Computational Approaches for Cancer-Fighting: From Gene Expression to Functional Foods
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Programmed Cell Death Candidate Genes and Their Response in Apoptosis-Inducing Nutrigenomics Treatments
3.2. Search for Gene Involvement in Cancer
3.3. Protein-Protein Interaction Patterns to Infer on Gene Product Functionality
3.4. Nutrients and Bioactive Compounds
4. Discussion
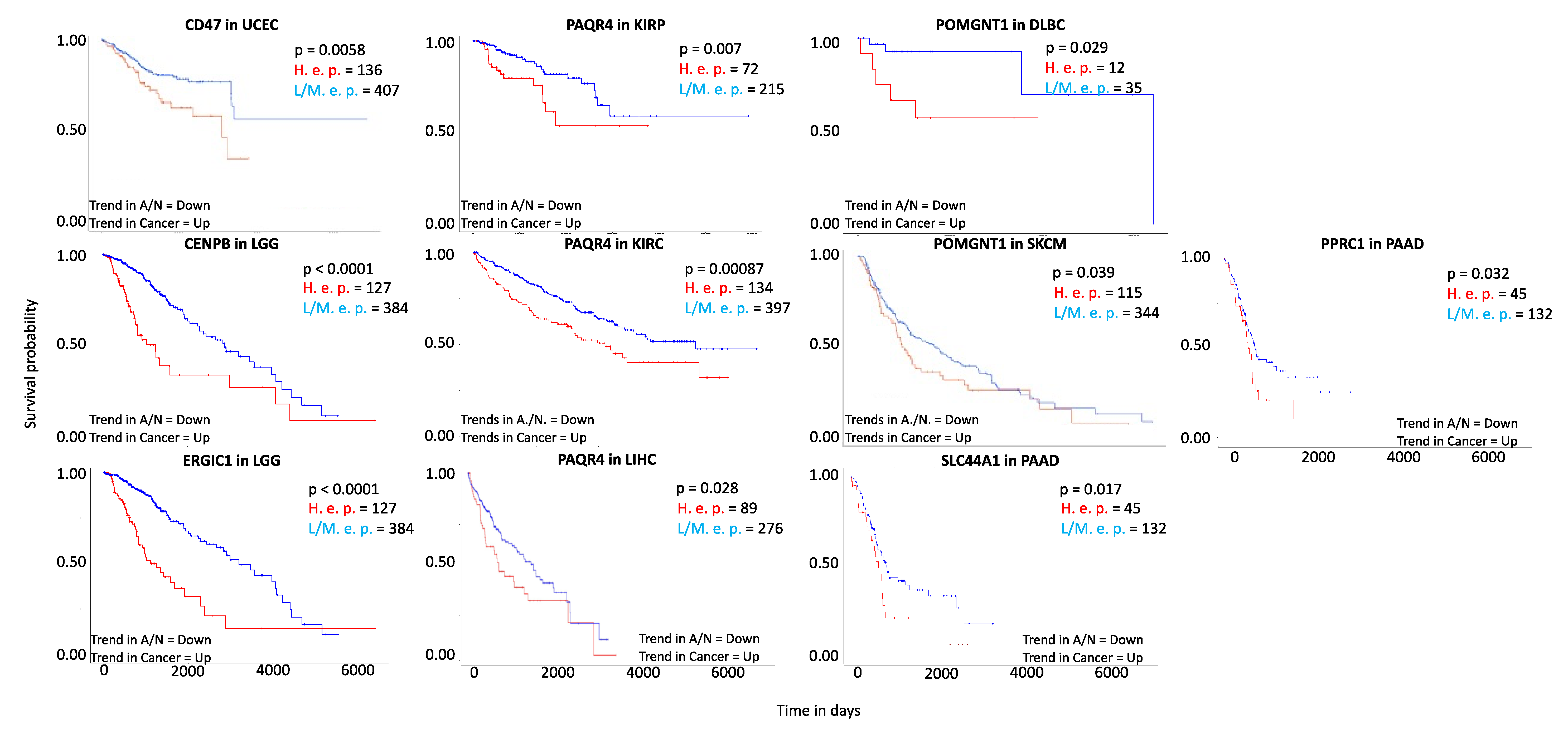
4.1. Novel Prognostic Genes in Different Cancer Types
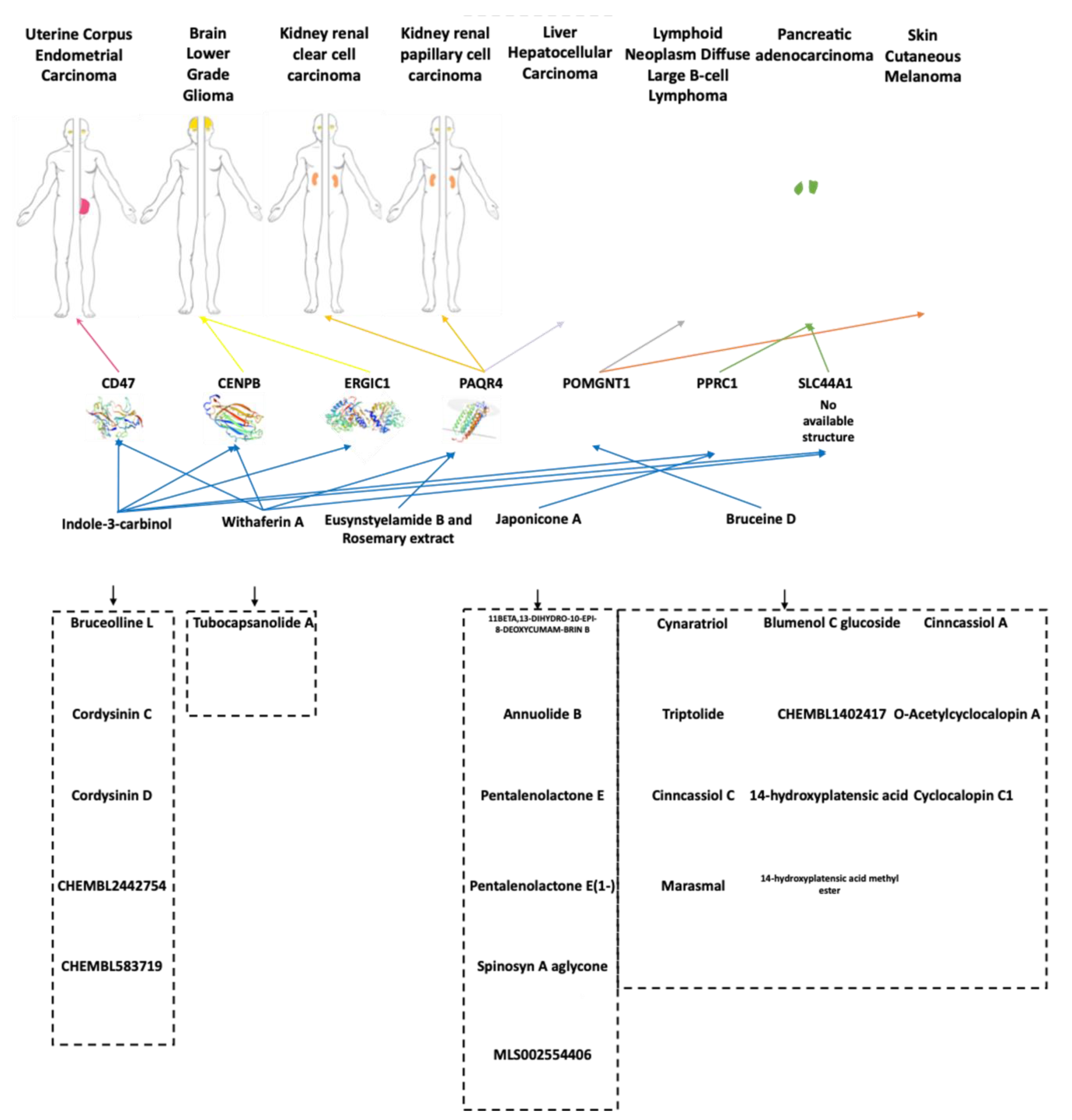
4.2. Candidate Markers and Associated Functional Partners
4.3. Putative Effectors from Nutritional Treatments
4.4. Opportunities from Functional Foods in Cancer Treatments
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Rock, C.L.; Thomson, C.; Gansler, T.; Gapstur, S.M.; McCullough, M.L.; Patel, A.V.; Andrews, K.S.; Bandera, E.V.; Spees, C.K.; Robien, K.; et al. American Cancer Society guideline for diet and physical activity for cancer prevention. CA Cancer J. Clin. 2020, 70, 245–271. [Google Scholar] [CrossRef] [PubMed]
- Hofmarcher, T.; Lindgren, P.; Wilking, N.; Jönsson, B. The cost of cancer in Europe 2018. Eur. J. Cancer 2020, 129, 41–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mariotto, A.B.; Enewold, L.; Zhao, J.; Zeruto, C.A.; Yabroff, K.R. Medical Care Costs Associated with Cancer Survivorship in the United States. Cancer Epidemiol. Biomark. Prev. 2020, 29, 1304–1312. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Bajari, R.; Andric, D.; Gerthoffert, F.; Lepsa, A.; Nahal-Bose, H.; Stein, L.D.; Ferretti, V. The International Cancer Genome Consortium Data Portal. Nat. Biotechnol. 2019, 37, 367–369. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Corrigendum: Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2013, 494, 506. [Google Scholar] [CrossRef]
- Hoadley, K.A.; Yau, C.; Wolf, D.M.; Cherniack, A.D.; Tamborero, D.; Ng, S.; Leiserson, M.D.M.; Niu, B.; McLellan, M.D.; Uzunangelov, V.; et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 2014, 158, 929–944. [Google Scholar] [CrossRef] [Green Version]
- Hoadley, K.A.; Yau, C.; Hinoue, T.; Wolf, D.M.; Lazar, A.J.; Drill, E.; Shen, R.; Taylor, A.M.; Cherniack, A.D.; Thorsson, V.; et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell 2018, 173, 291–304.e6. [Google Scholar] [CrossRef] [Green Version]
- Campbell, P.J.; Getz, G.; Korbel, J.O.; Stuart, J.M.; Jennings, J.L.; Stein, L.D.; Perry, M.D.; Nahal-Bose, H.K.; Ouellette, B.F.F.; Li, C.H.; et al. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef] [Green Version]
- Clinton, S.K.; Giovannucci, E.L.; Hursting, S.D. The World Cancer Research Fund/American Institute for Cancer Research Third Expert Report on Diet, Nutrition, Physical Activity, and Cancer: Impact and Future Directions. J. Nutr. 2020, 150, 663–671. [Google Scholar] [CrossRef]
- Azab, A.; Nassar, A.; Azab, A.N. Anti-Inflammatory Activity of Natural Products. Molecules 2016, 21, 1321. [Google Scholar] [CrossRef] [PubMed]
- Serafini, M.; Peluso, I.; Raguzzini, A. Flavonoids as anti-inflammatory agents. Proc. Nutr. Soc. 2010, 69, 273–278. [Google Scholar] [CrossRef] [Green Version]
- Arts, I.C.W.; Hollman, P.C.H. Polyphenols and disease risk in epidemiologic studies. Am. J. Clin. Nutr. 2005, 81, 317S–325S. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Ouyang, Y.Y.; Liu, J.; Zhao, G. Flavonoid intake and risk of CVD: A systematic review and meta-analysis of prospective cohort studies. Br. J. Nutr. 2014, 111, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monticolo, F.; Palomba, E.; De Santis, R.; Assentato, L.; Triscino, V.; Langella, M.C.; Lanzotti, V.; Chiusano, M.L. Anti-HCoV: A web resource to collect natural compounds against human coronaviruses. Trends Food Sci. Technol. 2020, 106, 1–11. [Google Scholar] [CrossRef]
- Baur, J.A.; Sinclair, D.A. Therapeutic potential of resveratrol: The in vivo evidence. Nat. Rev. Drug Discov. 2006, 5, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Astorg, P. Food carotenoids and cancer prevention: An overview of current research. Trends Food Sci. Technol. 1997, 8, 406–413. [Google Scholar] [CrossRef]
- Oyagbemi, A.A.; Saba, A.B.; Ibraheem, A.O. Curcumin: From food spice to cancer prevention. Asian Pac. J. Cancer Prev. 2009, 10, 963–967. [Google Scholar]
- Sotos-Prieto, M.; Bhupathiraju, S.N.; Mattei, J.; Fung, T.T.; Li, Y.; Pan, A.; Willett, W.C.; Rimm, E.B.; Hu, F.B. Association of Changes in Diet Quality with Total and Cause-Specific Mortality. N. Engl. J. Med. 2017, 377, 143–153. [Google Scholar] [CrossRef]
- Shefchek, K.A.; Harris, N.L.; Gargano, M.; Matentzoglu, N.; Unni, D.; Brush, M.; Keith, D.; Conlin, T.; Vasilevsky, N.; Zhang, X.A.; et al. The Monarch Initiative in 2019: An integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res. 2020, 48, D704–D715. [Google Scholar] [CrossRef] [Green Version]
- Davis, A.P.; Grondin, C.J.; Johnson, R.J.; Sciaky, D.; Wiegers, J.; Wiegers, T.C.; Mattingly, C.J. Comparative Toxicogenomics Database (CTD): Update 2021. Nucleic Acids Res. 2021, 49, D1138–D1143. [Google Scholar] [CrossRef] [PubMed]
- Chan, L.; Vasilevsky, N.; Thessen, A.; McMurry, J.; Haendel, M. The landscape of nutri-informatics: A review of current resources and challenges for integrative nutrition research. Database 2021, 2021, baab003. [Google Scholar] [CrossRef] [PubMed]
- Martín-Hernández, R.; Reglero, G.; Ordovás, J.M.; Dávalos, A. NutriGenomeDB: A nutrigenomics exploratory and analytical platform. Database 2019, 2019, baz097. [Google Scholar] [CrossRef] [Green Version]
- Monticolo, F.; Palomba, E.; Chiusano, M.L. Identification of Novel Potential Genes Involved in Cancer by Integrated Comparative Analyses. Int. J. Mol. Sci. 2020, 21, 9560. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [Green Version]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zoete, V.; Daina, A.; Bovigny, C.; Michielin, O. SwissSimilarity: A Web Tool for Low to Ultra High Throughput Ligand-Based Virtual Screening. J. Chem. Inf. Model. 2016, 56, 1399–1404. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rouhimoghadam, M.; Safarian, S.; Carroll, J.S.; Sheibani, N.; Bidkhori, G. Tamoxifen-Induced Apoptosis of MCF-7 Cells via GPR30/PI3K/MAPKs Interactions: Verification by ODE Modeling and RNA Sequencing. Front. Physiol. 2018, 9, 907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, Y.; Zhang, F.; Wang, Y.; Lee, E.M.; Choi, I.Y.; Lim, H.; Mirakhori, F.; Li, R.; Huang, L.; Xu, T.; et al. Zika virus directly infects peripheral neurons and induces cell death. Nat. Neurosci. 2017, 20, 1209–1212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sant, D.W.; Mustafi, S.; Gustafson, C.B.; Chen, J.; Slingerland, J.M.; Wang, G. Vitamin C promotes apoptosis in breast cancer cells by increasing TRAIL expression. Sci. Rep. 2018, 8, 5306. [Google Scholar] [CrossRef]
- Sun, G.; Guzman, E.; Balasanyan, V.; Conner, C.M.; Wong, K.; Zhou, H.R.; Kosik, K.S.; Montell, D.J. A molecular signature for anastasis, recovery from the brink of apoptotic cell death. J. Cell Biol. 2017, 216, 3355–3368. [Google Scholar] [CrossRef] [Green Version]
- Pulikkan, J.A.; Hegde, M.; Ahmad, H.M.; Belaghzal, H.; Illendula, A.; Yu, J.; O’Hagan, K.; Ou, J.; Muller-Tidow, C.; Wolfe, S.A.; et al. CBFβ-SMMHC Inhibition Triggers Apoptosis by Disrupting MYC Chromatin Dynamics in Acute Myeloid Leukemia. Cell 2018, 174, 1325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eriksson, M.; Peña-Martínez, P.; Ramakrishnan, R.; Chapellier, M.; Högberg, C.; Glowacki, G.; Orsmark-Pietras, C.; Velasco-Hernández, T.; Lazarević, V.L.; Juliusson, G.; et al. Agonistic targeting of TLR1/TLR2 induces p38 MAPK-dependent apoptosis and NFκB-dependent differentiation of AML cells. Blood Adv. 2017, 1, 2046–2057. [Google Scholar] [CrossRef] [Green Version]
- Sareddy, G.R.; Viswanadhapalli, S.; Surapaneni, P.; Suzuki, T.; Brenner, A.; Vadlamudi, R.K. Novel KDM1A inhibitors induce differentiation and apoptosis of glioma stem cells via unfolded protein response pathway. Oncogene 2017, 36, 2423–2434. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [Green Version]
- Dumont, P.; Ingrassia, L.; Rouzeau, S.; Ribaucour, F.; Thomas, S.; Roland, I.; Darro, F.; Lefranc, F.; Kiss, R. The Amaryllidaceae isocarbostyril narciclasine induces apoptosis by activation of the death receptor and/or mitochondrial pathways in cancer cells but not in normal fibroblasts. Neoplasia 2007, 9, 766–776. [Google Scholar] [CrossRef] [Green Version]
- Moore, J.; Yousef, M.; Tsiani, E. Anticancer Effects of Rosemary (Rosmarinus officinalis L.) Extract and Rosemary Extract Polyphenols. Nutrients 2016, 8, 731. [Google Scholar] [CrossRef]
- Mazzio, E.A.; Soliman, K.F.A. Whole-transcriptomic Profile of SK-MEL-3 Melanoma Cells Treated with the Histone Deacetylase Inhibitor: Trichostatin A. Cancer Genom. Proteom. 2018, 15, 349–364. [Google Scholar] [CrossRef]
- Szarc vel Szic, K.; Op de Beeck, K.; Ratman, D.; Wouters, A.; Beck, I.M.; Declerck, K.; Heyninck, K.; Fransen, E.; Bracke, M.; De Bosscher, K.; et al. Pharmacological levels of Withaferin A (Withania somnifera) trigger clinically relevant anticancer effects specific to triple negative breast cancer cells. PLoS ONE 2014, 9, e87850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caruso, J.A.; Campana, R.; Wei, C.; Su, C.-H.; Hanks, A.M.; Bornmann, W.G.; Keyomarsi, K. Indole-3-carbinol and its N-alkoxy derivatives preferentially target ERα-positive breast cancer cells. Cell Cycle 2014, 13, 2587–2599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agyeman, A.S.; Chaerkady, R.; Shaw, P.G.; Davidson, N.E.; Visvanathan, K.; Pandey, A.; Kensler, T.W. Transcriptomic and proteomic profiling of KEAP1 disrupted and sulforaphane-treated human breast epithelial cells reveals common expression profiles. Breast Cancer Res. Treat. 2012, 132, 175–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batova, A.; Altomare, D.; Creek, K.E.; Naviaux, R.K.; Wang, L.; Li, K.; Green, E.; Williams, R.; Naviaux, J.C.; Diccianni, M.; et al. Englerin A induces an acute inflammatory response and reveals lipid metabolism and ER stress as targetable vulnerabilities in renal cell carcinoma. PLoS ONE 2017, 12, e0172632. [Google Scholar] [CrossRef]
- Naciff, J.M.; Khambatta, Z.S.; Carr, G.J.; Tiesman, J.P.; Singleton, D.W.; Khan, S.A.; Daston, G.P. Dose- and Time-Dependent Transcriptional Response of Ishikawa Cells Exposed to Genistein. Toxicol. Sci. 2016, 151, 71–87. [Google Scholar] [CrossRef] [Green Version]
- Tang, J.-J.; He, Q.-R.; Dong, S.; Guo, X.; Wang, Y.-G.; Lei, B.-L.; Tian, J.-M.; Gao, J.-M. Diversity Modification and Structure-Activity Relationships of Two Natural Products 1β-hydroxy Alantolactone and Ivangustin as Potent Cytotoxic Agents. Sci. Rep. 2018, 8, 1722. [Google Scholar] [CrossRef] [Green Version]
- The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaulton, A.; Bellis, L.J.; Bento, A.P.; Chambers, J.; Davies, M.; Hersey, A.; Light, Y.; McGlinchey, S.; Michalovich, D.; Al-Lazikani, B.; et al. ChEMBL: A large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012, 40, D1100–D1107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Matos, P.; Alcántara, R.; Dekker, A.; Ennis, M.; Hastings, J.; Haug, K.; Spiteri, I.; Turner, S.; Steinbeck, C. Chemical Entities of Biological Interest: An update. Nucleic Acids Res. 2010, 38, D249–D254. [Google Scholar] [CrossRef] [Green Version]
- Chan, W.K.B.; Zhang, H.; Yang, J.; Brender, J.R.; Hur, J.; Özgür, A.; Zhang, Y. GLASS: A comprehensive database for experimentally validated GPCR-ligand associations. Bioinformatics 2015, 31, 3035–3042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef]
- de Oliveira Só, Y.A.; de Abreu Silva, M.; Carvalho, F.M.; Kiametis, A.S.; Gargano, R. Combining electronic properties and virtual screening for the development of new antioxidants: Trolox-like compounds as application example. Int. J. Quantum Chem. 2020, 120, e26194. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Egan, W.J.; Merz, K.M.J.; Baldwin, J.J. Prediction of drug absorption using multivariate statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef]
- Muegge, I.; Heald, S.L.; Brittelli, D. Simple selection criteria for drug-like chemical matter. J. Med. Chem. 2001, 44, 1841–1846. [Google Scholar] [CrossRef]
- Baell, J.B.; Holloway, G.A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010, 53, 2719–2740. [Google Scholar] [CrossRef] [Green Version]
- Brenk, R.; Schipani, A.; James, D.; Krasowski, A.; Gilbert, I.H.; Frearson, J.; Wyatt, P.G. Lessons learnt from assembling screening libraries for drug discovery for neglected diseases. ChemMedChem 2008, 3, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Teague, S.J.; Davis, A.M.; Leeson, P.D.; Oprea, T. The Design of Leadlike Combinatorial Libraries. Angew. Chem. Int. Ed. 1999, 38, 3743–3748. [Google Scholar] [CrossRef]
- Sick, E.; Jeanne, A.; Schneider, C.; Dedieu, S.; Takeda, K.; Martiny, L. CD47 update: A multifaceted actor in the tumour microenvironment of potential therapeutic interest. Br. J. Pharmacol. 2012, 167, 1415–1430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levkau, B.; Herren, B.; Koyama, H.; Ross, R.; Raines, E.W. Caspase-mediated cleavage of focal adhesion kinase pp125FAK and disassembly of focal adhesions in human endothelial cell apoptosis. J. Exp. Med. 1998, 187, 579–586. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Zhao, J.; Jia, C.; Zhou, L.; Cai, Y.; Ni, J.; Ma, J.; Zheng, M.; Lu, A. FPR2 enhances colorectal cancer progression by promoting EMT process. Neoplasma 2019, 66, 785–791. [Google Scholar] [CrossRef]
- Earnshaw, W.C.; Sullivan, K.F.; Machlin, P.S.; Cooke, C.A.; Kaiser, D.A.; Pollard, T.D.; Rothfield, N.F.; Cleveland, D.W. Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J. Cell Biol. 1987, 104, 817–829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fatokun, A.A.; Dawson, V.L.; Dawson, T.M. Parthanatos: Mitochondrial-linked mechanisms and therapeutic opportunities. Br. J. Pharmacol. 2014, 171, 2000–2016. [Google Scholar] [CrossRef] [Green Version]
- D’Amours, D.; Sallmann, F.R.; Dixit, V.M.; Poirier, G.G. Gain-of-function of poly(ADP-ribose) polymerase-1 upon cleavage by apoptotic proteases: Implications for apoptosis. J. Cell Sci. 2001, 114, 3771–3778. [Google Scholar] [CrossRef]
- Chaitanya, G.V.; Steven, A.J.; Babu, P.P. PARP-1 cleavage fragments: Signatures of cell-death proteases in neurodegeneration. Cell Commun. Signal. 2010, 8, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engbrecht, M.; Mangerich, A. The Nucleolus and PARP1 in Cancer Biology. Cancers 2020, 12, 1813. [Google Scholar] [CrossRef] [PubMed]
- Pascal, J.M. The comings and goings of PARP-1 in response to DNA damage. DNA Repair 2018, 71, 177–182. [Google Scholar] [CrossRef]
- Alomari, M. TRIM21—A potential novel therapeutic target in cancer. Pharmacol. Res. 2021, 165, 105443. [Google Scholar] [CrossRef]
- Breuza, L.; Halbeisen, R.; Jenö, P.; Otte, S.; Barlowe, C.; Hong, W.; Hauri, H.-P. Proteomics of endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membranes from brefeldin A-treated HepG2 cells identifies ERGIC-32, a new cycling protein that interacts with human Erv46. J. Biol. Chem. 2004, 279, 47242–47253. [Google Scholar] [CrossRef] [Green Version]
- Adélaïde, J.; Finetti, P.; Bekhouche, I.; Repellini, L.; Geneix, J.; Sircoulomb, F.; Charafe-Jauffret, E.; Cervera, N.; Desplans, J.; Parzy, D.; et al. Integrated profiling of basal and luminal breast cancers. Cancer Res. 2007, 67, 11565–11575. [Google Scholar] [CrossRef] [Green Version]
- Ameri, K.; Rajah, A.M.; Nguyen, V.; Sanders, T.A.; Jahangiri, A.; Delay, M.; Donne, M.; Choi, H.J.; Tormos, K.V.; Yeghiazarians, Y.; et al. Nuclear localization of the mitochondrial factor HIGD1A during metabolic stress. PLoS ONE 2013, 8, e62758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Y.T.; Hu, T.; Arterburn, M.; Boyle, B.; Bright, J.M.; Emtage, P.C.; Funk, W.D. PAQR proteins: A novel membrane receptor family defined by an ancient 7-transmembrane pass motif. J. Mol. Evol. 2005, 61, 372–380. [Google Scholar] [CrossRef]
- Guibal, F.C.; Moog-Lutz, C.; Smolewski, P.; Di Gioia, Y.; Darzynkiewicz, Z.; Lutz, P.G.; Cayre, Y.E. ASB-2 inhibits growth and promotes commitment in myeloid leukemia cells. J. Biol. Chem. 2002, 277, 218–224. [Google Scholar] [CrossRef] [Green Version]
- Muhammad, B.A.; Almozyan, S.; Babaei-Jadidi, R.; Onyido, E.K.; Saadeddin, A.; Kashfi, S.H.; Spencer-Dene, B.; Ilyas, M.; Lourdusamy, A.; Behrens, A.; et al. FLYWCH1, a Novel Suppressor of Nuclear β-Catenin, Regulates Migration and Morphology in Colorectal Cancer. Mol. Cancer Res. 2018, 16, 1977–1990. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, A.; Kobayashi, K.; Manya, H.; Taniguchi, K.; Kano, H.; Mizuno, M.; Inazu, T.; Mitsuhashi, H.; Takahashi, S.; Takeuchi, M.; et al. Muscular dystrophy and neuronal migration disorder caused by mutations in a glycosyltransferase, POMGnT1. Dev. Cell 2001, 1, 717–724. [Google Scholar] [CrossRef] [Green Version]
- Kuwabara, N.; Manya, H.; Yamada, T.; Tateno, H.; Kanagawa, M.; Kobayashi, K.; Akasaka-Manya, K.; Hirose, Y.; Mizuno, M.; Ikeguchi, M.; et al. Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of α-dystroglycan. Proc. Natl. Acad. Sci. USA 2016, 113, 9280–9285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sgambato, A.; Brancaccio, A. The dystroglycan complex: From biology to cancer. J. Cell Physiol. 2005, 205, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Côté, P.D.; Moukhles, H.; Lindenbaum, M.; Carbonetto, S. Chimaeric mice deficient in dystroglycans develop muscular dystrophy and have disrupted myoneural synapses. Nat. Genet. 1999, 23, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Andersson, U.; Scarpulla, R.C. Pgc-1-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor 1-dependent transcription in mammalian cells. Mol. Cell Biol. 2001, 21, 3738–3749. [Google Scholar] [CrossRef] [Green Version]
- Yoon, A.; Peng, G.; Brandenburger, Y.; Zollo, O.; Xu, W.; Rego, E.; Ruggero, D. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science 2006, 312, 902–906. [Google Scholar] [CrossRef]
- Bellodi, C.; Krasnykh, O.; Haynes, N.; Theodoropoulou, M.; Peng, G.; Montanaro, L.; Ruggero, D. Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis. Cancer Res. 2010, 70, 6026–6035. [Google Scholar] [CrossRef] [Green Version]
- Son, S.W.; Chau, G.C.; Kim, S.-T.; Um, S.H. Vacuolar H(+)-ATPase Subunit V0C Regulates Aerobic Glycolysis of Esophageal Cancer Cells via PKM2 Signaling. Cells 2019, 8, 1137. [Google Scholar] [CrossRef] [Green Version]
- Michel, V.; Bakovic, M. The solute carrier 44A1 is a mitochondrial protein and mediates choline transport. FASEB J. 2009, 23, 2749–2758. [Google Scholar] [CrossRef]
- Li, M.; Hao, S.; Li, C.; Xiao, H.; Sun, L.; Yu, Z.; Zhang, N.; Xiong, Y.; Zhao, D.; Yin, Y. Elevated SH3BP5 Correlates with Poor Outcome and Contributes to the Growth of Acute Myeloid Leukemia Cells. Biomolecules 2019, 9, 505. [Google Scholar] [CrossRef] [Green Version]
- Ding, L.; Bailey, M.H.; Porta-Pardo, E.; Thorsson, V.; Colaprico, A.; Bertrand, D.; Gibbs, D.L.; Weerasinghe, A.; Huang, K.-L.; Tokheim, C.; et al. Perspective on Oncogenic Processes at the End of the Beginning of Cancer Genomics. Cell 2018, 173, 305–320.e10. [Google Scholar] [CrossRef] [Green Version]
- Jaiswal, S.; Jamieson, C.H.M.; Pang, W.W.; Park, C.Y.; Chao, M.P.; Majeti, R.; Traver, D.; van Rooijen, N.; Weissman, I.L. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell 2009, 138, 271–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betancur, P.A.; Abraham, B.J.; Yiu, Y.Y.; Willingham, S.B.; Khameneh, F.; Zarnegar, M.; Kuo, A.H.; McKenna, K.; Kojima, Y.; Leeper, N.J.; et al. A CD47-associated super-enhancer links pro-inflammatory signalling to CD47 upregulation in breast cancer. Nat. Commun. 2017, 8, 14802. [Google Scholar] [CrossRef] [PubMed]
- Chao, M.P.; Alizadeh, A.A.; Tang, C.; Myklebust, J.H.; Varghese, B.; Gill, S.; Jan, M.; Cha, A.C.; Chan, C.K.; Tan, B.T.; et al. Anti-CD47 antibody synergizes with rituximab to promote phagocytosis and eradicate non-Hodgkin lymphoma. Cell 2010, 142, 699–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Xu, Z.; Guo, S.; Zhang, L.; Sharma, A.; Robertson, G.P.; Huang, L. Intravenous delivery of siRNA targeting CD47 effectively inhibits melanoma tumor growth and lung metastasis. Mol. Ther. 2013, 21, 1919–1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Chang, Y.; He, X.; Cai, Y.; Jiang, H.; Jia, R.; Leng, J. CD47 Enhances Cell Viability and Migration Ability but Inhibits Apoptosis in Endometrial Carcinoma Cells via the PI3K/Akt/mTOR Signaling Pathway. Front. Oncol. 2020, 10, 1525. [Google Scholar] [CrossRef] [PubMed]
- Tsai, R.K.; Discher, D.E. Inhibition of “self” engulfment through deactivation of myosin-II at the phagocytic synapse between human cells. J. Cell Biol. 2008, 180, 989–1003. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.-Y.; Ye, Z.-H.; Huang, M.-Y.; Lu, J.-J. Regulation of CD47 expression in cancer cells. Transl. Oncol. 2020, 13, 100862. [Google Scholar] [CrossRef]
- Anaganti, S.; Fernández-Cuesta, L.; Langerød, A.; Hainaut, P.; Olivier, M. p53-Dependent repression of focal adhesion kinase in response to estradiol in breast cancer cell-lines. Cancer Lett. 2011, 300, 215–224. [Google Scholar] [CrossRef]
- Mehlen, P.; Puisieux, A. Metastasis: A question of life or death. Nat. Rev. Cancer 2006, 6, 449–458. [Google Scholar] [CrossRef]
- Ye, R.D.; Boulay, F.; Wang, J.M.; Dahlgren, C.; Gerard, C.; Parmentier, M.; Serhan, C.N.; Murphy, P.M. International Union of Basic and Clinical Pharmacology. LXXIII. Nomenclature for the formyl peptide receptor (FPR) family. Pharmacol. Rev. 2009, 61, 119–161. [Google Scholar] [CrossRef]
- Coffelt, S.B.; Tomchuck, S.L.; Zwezdaryk, K.J.; Danka, E.S.; Scandurro, A.B. Leucine leucine-37 uses formyl peptide receptor-like 1 to activate signal transduction pathways, stimulate oncogenic gene expression, and enhance the invasiveness of ovarian cancer cells. Mol. Cancer Res. 2009, 7, 907–915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakravarti, N.; Peddareddigari, V.G.R.; Warneke, C.L.; Johnson, M.M.; Overwijk, W.W.; Hwu, P.; Prieto, V.G. Differential expression of the G-protein-coupled formyl Peptide receptor in melanoma associates with aggressive phenotype. Am. J. Dermatopathol. 2013, 35, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Yao, X.; Chen, K.; Wang, X.; Zhou, J.; Gong, W.; Yoshimura, T.; Huang, J.; Wang, R.; Wu, Y.; et al. The G-protein coupled chemoattractant receptor FPR2 promotes malignant phenotype of human colon cancer cells. Am. J. Cancer Res. 2016, 6, 2599–2610. [Google Scholar]
- Earnshaw, W.; Bordwell, B.; Marino, C.; Rothfield, N. Three human chromosomal autoantigens are recognized by sera from patients with anti-centromere antibodies. J. Clin. Investig. 1986, 77, 426–430. [Google Scholar] [CrossRef] [Green Version]
- Atalay, C.; Atalay, G.; Yilmaz, K.B.; Altinok, M. The role of anti-CENP-B and anti-SS-B antibodies in breast cancer. Neoplasma 2005, 52, 32–35. [Google Scholar]
- Briasoulis, E.; Kamposioras, K.; Tzovaras, V.; Pafitanis, G.; Kostoula, A.; Mavridis, A.; Pavlidis, N. CENP-B specific anti-centromere autoantibodies heralding small-cell lung cancer. A case study and review of the literature. Lung Cancer 2008, 60, 302–306. [Google Scholar] [CrossRef] [PubMed]
- Rajman, L.; Chwalek, K.; Sinclair, D.A. Therapeutic Potential of NAD-Boosting Molecules: The In Vivo Evidence. Cell Metab. 2018, 27, 529–547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibson, B.A.; Kraus, W.L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012, 13, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Bryant, H.E.; Schultz, N.; Thomas, H.D.; Parker, K.M.; Flower, D.; Lopez, E.; Kyle, S.; Meuth, M.; Curtin, N.J.; Helleday, T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005, 434, 913–917. [Google Scholar] [CrossRef] [PubMed]
- Farmer, H.; McCabe, N.; Lord, C.J.; Tutt, A.N.J.; Johnson, D.A.; Richardson, T.B.; Santarosa, M.; Dillon, K.J.; Hickson, I.; Knights, C.; et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005, 434, 917–921. [Google Scholar] [CrossRef]
- Vainio, P.; Mpindi, J.-P.; Kohonen, P.; Fey, V.; Mirtti, T.; Alanen, K.A.; Perälä, M.; Kallioniemi, O.; Iljin, K. High-throughput transcriptomic and RNAi analysis identifies AIM1, ERGIC1, TMED3 and TPX2 as potential drug targets in prostate cancer. PLoS ONE 2012, 7, e39801. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Sun, X.; Sun, B.; Li, T.; Wu, G.; Li, Y.; Chen, L.; Liu, Q.; Cui, M.; Zhou, Z. ARRDC3 suppresses colorectal cancer progression through destabilizing the oncoprotein YAP. FEBS Lett. 2018, 592, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Soung, Y.H.; Pruitt, K.; Chung, J. Epigenetic silencing of ARRDC3 expression in basal-like breast cancer cells. Sci. Rep. 2014, 4, 3846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Y.; Lin, Z.-Y.; Xie, J.-J.; Jiang, F.-N.; Chen, C.-J.; Li, J.-X.; Zhou, X.; Zhong, W.-D. ARRDC3 Inhibits the Progression of Human Prostate Cancer Through ARRDC3-ITGβ4 Pathway. Curr. Mol. Med. 2017, 17, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Arakaki, A.K.S.; Pan, W.-A.; Lin, H.; Trejo, J. The α-arrestin ARRDC3 suppresses breast carcinoma invasion by regulating G protein-coupled receptor lysosomal sorting and signaling. J. Biol. Chem. 2018, 293, 3350–3362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, J.; Shi, Q.; Li, W.; Mu, X.; Peng, J.; Li, M.; Chen, M.; Huang, H.; Wang, C.; Gao, K.; et al. ARRDC1 and ARRDC3 act as tumor suppressors in renal cell carcinoma by facilitating YAP1 degradation. Am. J. Cancer Res. 2018, 8, 132–143. [Google Scholar]
- An, H.-J.; Cho, G.; Lee, J.-O.; Paik, S.-G.; Kim, Y.S.; Lee, H. Higd-1a interacts with Opa1 and is required for the morphological and functional integrity of mitochondria. Proc. Natl. Acad. Sci. USA 2013, 110, 13014–13019. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Han, R.; Ling, Z.-Q.; Zhang, F.; Hou, Y.; You, X.; Huang, M.; Zhao, Z.; Wang, Z.; Chen, Y. PAQR4 has a tumorigenic effect in human breast cancers in association with reduced CDK4 degradation. Carcinogenesis 2018, 39, 439–446. [Google Scholar] [CrossRef]
- Pedersen, L.; Panahandeh, P.; Siraji, M.I.; Knappskog, S.; Lønning, P.E.; Gordillo, R.; Scherer, P.E.; Molven, A.; Teigen, K.; Halberg, N. Golgi-Localized PAQR4 Mediates Antiapoptotic Ceramidase Activity in Breast Cancer. Cancer Res. 2020, 80, 2163–2174. [Google Scholar] [CrossRef] [Green Version]
- Wu, B.; Liu, R. PAQR4 promotes cell proliferation and metastasis through the CDK4-pRB-E2F1 pathway in non-small-cell lung cancer. Onco. Targets Ther. 2019, 12, 3625–3633. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Gao, M.; Guo, X.; Zhang, H.; Jiang, F. Breviscapine suppresses the growth and metastasis of prostate cancer through regulating PAQR4-mediated PI3K/Akt pathway. Biomed. Pharmacother. 2020, 127, 110223. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Xue, Y.; Chen, Q.; Chen, H.; Zhang, X.; Wang, L.; Han, C.; Que, S.; Lou, M.; Lan, J. PomGnT1 enhances temozolomide resistance by activating epithelial-mesenchymal transition signaling in glioblastoma. Oncol. Rep. 2017, 38, 2911–2918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibraghimov-Beskrovnaya, O.; Ervasti, J.M.; Leveille, C.J.; Slaughter, C.A.; Sernett, S.W.; Campbell, K.P. Primary structure of dystrophin-associated glycoproteins linking dystrophin to the extracellular matrix. Nature 1992, 355, 696–702. [Google Scholar] [CrossRef] [PubMed]
- Savagner, F.; Mirebeau, D.; Jacques, C.; Guyetant, S.; Morgan, C.; Franc, B.; Reynier, P.; Malthièry, Y. PGC-1-related coactivator and targets are upregulated in thyroid oncocytoma. Biochem. Biophys. Res. Commun. 2003, 310, 779–784. [Google Scholar] [CrossRef]
- Safra, M.; Nir, R.; Farouq, D.; Vainberg Slutskin, I.; Schwartz, S. Corrigendum: TRUB1 is the predominant pseudouridine synthase acting on mammalian mRNA via a predictable and conserved code. Genome Res. 2017, 27, 1460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurimoto, R.; Chiba, T.; Ito, Y.; Matsushima, T.; Yano, Y.; Miyata, K.; Yashiro, Y.; Suzuki, T.; Tomita, K.; Asahara, H. The tRNA pseudouridine synthase TruB1 regulates the maturation of let-7 miRNA. EMBO J. 2020, 39, e104708. [Google Scholar] [CrossRef] [PubMed]
- Spacková, N.; Réblová, K.; Sponer, J. Structural dynamics of the box C/D RNA kink-turn and its complex with proteins: The role of the A-minor 0 interaction, long-residency water bridges, and structural ion-binding sites revealed by molecular simulations. J. Phys. Chem. B 2010, 114, 10581–10593. [Google Scholar] [CrossRef]
- Kiss, T.; Fayet-Lebaron, E.; Jády, B.E. Box H/ACA small ribonucleoproteins. Mol. Cell 2010, 37, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Li, Y.; Liu, C.-J.; Xiang, Y.; Li, C.; Ye, Y.; Zhang, Z.; Hawke, D.H.; Park, P.K.; Diao, L.; et al. A Pan-cancer Analysis of the Expression and Clinical Relevance of Small Nucleolar RNAs in Human Cancer. Cell Rep. 2017, 21, 1968–1981. [Google Scholar] [CrossRef] [Green Version]
- Utama, B.; Kennedy, D.; Ru, K.; Mattick, J.S. Isolation and characterization of a new nucleolar protein, Nrap, that is conserved from yeast to humans. Genes Cells 2002, 7, 115–132. [Google Scholar] [CrossRef]
- Dong, D.; Song, M.; Wu, X.; Wang, W. NOL6, a new founding oncogene in human prostate cancer and targeted by miR-590-3p. Cytotechnology 2020, 72, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Zou, P.; Yang, Y.; Xu, X.; Liu, B.; Mei, F.; You, J.; Liu, Q.; Pei, F. Silencing of vacuolar ATPase c subunit ATP6V0C inhibits the invasion of prostate cancer cells through a LASS2/TMSG1-independent manner. Oncol. Rep. 2018, 39, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Penet, M.-F.; Shah, T.; Bharti, S.; Krishnamachary, B.; Artemov, D.; Mironchik, Y.; Wildes, F.; Maitra, A.; Bhujwalla, Z.M. Metabolic imaging of pancreatic ductal adenocarcinoma detects altered choline metabolism. Clin. Cancer Res. 2015, 21, 386–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, K.; Yamaguchi, M.; Miyazaki, K.; Imai, H.; Yokoe, K.; Ono, R.; Nosaka, T.; Katayama, N. Expressions of SH3BP5, LMO3, and SNAP25 in diffuse large B-cell lymphoma cells and their association with clinical features. Cancer Med. 2016, 5, 1802–1809. [Google Scholar] [CrossRef] [PubMed]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Supuran, C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef] [PubMed]
- Sliwoski, G.; Kothiwale, S.; Meiler, J.; Lowe, E.W.J. Computational methods in drug discovery. Pharmacol. Rev. 2014, 66, 334–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, E.W.; Cheng, S.C.; Sin, F.W.; Xie, Y. Triptolide induced cytotoxic effects on human promyelocytic leukemia, T cell lymphoma and human hepatocellular carcinoma cell lines. Toxicol. Lett. 2001, 122, 81–87. [Google Scholar] [CrossRef]
- Tao, Y.; Zhang, M.-L.; Ma, P.-C.; Sun, J.-F.; Zhou, W.-Q.; Cao, Y.-P.; Li, L.-J. Triptolide inhibits proliferation and induces apoptosis of human melanoma A375 cells. Asian Pac. J. Cancer Prev. 2012, 13, 1611–1615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, H.-C.; Chang, F.-R.; Wang, Y.-C.; Pan, M.-R.; Hung, W.-C.; Wu, Y.-C. A bioactive withanolide Tubocapsanolide A inhibits proliferation of human lung cancer cells via repressing Skp2 expression. Mol. Cancer Ther. 2007, 6, 1572–1578. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.X.; Wang, S.; Nie, S.; Marcone, M. Properties of Cordyceps Sinensis: A review. J. Funct. Foods 2013, 5, 550–569. [Google Scholar] [CrossRef] [PubMed]
- Sloan, E.A. Functional Food Trends; 2014; Available online: https://www.ift.org/~/media/food%20technology/pdf/2018/04/0418_feat1_topten.pdf (accessed on 1 August 2021).
- Aghajanpour, M.; Nazer, M.R.; Obeidavi, Z.; Akbari, M.; Ezati, P.; Kor, N.M. Functional foods and their role in cancer prevention and health promotion: A comprehensive review. Am. J. Cancer Res. 2017, 7, 740–769. [Google Scholar] [PubMed]
- Islam, S.M.R.; Siddiqua, T.J. 20-Functional foods in cancer prevention and therapy: Recent epidemiological findings. In Functional Foods in Cancer Prevention and Therapy; Kabir, Y., Ed.; Academic Press: Cambridge, MA, USA, 2020; pp. 405–433. ISBN 978-0-12-816151-7. [Google Scholar]
- Meerovich, I.G.; Yang, M.; Jiang, P.; Hoffman, R.M.; Gerasimenya, V.P.; Orlov, A.E.; Savitsky, A.P.; Popov, V.O. Study of action of cyclophosphamide and extract of mycelium of Pleurotus ostreatus in vivo on mice, bearing melanoma B16-F0-GFP. In Proceedings of the Genetically Engineered and Optical Probes for Biomedical Applications III, 30 April 2005; Bornhop, D.J., Achilefu, S.I., Raghavachari, R., Savitsky, A.P., Eds.; SPIE: Bellingham, WA, USA, 2005; Volume 5704, pp. 214–221. [Google Scholar]
- Maiti, S.; Mallick, S.K.; Bhutia, S.K.; Behera, B.; Mandal, M.; Maiti, T.K. Antitumor effect of culinary-medicinal oyster mushroom, Pleurotus ostreatus (Jacq.: Fr.) P. Kumm., derived protein fraction on tumor-bearing mice models. Int. J. Med. Mushrooms 2011, 13, 427–440. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, S. Cytotoxicity Assay of Agaricus Bisporus Extract, Oxaliplatin and Combination of both on Melanoma-B16 and Vero-101 Cell line:An in Vitro study. Univ. Thi-Qar J. Sci. 2020, 7. [Google Scholar]
- Zayed, A.; Serag, A.; Farag, M.A. Cynara cardunculus L.: Outgoing and potential trends of phytochemical, industrial, nutritive and medicinal merits. J. Funct. Foods 2020, 69, 103937. [Google Scholar] [CrossRef]


| Treatments | Cancers | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Apoptotic | Nutrigenomic | |||||||||||||||||
| TMX | Al-10-49 | Rosemary | Withaferin A | Bruceine D | Japonicone A | Indole3carbinol | Indole3carbinol | Indole3carbinol | Eusynstyelamide B | DLBC | KIRC | KIRP | LGG | LIHC | PAAD | SKCM | UCEC | |
| Cell line | MCF7 | ME1 | SW620 | MDA | MCF7 | MCF7 | MCF7 | T47D | ZR75 | LNCaP | ||||||||
| Gene Symbol | ||||||||||||||||||
| CENPB | - | −1.3 | - | −1.5 | - | - | −1.5 | - | - | - | Up | - | - | Up/* | - | Up | - | - |
| ERGIC1 | −1.2 | - | - | - | - | - | - | −1.7 | - | - | - | Up | - | Up/* | - | Up | - | - |
| CD47 | −1.3 | - | - | −2.1 | - | - | −1.6 | −1.8 | −1.9 | - | Up | - | - | - | - | Up | - | Up/* |
| PAQR4 | −1.5 | −2.3 | −3.1 | −1.8 | - | - | - | - | - | −1.9 | Up | Up/* | Up/* | - | Up/* | Up | Up | Up |
| POMGNT1 | - | −1.1 | - | - | −1.6 | - | - | - | - | - | Up/* | - | - | - | - | Up | Up/* | - |
| PPRC1 | - | −1.9 | - | - | - | −1.5 | −2.1 | - | - | - | Up | - | - | - | - | Up/* | - | - |
| SLC44A1 | −1.1 | - | - | −1.8 | - | - | - | −1.9 | - | - | Up | - | - | Up | - | Up/* | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monticolo, F.; Chiusano, M.L. Computational Approaches for Cancer-Fighting: From Gene Expression to Functional Foods. Cancers 2021, 13, 4207. https://doi.org/10.3390/cancers13164207
Monticolo F, Chiusano ML. Computational Approaches for Cancer-Fighting: From Gene Expression to Functional Foods. Cancers. 2021; 13(16):4207. https://doi.org/10.3390/cancers13164207
Chicago/Turabian StyleMonticolo, Francesco, and Maria Luisa Chiusano. 2021. "Computational Approaches for Cancer-Fighting: From Gene Expression to Functional Foods" Cancers 13, no. 16: 4207. https://doi.org/10.3390/cancers13164207
APA StyleMonticolo, F., & Chiusano, M. L. (2021). Computational Approaches for Cancer-Fighting: From Gene Expression to Functional Foods. Cancers, 13(16), 4207. https://doi.org/10.3390/cancers13164207







