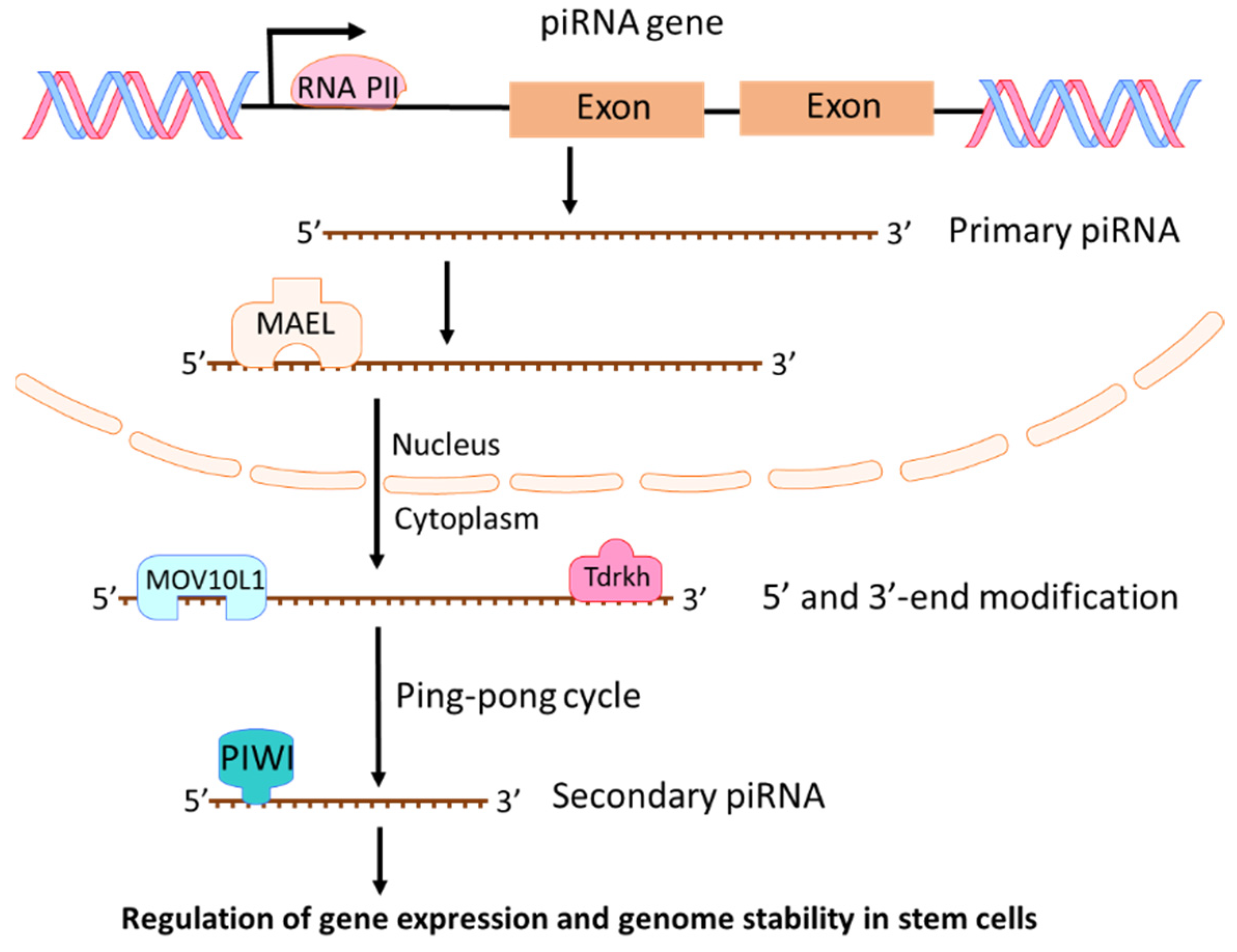
2.3. piRNAs
3. Major lncRNAs in Pancreatic Cancer
3.1. HOTAIR
3.2. MALAT1
3.3. GAS5
3.4. MEG3
3.5. H19
3.6. PVT1
3.7. HOTTIP
4. circRNAs in Pancreatic Cancer
4.1. circRNA Expression Profiling in Pancreatic Cancer
4.2. Oncogenic circRNAs
4.2.1. ciRS-7
4.2.2. CircEIF6
4.2.3. CircFOXK2
4.2.4. circRNA_100782 and circ_001653
4.2.5. hsa_circ_0071036 and hsa_circ_0007534
4.2.6. circBFAR
4.2.7. circ-ASH2L
4.2.8. circRHOT1
4.3. Tumor Suppressive circRNAs
4.3.1. circNFIB1
4.3.2. hsa_circ_001587
4.3.3. hsa_circ_0001649
5. piRNA in Pancreatic Cancer
5.1. Genotyping and piRNA Profiling in Pancreatic Cancer
5.2. Exosome piRNAs in Pancreatic Cancer
5.3. piR-017061
6. The Role of Non-Coding RNAs in the Diagnosis of Pancreatic Cancer
6.1. SNHG15
6.2. ABHD11-AS1
6.3. HULC
6.4. UFC1
6.5. circ_001569
6.6. circ-LDLRAD3
7. The Role of Non-Coding RNAs in the Therapy of Pancreatic Cancer
7.1. ncRNAs Regulated Drug Sensitivity in Pancreatic Cancer
7.2. ncRNAs for the Prognosis of Pancreatic Cancer
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Toden, S.; Zumwalt, T.J.; Goel, A. Non-coding RNAs and potential therapeutic targeting in cancer. Biochim. Biophys Acta Rev. Cancer 2021, 1875, 188491. [Google Scholar] [CrossRef]
- Eddy, S.R. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001, 2, 919–929. [Google Scholar] [CrossRef]
- Slack, F.J.; Chinnaiyan, A.M. The role of non-coding RNAs in oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Cheng, G. Circulating miRNAs: Roles in cancer diagnosis, prognosis and therapy. Adv. Drug Deliv. Rev. 2015, 81, 75–93. [Google Scholar] [CrossRef]
- Verduci, L.; Tarcitano, E.; Strano, S.; Yarden, Y.; Blandino, G. CircRNAs: Role in human diseases and potential use as biomarkers. Cell Death Dis. 2021, 12, 468. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Huang, Q.Y.; Liu, G.F.; Qian, X.L.; Tang, L.B.; Huang, Q.Y.; Xiong, L.X. Long non-coding RNA: Dual Effects on breast cancer metastasis and clinical applications. Cancers 2019, 11, 1802. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Meng, X.M.; Huang, C.; Wu, B.M.; Zhang, L.; Lv, X.W.; Li, J. Long noncoding RNAs: Novel insights into hepatocelluar carcinoma. Cancer Lett. 2014, 344, 20–27. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Brannan, C.I.; Dees, E.C.; Ingram, R.S.; Tilghman, S.M. The product of the H19 gene may function as an RNA. Mol. Cell Biol. 1990, 10, 28–36. [Google Scholar] [CrossRef] [Green Version]
- Brown, C.J.; Ballabio, A.; Rupert, J.L.; Lafreniere, R.G.; Grompe, M.; Tonlorenzi, R.; Willard, H.F. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 1991, 349, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, Y.; Furuno, M.; Kasukawa, T.; Adachi, J.; Bono, H.; Kondo, S.; Nikaido, I.; Osato, N.; Saito, R.; Suzuki, H.; et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 2002, 420, 563–573. [Google Scholar]
- Kung, J.T.; Colognori, D.; Lee, J.T. Long noncoding RNAs: Past, present, and future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, R.W.; Wang, Y.; Chen, L.L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.J.; Dang, H.X.; Lim, D.A.; Feng, F.Y.; Maher, C.A. Long noncoding RNAs in cancer metastasis. Nat. Rev. Cancer 2021, 21, 446–460. [Google Scholar] [CrossRef]
- Chen, L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016, 17, 205–211. [Google Scholar] [CrossRef]
- Hentze, M.W.; Preiss, T. Circular RNAs: Splicing’s enigma variations. EMBO J. 2013, 32, 923–925. [Google Scholar] [CrossRef] [PubMed]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef] [Green Version]
- Vicens, Q.; Westhof, E. Biogenesis of Circular RNAs. Cell 2014, 159, 13–14. [Google Scholar] [CrossRef] [Green Version]
- Cocquerelle, C.; Mascrez, B.; Hétuin, D.; Bailleul, B. Mis-splicing yields circular RNA molecules. FASEB J. 1993, 7, 155–160. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.L.; Bao, Y.; Yee, M.C.; Barrett, S.P.; Hogan, G.J.; Olsen, M.N.; Dinneny, J.R.; Brown, P.O.; Salzman, J. Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE 2014, 9, e90859. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Jiang, J.; Shi, H.; Qian, H.; Zhang, X.; Xu, W. CircRNA: A rising star in gastric cancer. Cell Mol. Life Sci. 2020, 77, 1661–1680. [Google Scholar] [CrossRef]
- Su, M.; Xiao, Y.; Ma, J.; Tang, Y.; Tian, B.; Zhang, Y.; Li, X.; Wu, Z.; Yang, D.; Zhou, Y.; et al. Circular RNAs in cancer: Emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol. Cancer 2019, 18, 90. [Google Scholar] [CrossRef]
- Ernst, C.; Odom, D.T.; Kutter, C. The emergence of piRNAs against transposon invasion to preserve mammalian genome integrity. Nat. Commun. 2017, 8, 1411. [Google Scholar] [CrossRef] [Green Version]
- Lin, H. piRNAs in the germ line. Science 2007, 316, 397. [Google Scholar] [CrossRef]
- Teixeira, F.K.; Okuniewska, M.; Malone, C.D.; Coux, R.X.; Rio, D.C.; Lehmann, R. piRNA-mediated regulation of transposon alternative splicing in the soma and germ line. Nature 2017, 552, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Grivna, S.T.; Beyret, E.; Wang, Z.; Lin, H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006, 20, 1709–1714. [Google Scholar] [CrossRef] [Green Version]
- Ross, R.J.; Weiner, M.M.; Lin, H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature 2014, 505, 353–359. [Google Scholar] [CrossRef] [Green Version]
- Aravin, A.; Gaidatzis, D.; Pfeffer, S.; Lagos-Quintana, M.; Landgraf, P.; Iovino, N.; Morris, P.; Brownstein, M.J.; Kuramochi-Miyagawa, S.; Nakano, T.; et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 2006, 442, 203–207. [Google Scholar] [CrossRef]
- Guo, B.; Li, D.; Du, L.; Zhu, X. piRNAs: Biogenesis and their potential roles in cancer. Cancer Metastasis Rev. 2020, 39, 567–575. [Google Scholar] [CrossRef]
- Wu, W.; Lu, B.F.; Jiang, R.Q.; Chen, S. The function and regulation mechanism of piRNAs in human cancers. Histol. Histopathol. 2021. [Google Scholar] [CrossRef]
- Ipsaro, J.J.; Haase, A.D.; Knott, S.R.; Joshua-Tor, L.; Hannon, G.J. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature 2012, 491, 279–283. [Google Scholar] [CrossRef] [Green Version]
- Saxe, J.P.; Chen, M.; Zhao, H.; Lin, H. Tdrkh is essential for spermatogenesis and participates in primary piRNA biogenesis in the germline. EMBO J. 2013, 32, 1869–1885. [Google Scholar] [CrossRef]
- Lim, S.L.; Qu, Z.P.; Kortschak, R.D.; Lawrence, D.M.; Geoghegan, J.; Hempfling, A.L.; Bergmann, M.; Goodnow, C.C.; Ormandy, C.J.; Wong, L.; et al. HENMT1 and piRNA Stability are required for adult male germ cell transposon repression and to define the spermatogenic program in the mouse. PLoS Genet. 2015, 11, e1005620. [Google Scholar]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Tu, S.; Stubna, M.; Wu, W.S.; Huang, W.C.; Weng, Z.; Lee, H.C. The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science 2018, 359, 587–592. [Google Scholar] [CrossRef] [Green Version]
- Rojas-Rios, P.; Simonelig, M. piRNAs and PIWI proteins: Regulators of gene expression in development and stem cells. Development 2018, 145. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Kang, J.Y.; Gou, L.T.; Wang, J.; Xue, Y.; Skogerboe, G.; Dai, P.; Huang, D.W.; Chen, R.; Fu, X.D.; et al. MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell Res. 2015, 25, 193–207. [Google Scholar] [CrossRef]
- Goh, W.S.; Falciatori, I.; Tam, O.H.; Burgess, R.; Meikar, O.; Kotaja, N.; Hammell, M.; Hannon, G.J. piRNA-directed cleavage of meiotic transcripts regulates spermatogenesis. Genes Dev. 2015, 29, 1032–1044. [Google Scholar] [CrossRef] [Green Version]
- Xie, J.; Xing, S.; Shen, B.Y.; Chen, H.T.; Sun, B.; Wang, Z.T.; Wang, J.W.; Lu, X.X. PIWIL1 interacting RNA piR-017061 inhibits pancreatic cancer growth via regulating EFNA5. Hum. Cell 2021, 34, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Fan, G.; Song, S.; Jiang, Y.; Qian, C.; Zhang, W.; Su, Q.; Xue, X.; Zhuang, W.; Li, B. piRNA-30473 contributes to tumorigenesis and poor prognosis by regulating m6A RNA methylation in DLBCL. Blood 2021, 137, 1603–1614. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Zheng, J.; Lin, D. PIWI-interacting RNAs in human cancer. Semin Cancer Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, N.; Zhang, F.; Jin, S.; Dong, Y.; Dong, X.; Chen, Y.; Kong, X.; Tong, Y.; Mi, Q.; et al. PIWI-interacting RNAs are aberrantly expressed and may serve as novel biomarkers for diagnosis of lung adenocarcinoma. Thorac. Cancer 2021. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Wang, J.; Sun, L.; Li, M.; He, X.; Jiang, J.; Zhou, Q. Piwi-interacting RNA-651 promotes cell proliferation and migration and inhibits apoptosis in breast cancer by facilitating DNMT1-mediated PTEN promoter methylation. Cell Cycle 2021. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Dong, Y.; He, X.; Gong, A.; Gao, J.; Hao, X.; Wang, S.; Fan, Y.; Wang, Z.; Li, M.; et al. piR-hsa-211106 inhibits the progression of lung adenocarcinoma through pyruvate carboxylase and enhances chemotherapy sensitivity. Front. Oncol. 2021, 11, 651915. [Google Scholar] [CrossRef]
- Ding, X.; Li, Y.; Lü, J.; Zhao, Q.; Guo, Y.; Lu, Z.; Ma, W.; Liu, P.; Pestell, R.G.; Liang, C.; et al. piRNA-823 is involved in cancer stem cell regulation through altering dna methylation in association with luminal breast cancer. Front. Cell Dev. Biol. 2021, 9, 641052. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Meng, X.; Li, D.; Han, X. piR-001773 and piR-017184 promote prostate cancer progression by interacting with PCDH9. Cell Signal. 2020, 76, 109780. [Google Scholar] [CrossRef]
- Jiang, D.; Xu, L.; Ni, J.; Zhang, J.; Cai, M.; Shen, L. Functional polymorphisms in LncRNA HOTAIR contribute to susceptibility of pancreatic cancer. Cancer Cell Int. 2019, 19, 47. [Google Scholar] [CrossRef]
- Kim, K.; Jutooru, I.; Chadalapaka, G.; Johnson, G.; Frank, J.; Burghardt, R.; Kim, S.; Safe, S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene 2013, 32, 1616–1625. [Google Scholar] [CrossRef] [Green Version]
- Li, C.H.; Xiao, Z.; Tong, J.H.; To, K.F.; Fang, X.; Cheng, A.S.; Chen, Y. EZH2 coupled with HOTAIR to silence MicroRNA-34a by the induction of heterochromatin formation in human pancreatic ductal adenocarcinoma. Int. J. Cancer 2017, 140, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; He, Y.; Ye, X.; Song, J.; Wang, Q.; Li, Y.; Xie, X. High expression of long noncoding RNA HOTAIRM1 is associated with the proliferation and migration in pancreatic ductal adenocarcinoma. Pathol. Oncol. Res. 2019, 25, 1567–1577. [Google Scholar] [CrossRef]
- Wang, L.; Dong, P.; Wang, W.; Huang, M.; Tian, B. Gemcitabine treatment causes resistance and malignancy of pancreatic cancer stem-like cells via induction of lncRNA HOTAIR. Exp. Med. 2017, 14, 4773–4780. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.Z.; Xu, F.; Zhou, T.; Zhao, X.; McDonald, J.M.; Chen, Y. The long non-coding RNA HOTAIR enhances pancreatic cancer resistance to TNF-related apoptosis-inducing ligand. J. Biol. Chem. 2017, 292, 10390–10397. [Google Scholar] [CrossRef] [Green Version]
- Han, T.; Jiao, F.; Hu, H.; Yuan, C.; Wang, L.; Jin, Z.L.; Song, W.F.; Wang, L.W. EZH2 promotes cell migration and invasion but not alters cell proliferation by suppressing E-cadherin, partly through association with MALAT-1 in pancreatic cancer. Oncotarget 2016, 7, 11194–11207. [Google Scholar] [CrossRef] [Green Version]
- Jiao, F.; Hu, H.; Yuan, C.; Wang, L.; Jiang, W.; Jin, Z.; Guo, Z.; Wang, L. Elevated expression level of long noncoding RNA MALAT-1 facilitates cell growth, migration and invasion in pancreatic cancer. Oncol. Rep. 2014, 32, 2485–2492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiao, F.; Hu, H.; Han, T.; Yuan, C.; Wang, L.; Jin, Z.; Guo, Z.; Wang, L. Long noncoding RNA MALAT-1 enhances stem cell-like phenotypes in pancreatic cancer cells. Int. J. Mol. Sci. 2015, 16, 6677–6693. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Chen, H.; Gao, Y.; Wang, Y.W.; Zhang, G.Q.; Pan, S.H.; Ji, L.; Kong, R.; Wang, G.; Jia, Y.H.; et al. Long noncoding RNA MALAT1 promotes aggressive pancreatic cancer proliferation and metastasis via the stimulation of autophagy. Mol. Cancer 2016, 15, 2232–2243. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Yang, H.; Zhang, J.; Peng, X.; Lu, Z.; Tong, W.; Chen, J. The lncRNA MALAT1 acts as a competing endogenous RNA to regulate KRAS expression by sponging miR-217 in pancreatic ductal adenocarcinoma. Sci. Rep. 2017, 7, 5186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, E.J.; Yang, R.; Fu, X.B.; Liu, Y.F. Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol. 2015, 36, 2403–2407. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tang, X.; Shi, M.; Wen, C.; Shen, B. MiR-216a decreases MALAT1 expression, induces G2/M arrest and apoptosis in pancreatic cancer cells. Biochem. Biophys. Res. Commun. 2017, 483, 816–822. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Shan, T.; Ding, W.; Hua, Z.; Shen, Y.; Lu, Z.; Chen, B.; Dai, T. Study on mechanism about long noncoding RNA MALAT1 affecting pancreatic cancer by regulating hippo-YAP signaling. J. Cell Physiol. 2018, 233, 5805–5814. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, M.; Yuan, C.; Han, T.; Cui, J.; Jiao, F.; Wang, L. A novel feedback loop between high MALAT-1 and low miR-200c-3p promotes cell migration and invasion in pancreatic ductal adenocarcinoma and is predictive of poor prognosis. BMC Cancer 2018, 18, 1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, C.; Nong, K.; Zhu, H.; Wang, W.; Huang, X.; Yuan, Z.; Ai, K. H19 promotes pancreatic cancer metastasis by derepressing let-7’s suppression on its target HMGA2-mediated EMT. Tumour Biol. 2014, 35, 9163–9169. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Tian, X.; Wang, F.; Zhang, Z.; Du, C.; Xie, X.; Kornmann, M.; Yang, Y. The long noncoding RNA H19 promotes cell proliferation via E2F-1 in pancreatic ductal adenocarcinoma. Cancer Biol. 2016, 17, 1051–1061. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Tian, X.; Guo, H.; Zhang, Z.; Du, C.; Wang, F.; Xie, X.; Gao, H.; Zhuang, Y.; Kornmann, M.; et al. Long noncoding RNA H19 derived miR-675 regulates cell proliferation by down-regulating E2F-1 in human pancreatic ductal adenocarcinoma. J. Cancer 2018, 9, 389–399. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Zhu, Q.; Yang, W.; Shan, Y.; Yu, Z.; Zhang, Q.; Wu, H. LncRNA H19/miR-194/PFTK1 axis modulates the cell proliferation and migration of pancreatic cancer. J. Cell Biochem. 2019, 120, 3874–3886. [Google Scholar] [CrossRef]
- Wang, F.; Rong, L.; Zhang, Z.; Li, M.; Ma, L.; Ma, Y.; Xie, X.; Tian, X.; Yang, Y. LncRNA H19-derived miR-675-3p promotes epithelial-mesenchymal transition and stemness in human pancreatic cancer cells by targeting the STAT3 pathway. J. Cancer 2020, 11, 4771–4782. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, H.; Matsuda, Y.; Yamamoto, M.; Michishita, M.; Takahashi, K.; Sasaki, N.; Ishikawa, N.; Aida, J.; Takubo, K.; Arai, T.; et al. Reduced expression of the H19 long non-coding RNA inhibits pancreatic cancer metastasis. Lab. Investig. 2018, 98, 814–824. [Google Scholar] [CrossRef]
- Huang, C.; Yu, W.; Wang, Q.; Cui, H.; Wang, Y.; Zhang, L.; Han, F.; Huang, T. Increased expression of the lncRNA PVT1 is associated with poor prognosis in pancreatic cancer patients. Minerva Med. 2015, 106, 143–149. [Google Scholar]
- Huang, F.; Chen, W.; Peng, J.; Li, Y.; Zhuang, Y.; Zhu, Z.; Shao, C.; Yang, W.; Yao, H.; Zhang, S. LncRNA PVT1 triggers cyto-protective autophagy and promotes pancreatic ductal adenocarcinoma development via the miR-20a-5p/ULK1 Axis. Mol. Cancer 2018, 17, 98. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Wang, P.; Dong, W.; Liu, H.; Sun, J.; Zhao, L. LncRNA PVT1 promotes exosome secretion through YKT6, RAB7, and VAMP3 in pancreatic cancer. Aging 2020, 12, 10427–10440. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhang, P.; Yin, T.; Zhang, F.; Wang, W. Upregulation of LncRNA PVT1 facilitates pancreatic ductal adenocarcinoma cell progression and glycolysis by regulating MiR-519d-3p and HIF-1A. J. Cancer 2020, 11, 2572–2579. [Google Scholar] [CrossRef]
- Wang, C.J.; Shi, S.B.; Tian, J.; Xu, J.; Niu, Z.X. lncRNA MALAT1, HOTTIP and PVT1 as predictors for predicting the efficacy of GEM based chemotherapy in first-line treatment of pancreatic cancer patients. Oncotarget 2017, 8, 95108–95115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, B.Q.; Jiang, Y.; Zhu, F.; Sun, D.L.; He, X.Z. Long noncoding RNA PVT1 promotes EMT and cell proliferation and migration through downregulating p21 in pancreatic cancer cells. Technol. Cancer Res. Treat. 2017, 16, 819–827. [Google Scholar] [CrossRef] [Green Version]
- You, L.; Chang, D.; Du, H.Z.; Zhao, Y.P. Genome-wide screen identifies PVT1 as a regulator of gemcitabine sensitivity in human pancreatic cancer cells. Biochem. Biophys. Res. Commun. 2011, 407, 1–6. [Google Scholar] [CrossRef]
- Zhao, L.; Kong, H.; Sun, H.; Chen, Z.; Chen, B.; Zhou, M. LncRNA-PVT1 promotes pancreatic cancer cells proliferation and migration through acting as a molecular sponge to regulate miR-448. J. Cell Physiol. 2018, 233, 4044–4055. [Google Scholar] [CrossRef]
- Zhou, C.; Yi, C.; Yi, Y.; Qin, W.; Yan, Y.; Dong, X.; Zhang, X.; Huang, Y.; Zhang, R.; Wei, J.; et al. LncRNA PVT1 promotes gemcitabine resistance of pancreatic cancer via activating Wnt/β-catenin and autophagy pathway through modulating the miR-619-5p/Pygo2 and miR-619-5p/ATG14 axes. Mol. Cancer 2020, 19, 118. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Chen, C.; Zhou, Q.; Wang, Y.; Zhao, Y.; Zhao, X.; Li, W.; Zheng, S.; Ye, H.; Wang, L.; et al. LncRNA HOTTIP modulates cancer stem cell properties in human pancreatic cancer by regulating HOXA9. Cancer Lett. 2017, 410, 68–81. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, X.; Zhou, Y.; Liu, Y.; Zhou, Q.; Ye, H.; Wang, Y.; Zeng, J.; Song, Y.; Gao, W.; et al. The long non-coding RNA HOTTIP promotes progression and gemcitabine resistance by regulating HOXA13 in pancreatic cancer. J. Transl. Med. 2015, 13, 84. [Google Scholar] [CrossRef] [Green Version]
- Wong, C.H.; Li, C.H.; He, Q.; Chan, S.L.; Tong, J.H.; To, K.F.; Lin, L.Z.; Chen, Y. Ectopic HOTTIP expression induces noncanonical transactivation pathways to promote growth and invasiveness in pancreatic ductal adenocarcinoma. Cancer Lett. 2020, 477, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yin, F.; Zhang, Q.; Dong, Z.; Hu, J.; Ma, Z. LncRNA HOTTIP Participates in cisplatin resistance of tumor cells by regulating miR-137 expression in pancreatic cancer. Onco Targets Ther. 2020, 13, 2689–2699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Z.Q.; Wang, J.F.; Chen, D.H.; Ma, X.S.; Wu, Y.; Tang, Z.; Dang, X.W. Long non-coding RNA GAS5 suppresses pancreatic cancer metastasis through modulating miR-32-5p/PTEN axis. Cell Biosci. 2017, 7, 66. [Google Scholar] [CrossRef] [Green Version]
- Gao, Z.Q.; Wang, J.F.; Chen, D.H.; Ma, X.S.; Yang, W.; Zhe, T.; Dang, X.W. Long non-coding RNA GAS5 antagonizes the chemoresistance of pancreatic cancer cells through down-regulation of miR-181c-5p. Biomed. Pharm. 2018, 97, 809–817. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Wu, S.; Ma, J.; Yan, S.; Xiao, Z.; Wan, L.; Zhang, F.; Shang, M.; Mao, A. lncRNA GAS5 reverses EMT and tumor stem cell-mediated gemcitabine resistance and metastasis by targeting miR-221/SOCS3 in pancreatic cancer. Mol. Nucleic Acids 2018, 13, 472–482. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Fang, Y.; Wang, Z.; Xie, J.; Zhan, Q.; Deng, X.; Chen, H.; Jin, J.; Peng, C.; Li, H.; et al. Downregulation of gas5 increases pancreatic cancer cell proliferation by regulating CDK6. Cell Tissue Res. 2013, 354, 891–896. [Google Scholar] [CrossRef]
- Gu, L.; Zhang, J.; Shi, M.; Zhan, Q.; Shen, B.; Peng, C. lncRNA MEG3 had anti-cancer effects to suppress pancreatic cancer activity. Biomed. Pharm. 2017, 89, 1269–1276. [Google Scholar] [CrossRef]
- Iyer, S.; Modali, S.D.; Agarwal, S.K. Long noncoding RNA MEG3 is an epigenetic determinant of oncogenic signaling in functional pancreatic neuroendocrine tumor cells. Mol. Cell Biol. 2017, 37, e00278-17. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Wang, F.; Du, C.; Zhang, Z.; Guo, H.; Xie, X.; Gao, H.; Zhuang, Y.; Kornmann, M.; Gao, H.; et al. Long non-coding RNA MEG3 functions as a tumour suppressor and has prognostic predictive value in human pancreatic cancer. Oncol. Rep. 2018, 39, 1132–1140. [Google Scholar] [CrossRef]
- Modali, S.D.; Parekh, V.I.; Kebebew, E.; Agarwal, S.K. Epigenetic regulation of the lncRNA MEG3 and its target c-MET in pancreatic neuroendocrine tumors. Mol. Endocrinol. 2015, 29, 224–237. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.Y.; Feng, H.M. MEG3 suppresses human pancreatic neuroendocrine tumor cells growth and metastasis by down-regulation of Mir-183. Cell Physiol. Biochem. 2017, 44, 345–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Li, Z.; Zheng, S.; Chen, H.; Zhao, X.; Gao, W.; Bi, Z.; You, K.; Wang, Y.; Li, W.; et al. The long non-coding RNA HOTAIR affects the radiosensitivity of pancreatic ductal adenocarcinoma by regulating the expression of Wnt inhibitory factor 1. Tumour Biol. 2016, 37, 3957–3967. [Google Scholar] [CrossRef]
- Ji, P.; Diederichs, S.; Wang, W.; Böing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef] [Green Version]
- Muller-Tidow, C.; Diederichs, S.; Thomas, M.; Serve, H. Genome-wide screening for prognosis-predicting genes in early-stage non-small-cell lung cancer. Lung Cancer 2004, 45 (Suppl. 2), S145–S150. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Wang, Y.; Li, H.; Chen, L.; Liu, Q. Regulatory networks of LncRNA MALAT-1 in cancer. Cancer Manag. Res. 2020, 12, 10181–10198. [Google Scholar] [CrossRef] [PubMed]
- Pei, C.; Gong, X.; Zhang, Y. LncRNA MALAT-1 promotes growth and metastasis of epithelial ovarian cancer via sponging microrna-22. Am. J. Transl. Res. 2020, 12, 6977–6987. [Google Scholar] [PubMed]
- Liu, J.H.; Chen, G.; Dang, Y.W.; Li, C.J.; Luo, D.Z. Expression and prognostic significance of lncRNA MALAT1 in pancreatic cancer tissues. Asian Pac. J. Cancer Prev. 2014, 15, 2971–2977. [Google Scholar] [CrossRef] [PubMed]
- Filippova, E.A.; Fridman, M.V.; Burdennyy, A.M.; Loginov, V.I.; Pronina, I.V.; Lukina, S.S.; Dmitriev, A.A.; Braga, E.A. Long noncoding RNA GAS5 in breast cancer: Epigenetic mechanisms and biological functions. Int. J. Mol. Sci. 2021, 22, 6810. [Google Scholar] [CrossRef]
- Pickard, M.R.; Williams, G.T. Molecular and cellular mechanisms of action of tumour suppressor GAS5 LncRNA. Genes 2015, 6, 484–499. [Google Scholar] [CrossRef] [Green Version]
- Ghafouri-Fard, S.; Taheri, M. Maternally expressed gene 3 (MEG3): A tumor suppressor long non coding RNA. Biomed. Pharm. 2019, 118, 109129. [Google Scholar] [CrossRef]
- Kallen, A.N.; Zhou, X.B.; Xu, J.; Qiao, C.; Ma, J.; Yan, L.; Lu, L.; Liu, C.; Yi, J.S.; Zhang, H.; et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell 2013, 52, 101–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onagoruwa, O.T.; Pal, G.; Ochu, C.; Ogunwobi, O.O. Oncogenic role of PVT1 and therapeutic implications. Front. Oncol. 2020, 10, 17. [Google Scholar] [CrossRef] [Green Version]
- Ghafouri-Fard, S.; Dashti, S.; Taheri, M. The HOTTIP (HOXA transcript at the distal tip) lncRNA: Review of oncogenic roles in human. Biomed. Pharm. 2020, 127, 110158. [Google Scholar] [CrossRef]
- Shang, Q.; Yang, Z.; Jia, R.; Ge, S. The novel roles of circRNAs in human cancer. Mol. Cancer 2019, 18, 6. [Google Scholar] [CrossRef] [PubMed]
- Qu, S.; Song, W.; Yang, X.; Wang, J.; Zhang, R.; Zhang, Z.; Zhang, H.; Li, H. Microarray expression profile of circular RNAs in human pancreatic ductal adenocarcinoma. Genom Data 2015, 5, 385–387. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Xu, X.; Ouyang, Y.; Wang, Y.; Yang, J.; Yin, L.; Ge, J.; Wang, H. Microarray expression profile analysis of circular RNAs in pancreatic cancer. Mol. Med. Rep. 2018, 17, 7661–7671. [Google Scholar] [CrossRef]
- Liu, L.; Liu, F.B.; Huang, M.; Xie, K.; Xie, Q.S.; Liu, C.H.; Shen, M.J.; Huang, Q. Circular RNA ciRS-7 promotes the proliferation and metastasis of pancreatic cancer by regulating miR-7-mediated EGFR/STAT3 signaling pathway. Hepatobiliary Pancreat. Dis. Int. 2019, 18, 580–586. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Li, M.; Lu, H.; Peng, T. Up-regulation of circEIF6 contributes to pancreatic cancer development through targeting miR-557/SLC7A11/PI3K/AKT signaling. Cancer Manag. Res. 2021, 13, 247–258. [Google Scholar] [CrossRef]
- Wong, C.H.; Lou, U.K.; Li, Y.; Chan, S.L.; Tong, J.H.; To, K.F.; Chen, Y. CircFOXK2 promotes growth and metastasis of pancreatic ductal adenocarcinoma by complexing with rna-binding proteins and sponging MiR-942. Cancer Res. 2020, 80, 2138–2149. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Zhou, Q.; Su, D.; Luo, Y.; Fu, Z.; Huang, L.; Li, Z.; Jiang, D.; Kong, Y.; Li, Z.; et al. Circular RNA circBFAR promotes the progression of pancreatic ductal adenocarcinoma via the miR-34b-5p/MET/Akt axis. Mol. Cancer 2020, 19, 83. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, Z.; Zhang, M.; Wang, B.; Ye, J.; Zhang, Y.; Tang, D.; Ma, D.; Jin, W.; Li, X.; et al. Circ-ASH2L promotes tumor progression by sponging miR-34a to regulate Notch1 in pancreatic ductal adenocarcinoma. J. Exp. Clin. Cancer Res. 2019, 38, 466. [Google Scholar] [CrossRef] [Green Version]
- Ling, S.; He, Y.; Li, X.; Hu, M.; Ma, Y.; Li, Y.; Lu, Z.; Shen, S.; Kong, B.; Zou, X.; et al. CircRHOT1 mediated cell proliferation, apoptosis and invasion of pancreatic cancer cells by sponging miR-125a-3p. J. Cell Mol. Med. 2020, 24, 9881–9889. [Google Scholar] [CrossRef]
- Qu, S.; Hao, X.; Song, W.; Niu, K.; Yang, X.; Zhang, X.; Shang, R.; Wang, Q.; Li, H.; Liu, Z. Circular RNA circRHOT1 is upregulated and promotes cell proliferation and invasion in pancreatic cancer. Epigenomics 2019, 11, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Shi, Y.; Zhang, Y.; Sun, J. CircRNA_100782 regulates pancreatic carcinoma proliferation through the IL6-STAT3 pathway. Onco Targets Ther. 2017, 10, 5783–5794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, X.; Fang, Y.; Chen, P.; Xu, Y.; Zhou, W.; Rong, Y.; Li, J.A.; Chen, W.; Lou, W. Upregulated circRNA hsa_circ_0071036 promotes tumourigenesis of pancreatic cancer by sponging miR-489 and predicts unfavorable characteristics and prognosis. Cell Cycle 2021, 4, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Hao, L.; Rong, W.; Bai, L.; Cui, H.; Zhang, S.; Li, Y.; Chen, D.; Meng, X. Upregulated circular RNA circ_0007534 indicates an unfavorable prognosis in pancreatic ductal adenocarcinoma and regulates cell proliferation, apoptosis, and invasion by sponging miR-625 and miR-892b. J. Cell Biochem. 2019, 120, 3780–3789. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Li, Y.; Luo, Y.; Zhu, J.; Zheng, H.; Gao, B.; Guo, X.; Li, Z.; Chen, R.; Chen, C. circNFIB1 inhibits lymphangiogenesis and lymphatic metastasis via the miR-486-5p/PIK3R1/VEGF-C axis in pancreatic cancer. Mol. Cancer 2020, 19, 82. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Tan, P.; Zhuang, Y.; Du, L. hsa_circRNA_001587 upregulates SLC4A4 expression to inhibit migration, invasion, and angiogenesis of pancreatic cancer cells via binding to microRNA-223. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 319, G703–G717. [Google Scholar] [CrossRef]
- Li, H.; Hao, X.; Wang, H.; Liu, Z.; He, Y.; Pu, M.; Zhang, H.; Yu, H.; Duan, J.; Qu, S. Circular RNA expression profile of pancreatic ductal adenocarcinoma revealed by microarray. Cell Physiol. Biochem. 2016, 40, 1334–1344. [Google Scholar] [CrossRef]
- Qian, Y.; Gong, Y.; Fan, Z.; Luo, G.; Huang, Q.; Deng, S.; Cheng, H.; Jin, K.; Ni, Q.; Yu, X.; et al. Molecular alterations and targeted therapy in pancreatic ductal adenocarcinoma. J. Hematol. Oncol. 2020, 13, 130. [Google Scholar] [CrossRef]
- Buscail, L.; Bournet, B.; Cordelier, P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 153–168. [Google Scholar] [CrossRef] [PubMed]
- Van Mackelenbergh, M.G.; Stroes, C.I.; Spijker, R.; van Eijck, C.H.J.; Wilmink, J.W.; Bijlsma, M.F.; van Laarhoven, H.W.M. Clinical trials targeting the stroma in pancreatic cancer: A systematic review and meta-analysis. Cancers 2019, 11, 588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fathi, M.; Ghafouri-Fard, S.; Abak, A.; Taheri, M. Emerging roles of miRNAs in the development of pancreatic cancer. Biomed. Pharm. 2021, 141, 111914. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ju, J.; Ni, B.; Wang, H. The emerging role of miR-506 in cancer. Oncotarget 2016, 7, 62778–62788. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, J.Y.; Zhou, S.Y.; Yang, S.J.; Zhong, S.L. Circular RNA expression in pancreatic ductal adenocarcinoma. Oncol. Lett. 2019, 18, 2923–2930. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Ye, T.; Liu, H.; Lv, P.; Duan, C.; Wu, X.; Jiang, K.; Lu, H.; Xia, D.; Peng, E.; et al. Expression profiles, biological functions and clinical significance of circRNAs in bladder cancer. Mol. Cancer 2021, 20, 4. [Google Scholar] [CrossRef]
- Gao, Y.; Zhao, F. Computational strategies for exploring circular RNAs. Trends Genet. 2018, 34, 389–400. [Google Scholar] [CrossRef]
- Xu, Z.; Shen, J.; Hua, S.; Wan, D.; Chen, Q.; Han, Y.; Ren, R.; Liu, F.; Du, Z.; Guo, X.; et al. High-throughput sequencing of circRNAs reveals novel insights into mechanisms of nigericin in pancreatic cancer. BMC Genom. 2019, 20, 716. [Google Scholar] [CrossRef] [Green Version]
- Hansen, T.B.; Kjems, J.; Damgaard, C.K. Circular RNA and miR-7 in cancer. Cancer Res. 2013, 73, 5609–5612. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Li, X.; Xu, Q.; Lv, S.; Li, J.; Ma, Q. Role of glial cell line-derived neurotrophic factor in perineural invasion of pancreatic cancer. Biochim. Biophys. Acta 2012, 1826, 112–120. [Google Scholar] [CrossRef]
- Van, D.G.; Griffin, M.D.W.; Putoczki, T.L. Emerging roles for the IL-6 family of cytokines in pancreatic cancer. Clin. Sci. 2020, 134, 2091–2115. [Google Scholar]
- Shi, H.; Li, H.; Zhen, T.; Dong, Y.; Pei, X.; Zhang, X. hsa_circ_001653 implicates in the development of pancreatic ductal adenocarcinoma by regulating microrna-377-mediated HOXC6 Axis. Mol. Nucleic Acids 2020, 20, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Wang, T.; Yan, L.; Qu, L. A novel prognostic biomarker for pancreatic ductal adenocarcinoma: Hsa_circ_0001649. Gene 2018, 675, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Xie, H.; Zhang, L.; Zhao, Q.; Lü, J.; Yu, Z. Piwi-interacting RNAs: A new class of regulator in human breast cancer. Front. Oncol. 2021, 11, 695077. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, C.G.; Azevedo Dos Santos, P.J.; Vidal, A.F.; Santos, S.; Ribeiro-Dos-Santos, Ã. piRNAs in gastric cancer: A new approach towards translational research. Int. J. Mol. Sci. 2020, 21, 2126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, J.; Deng, H.; Xiao, B.; Zhou, H.; Zhou, F.; Shen, Z.; Guo, J. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett. 2012, 315, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Xin, J.; Du, M.; Jiang, X.; Wu, Y.; Ben, S.; Zheng, R.; Chu, H.; Li, S.; Zhang, Z.; Wang, M. Systematic evaluation of the effects of genetic variants on PIWI-interacting RNA expression across 33 cancer types. Nucleic Acids Res. 2021, 49, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.R.; Kimchi, E.T.; Manjunath, Y.; Gajagowni, S.; Stuckel, A.J.; Kaifi, J.T. RNA cargos in extracellular vesicles derived from blood serum in pancreas associated conditions. Sci. Rep. 2020, 10, 2800. [Google Scholar] [CrossRef] [Green Version]
- Yee, N.S.; Zhang, S.; He, H.Z.; Zheng, S.Y. Extracellular vesicles as potential biomarkers for early detection and diagnosis of pancreatic cancer. Biomedicines 2020, 8, 581. [Google Scholar] [CrossRef] [PubMed]
- Yuan, T.; Huang, X.; Woodcock, M.; Du, M.; Dittmar, R.; Wang, Y.; Tsai, S.; Kohli, M.; Boardman, L.; Patel, T.; et al. Plasma extracellular RNA profiles in healthy and cancer patients. Sci. Rep. 2016, 6, 19413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muller, S.; Raulefs, S.; Bruns, P.; Afonso-Grunz, F.; Plotner, A.; Thermann, R.; Jäger, C.; Schlitter, A.M.; Kong, B.; Regel, I.; et al. Next-generation sequencing reveals novel differentially regulated mRNAs, lncRNAs, miRNAs, sdRNAs and a piRNA in pancreatic cancer. Mol. Cancer 2015, 14, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sukowati, C.H.C.; Cabral, L.K.D.; Tiribelli, C.; Pascut, D. Circulating long and circular noncoding RNA as non-invasive diagnostic tools of hepatocellular carcinoma. Biomedicines 2021, 9, 90. [Google Scholar] [CrossRef] [PubMed]
- Mu, H.; Zhang, S.; Yao, Z.; Liu, Y.; Lin, K.; Zhao, Z.; Zhu, Y. The diagnostic and prognostic value of exosome-derived long non-coding RNAs in cancer patients: A meta-analysis. Clin. Exp. Med. 2020, 20, 339–348. [Google Scholar] [CrossRef]
- Galamb, O.; Barták, B.K.; Kalmár, A.; Nagy, Z.B.; Szigeti, K.A.; Tulassay, Z.; Igaz, P.; Molnár, B. Diagnostic and prognostic potential of tissue and circulating long non-coding RNAs in colorectal tumors. World J. Gastroenterol. 2019, 25, 5026–5048. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.J.; Ying, X.L.; Jiang, J.H.; Xu, Y.H. Prostate cancer antigen 3 as a biomarker in the urine for prostate cancer diagnosis: A meta-analysis. J. Cancer Res. 2014, 10, C218–C221. [Google Scholar] [CrossRef]
- Ou, Z.L.; Luo, Z.; Lu, Y.B. Long non-coding RNA HULC as a diagnostic and prognostic marker of pancreatic cancer. World J. Gastroenterol. 2019, 25, 6728–6742. [Google Scholar] [CrossRef]
- Takahashi, K.; Ota, Y.; Kogure, T.; Suzuki, Y.; Iwamoto, H.; Yamakita, K.; Kitano, Y.; Fujii, S.; Haneda, M.; Patel, T.; et al. Circulating extracellular vesicle-encapsulated HULC is a potential biomarker for human pancreatic cancer. Cancer Sci. 2020, 111, 98–111. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Feng, W.; Liu, W.; Kong, X.; Li, L.; He, J.; Wang, D.; Zhang, M.; Zhou, G.; Xu, W.; et al. Circulating lncRNA ABHD11-AS1 serves as a biomarker for early pancreatic cancer diagnosis. J. Cancer 2019, 10, 3746–3756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mortoglou, M.; Tabin, Z.K.; Arisan, E.D.; Kocher, H.M.; Uysal-Onganer, P. Non-coding RNAs in pancreatic ductal adenocarcinoma: New approaches for better diagnosis and therapy. Transl. Oncol. 2021, 14, 101090. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, L.; Jiang, J.; Qiao, Z. The lncRNA SNHG15/miR-18a-5p axis promotes cell proliferation in ovarian cancer through activating Akt/mTOR signaling pathway. J. Cell Biochem. 2020, 121, 4699–4710. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Feng, Y.; Wang, J.; Liang, Y.; Zou, W. Long non-coding RNA SNHG15 in various cancers: A meta and bioinformatic analysis. BMC Cancer 2020, 20, 1156. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Lin, H.; Kang, L.; Huang, P.; Huang, J.; Cai, J.; Xian, Z.; Zhu, P.; Huang, M.; Wang, L.; et al. Aberrant expression of long noncoding RNA SNHG15 correlates with liver metastasis and poor survival in colorectal cancer. J. Cell Physiol. 2019, 234, 7032–7039. [Google Scholar] [CrossRef]
- Zhang, J.H.; Wei, H.W.; Yang, H.G. Long noncoding RNA SNHG15, a potential prognostic biomarker for hepatocellular carcinoma. Eur. Rev. Med. Pharm. Sci. 2016, 20, 1720–1724. [Google Scholar]
- Guo, X.B.; Yin, H.S.; Wang, J.Y. Evaluating the diagnostic and prognostic value of long non-coding RNA SNHG15 in pancreatic ductal adenocarcinoma. Eur. Rev. Med. Pharm. Sci. 2018, 22, 5892–5898. [Google Scholar]
- Li, S.P.; Xu, H.X.; Yu, Y.; He, J.D.; Wang, Z.; Xu, Y.J.; Wang, C.Y.; Zhang, H.M.; Zhang, R.X.; Zhang, J.J.; et al. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget 2016, 7, 42431–42446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, T.; Wu, J.; Hu, Y.; Zhang, M.; He, J. Long non-coding RNA HULC stimulates the epithelial-mesenchymal transition process and vasculogenic mimicry in human glioblastoma. Cancer Med. 2021, 10, 5270–5282. [Google Scholar] [CrossRef]
- Jin, C.; Shi, W.; Wang, F.; Shen, X.; Qi, J.; Cong, H.; Yuan, J.; Shi, L.; Zhu, B.; Luo, X.; et al. Long non-coding RNA HULC as a novel serum biomarker for diagnosis and prognosis prediction of gastric cancer. Oncotarget 2016, 7, 51763–51772. [Google Scholar] [CrossRef] [Green Version]
- Zang, X.; Gu, J.; Zhang, J.; Shi, H.; Hou, S.; Xu, X.; Chen, Y.; Zhang, Y.; Mao, F.; Qian, H.; et al. Exosome-transmitted lncRNA UFC1 promotes non-small-cell lung cancer progression by EZH2-mediated epigenetic silencing of PTEN expression. Cell Death Dis. 2020, 11, 215. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Sun, Q.Q.; Liu, T.X.; Lu, K.; Zhang, N.; Zhu, Y.; Chen, M. Serum lncRNA-UFC1 as a potential biomarker for diagnosis and prognosis of pancreatic cancer. Int. J. Clin. Exp. Pathol. 2019, 12, 4125–4129. [Google Scholar]
- Xu, J.H.; Wang, Y.; Xu, D. Hsa_circ_001569 is an unfavorable prognostic factor and promotes cell proliferation and metastasis by modulating PI3K-AKT pathway in breast cancer. Cancer Biomark. 2019, 25, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xue, L.; Song, C.; Liu, F.; Jiang, T.; Yang, X. Overexpression of circular RNA circ_001569 indicates poor prognosis in hepatocellular carcinoma and promotes cell growth and metastasis by sponging miR-411-5p and miR-432-5p. Biochem. Biophys. Res. Commun. 2018, 503, 2659–2665. [Google Scholar] [CrossRef]
- Xie, H.; Ren, X.; Xin, S.; Lan, X.; Lu, G.; Lin, Y.; Yang, S.; Zeng, Z.; Liao, W.; Ding, Y.Q.; et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget 2016, 7, 26680–26691. [Google Scholar] [CrossRef] [PubMed]
- Mai, S.; Zhang, Z.; Mi, W. Upregulation of circ_PVT1 and circ_001569 indicate unfavorable prognosis in colorectal cancer. Ann. Clin. Lab. Sci. 2021, 51, 55–60. [Google Scholar] [PubMed]
- Shen, X.; Chen, Y.; Li, J.; Huang, H.; Liu, C.; Zhou, N. Identification of Circ_001569 as a potential biomarker in the diagnosis and prognosis of pancreatic cancer. Technol. Cancer Res. Treat. 2021, 20, 1533033820983302. [Google Scholar] [CrossRef]
- Yang, F.; Liu, D.Y.; Guo, J.T.; Ge, N.; Zhu, P.; Liu, X.; Wang, S.; Wang, G.X.; Sun, S.Y. Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J. Gastroenterol. 2017, 23, 8345–8354. [Google Scholar] [CrossRef]
- Davis, M.E.; Zuckerman, J.E.; Choi, C.H.; Seligson, D.; Tolcher, A.; Alabi, C.A.; Yen, Y.; Heidel, J.D.; Ribas, A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature 2010, 464, 1067–1070. [Google Scholar] [CrossRef]
- Michalik, K.M.; You, X.; Manavski, Y.; Doddaballapur, A.; Zörnig, M.; Braun, T.; John, D.; Ponomareva, Y.; Chen, W.; Uchida, S.; et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ. Res. 2014, 114, 1389–1397. [Google Scholar] [CrossRef] [PubMed]
- Munagala, R.; Aqil, F.; Jeyabalan, J.; Kandimalla, R.; Wallen, M.; Tyagi, N.; Wilcher, S.; Yan, J.; Schultz, D.J.; Spencer, W.; et al. Exosome-mediated delivery of RNA and DNA for gene therapy. Cancer Lett. 2021, 505, 58–72. [Google Scholar] [CrossRef] [PubMed]
- Orefice, N.S. Development of new strategies using extracellular vesicles loaded with exogenous nucleic acid. Pharmaceutics 2020, 12, 705. [Google Scholar] [CrossRef]
- Maheshwari, R.; Tekade, M.; Gondaliya, P.; Kalia, K.; D’Emanuele, A.; Tekade, R.K. Recent advances in exosome-based nanovehicles as RNA interference therapeutic carriers. Nanomedicine 2017, 12, 2653–2675. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Zhang, G.; Zhang, L.; Hu, Y.; Zhang, K.; Sun, X.; Zhao, C.; Li, H.; Li, Y.M.; Zhao, J. Mesenchymal stem cells deliver exogenous miR-21 via exosomes to inhibit nucleus pulposus cell apoptosis and reduce intervertebral disc degeneration. J. Cell Mol. Med. 2018, 22, 261–276. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, D.; Hua, R.; Zhang, J.; Liu, W.; Huo, Y.; Cheng, Y.; Hong, J.; Sun, Y. Long non-coding RNAs expressed in pancreatic ductal adenocarcinoma and lncRNA BC008363 an independent prognostic factor in PDAC. Pancreatology 2014, 14, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Shao, F.; Huang, M.; Meng, F.; Huang, Q. Circular RNA Signature predicts gemcitabine resistance of pancreatic ductal adenocarcinoma. Front. Pharm. 2018, 9, 584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Yao, Y.; Gao, P.; Cui, Y. Upregulated circular RNA circ_0030235 predicts unfavorable prognosis in pancreatic ductal adenocarcinoma and facilitates cell progression by sponging miR-1253 and miR-1294. Biochem. Biophys. Res. Commun. 2019, 509, 138–142. [Google Scholar] [CrossRef]
- Zhou, M.; Ye, Z.; Gu, Y.; Tian, B.; Wu, B.; Li, J. Genomic analysis of drug resistant pancreatic cancer cell line by combining long non-coding RNA and mRNA expression profling. Int. J. Clin. Exp. Pathol. 2015, 8, 38–52. [Google Scholar]
- Zhan, H.X.; Wang, Y.; Li, C.; Xu, J.W.; Zhou, B.; Zhu, J.K.; Han, H.F.; Wang, L.; Wang, Y.S.; Hu, S.Y. LincRNA-ROR promotes invasion, metastasis and tumor growth in pancreatic cancer through activating ZEB1 pathway. Cancer Lett. 2016, 374, 261–271. [Google Scholar] [CrossRef]
- Li, C.; Zhao, Z.; Zhou, Z.; Liu, R. Linc-ROR confers gemcitabine resistance to pancreatic cancer cells via inducing autophagy and modulating the miR-124/PTBP1/PKM2 axis. Cancer Chemother. Pharm. 2016, 78, 1199–1207. [Google Scholar] [CrossRef]
- Liu, Y.; Xia, L.; Dong, L.; Wang, J.; Xiao, Q.; Yu, X.; Zhu, H. CircHIPK3 promotes gemcitabine (GEM) Resistance in pancreatic cancer cells by sponging miR-330-5p and targets RASSF1. Cancer Manag. Res. 2020, 12, 921–929. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Yu, Y.; Ding, F. Microarray analysis of circular RNA expression profiles associated with gemcitabine resistance in pancreatic cancer cells. Oncol. Rep. 2018, 40, 395–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef]
- Cheng, C.J.; Bahal, R.; Babar, I.A.; Pincus, Z.; Barrera, F.; Liu, C.; Svoronos, A.; Braddock, D.T.; Glazer, P.M.; Engelman, D.M.; et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature 2015, 518, 107–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, S.; Ansarullah; Kumar, D.; Jaggi, M.; Chauhan, S.C. Targeting microRNAs in pancreatic cancer: Microplayers in the big game. Cancer Res. 2013, 73, 6541–6547. [Google Scholar] [CrossRef] [Green Version]
- Fu, X.L.; Liu, D.J.; Yan, T.T.; Yang, J.Y.; Yang, M.W.; Li, J.; Huo, Y.M.; Liu, W.; Zhang, J.F.; Hong, J.; et al. Analysis of long non-coding RNA expression profiles in pancreatic ductal adenocarcinoma. Sci. Rep. 2016, 6, 33535. [Google Scholar] [CrossRef] [Green Version]
- Ye, Y.; Chen, J.; Zhou, Y.; Fu, Z.; Zhou, Q.; Wang, Y.; Gao, W.; Zheng, S.; Zhao, X.; Chen, T.; et al. High expression of AFAP1-AS1 is associated with poor survival and short-term recurrence in pancreatic ductal adenocarcinoma. J. Transl. Med. 2015, 13, 137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, C.; Ye, H.; Wang, W.; Sun, M.; Zhang, J.; Zhao, Z.; Jiang, G. Circular RNA ADAM9 facilitates the malignant behaviours of pancreatic cancer by sponging miR-217 and upregulating PRSS3 expression. Artif. Cells Nanomed. 2019, 47, 3920–3928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, J.; Zhang, C.; Chen, Y.; Gao, S. Downregulation of circular RNA circ-LDLRAD3 suppresses pancreatic cancer progression through miR-137-3p/PTN axis. Life Sci. 2019, 239, 116871. [Google Scholar] [CrossRef]
- Li, J.; Li, Z.; Jiang, P.; Peng, M.; Zhang, X.; Chen, K.; Liu, H.; Bi, H.; Liu, X.; Li, X. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J. Exp. Clin. Cancer Res. 2018, 37, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Yanfang, W.; Li, J.; Jiang, P.; Peng, T.; Chen, K.; Zhao, X.; Zhang, Y.; Zhen, P.; Zhu, J.; et al. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 2018, 432, 237–250. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 3, 209–249. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Xu, R.; Wang, C.; Qiu, J.; Ren, B.; You, L. Early screening and diagnosis strategies of pancreatic cancer: A comprehensive review. Cancer Commun. 2021. [Google Scholar] [CrossRef] [PubMed]


| lncRNA | Role | Functions | Related Signaling | Ref. |
|---|---|---|---|---|
| HOTAIR | Oncogenic. Up-regulated in cancer. | Promote proliferation and drug resistance. | EZH2, miR-34a. | [52,53,54,55,56,57] |
| MALAT-1 | Oncogenic. Up-regulated in cancer. | Promote cell growth, migration, invasion and metastasis. | Sox-2, EZH2, miR-216a, miR-217, miR-200c, Hippo-YAP. | [58,59,60,61,62,63,64,65,66] |
| H19 | Oncogenic. Up-regulated in cancer. | Promote cell proliferation, tumor growth and metastasis. | let-7, HMGA2, E2F, miR-675, miR194. | [67,68,69,70,71,72] |
| PVT1 | Oncogenic. Up-regulated in cancer. | Promote cell proliferation, migration, drug resistance. | p21, miR-448, miR-20a-5p, ULK1, miR519, HIF-1, YKT6, RAB7, VAMP3. | [73,74,75,76,77,78,79,80,81] |
| HOTTIP | Oncogenic. Up-regulated in cancer. | Promote cell growth, invasiveness and drug resistance. Modulate stem cells. | HOXA9, miR-137, HOXA13. | [82,83,84,85] |
| GAS5 | Tumor suppressive. Down-regulated in cancer. | Reverse EMT, inhibit metastasis and increase drug sensitivity. | CDK6, miR-221/SOCS3, miR-32, miR-181c. | [86,87,88,89] |
| MEG | Tumor suppressive. Down-regulated in cancer. | Inhibit cell proliferation, migration and invasion. | c-Met, PI3K/AKT. | [90,91,92,93,94] |
| circRNA | Role in Pancreatic Cancer | Functions | Related miRNA and Signaling | Ref. |
|---|---|---|---|---|
| ciRS-7 | Oncogenic. Up-regulated in cancer. | Promote cell proliferation and invasion. | Sponge miR-7. Regulate EGFR and STAT3. | [110] |
| circEIF6 | Oncogenic. Up-regulated in cancer. | Promote cell proliferation and inhibit apoptosis. | Sponge miR-557. Regulate SLC7A11 and PI3K/AKT. | [111] |
| circFOXK2 | Oncogenic. Up-regulated in cancer. | Promote cell proliferation, migration and invasion. | Sponge miR-942. Regulate ANK1, GDNF, PAX6, NUF2 and PDXK | [112] |
| circBFAR | Oncogenic. Up-regulated in cancer. | Promote cell proliferation and motility. | Sponge miR-34b-5p. Regulate MET/PI3K/Akt. | [113] |
| circ-ASH2L | Oncogenic. Up-regulated in cancer. | Induce cell proliferation, tumor invasion and angiogenesis. | Sponge miR-34a. Regulate Notch 1. | [114] |
| circRHOT1 | Oncogenic. Up-regulated in cancer. | Promote cell proliferation, migration and invasion. | Sponge miR-125a, miR-330, miR-26b and miR-382. Regulate E2F3. | [115,116] |
| circRNA_100782 | Oncogenic. Up-regulated in cancer. | Promote cell proliferation and tumor growth. | Sponge miR-124. Regulate IL6 and STAT3. | [117] |
| hsa_circ_0071036 | Oncogenic. Up-regulated in cancer. | Promote cell proliferation, invasion and tumor growth. | Sponge miR-489. | [118] |
| hsa_circ_0007534 | Oncogenic. Up-regulated in cancer. | Inhibit apoptosis. | Sponge miR-625 and miR-892b. Regulate Bcl-2 and caspase-3. | [119] |
| circNFIB1 | Tumor suppressor. Down-regulated in cancer. | Inhibit lymph node metastasis. | Sponge miR-486-5p. Regulate PIK3R1 and VEGF-C. | [120] |
| hsa_circ_001587 | Tumor suppressor. Down-regulated in cancer. | Inhibit cell proliferation, migration, invasion and angiogenesis. | Sponge miR-223. Regulate SLC4A4, MMP-2, MMP-9, MCM2 and VEGF. | [121] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Al Hallak, M.N.; Philip, P.A.; Azmi, A.S.; Mohammad, R.M. Non-Coding RNAs in Pancreatic Cancer Diagnostics and Therapy: Focus on lncRNAs, circRNAs, and piRNAs. Cancers 2021, 13, 4161. https://doi.org/10.3390/cancers13164161
Li Y, Al Hallak MN, Philip PA, Azmi AS, Mohammad RM. Non-Coding RNAs in Pancreatic Cancer Diagnostics and Therapy: Focus on lncRNAs, circRNAs, and piRNAs. Cancers. 2021; 13(16):4161. https://doi.org/10.3390/cancers13164161
Chicago/Turabian StyleLi, Yiwei, Mohammed Najeeb Al Hallak, Philip A. Philip, Asfar S. Azmi, and Ramzi M. Mohammad. 2021. "Non-Coding RNAs in Pancreatic Cancer Diagnostics and Therapy: Focus on lncRNAs, circRNAs, and piRNAs" Cancers 13, no. 16: 4161. https://doi.org/10.3390/cancers13164161
APA StyleLi, Y., Al Hallak, M. N., Philip, P. A., Azmi, A. S., & Mohammad, R. M. (2021). Non-Coding RNAs in Pancreatic Cancer Diagnostics and Therapy: Focus on lncRNAs, circRNAs, and piRNAs. Cancers, 13(16), 4161. https://doi.org/10.3390/cancers13164161







