Diagnostic and Prognostic Performance of Liquid Biopsy-Derived Exosomal MicroRNAs in Thyroid Cancer Patients: A Systematic Review and Meta-Analysis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Literature Search Strategy
2.2. Inclusion and Exclusion Criteria
2.3. Quality Assessment
2.4. Data Extraction
2.5. Statistical Analysis
3. Results
3.1. Characteristics of Included Studies
3.2. Diagnostic Value of Exosome-Derived miRNAs
3.3. Prognostic Value of Exosome-Derived miRNAs
3.4. Functional Enrichment Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [Green Version]
- Yoon, R.G.; Baek, J.H.; Lee, J.H.; Choi, Y.J.; Hong, M.J.; Song, D.E.; Kim, J.K.; Yoon, J.H.; Kim, W.B. Diagnosis of Thyroid Follicular Neoplasm: Fine-Needle Aspiration Versus Core-Needle Biopsy. Thyroid 2014, 24, 1612–1617. [Google Scholar] [CrossRef]
- Guille, J.T.; Opoku-Boateng, A.; Thibeault, S.L.; Chen, H. Evaluation and Management of the Pediatric Thyroid Nodule. Oncologist 2014, 20, 19–27. [Google Scholar] [CrossRef] [Green Version]
- Papini, E.; Guglielmi, R.; Bianchini, A.; Crescenzi, A.; Taccogna, S.; Nardi, F.; Panunzi, C.; Rinaldi, R.; Toscano, V.; Pacella, C.M. Risk of Malignancy in Nonpalpable Thyroid Nodules: Predictive Value of Ultrasound and Color-Doppler Features. J. Clin. Endocrinol. Metab. 2002, 87, 1941–1946. [Google Scholar] [CrossRef]
- Caraway, N.P.; Sneige, N.; Samaan, N.A. Diagnostic pitfalls in thyroid fine-needle aspiration: A review of 394 cases. Diagn. Cytopathol. 1993, 9, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Mongelli, M.N.; Giri, S.; Peipert, B.J.; Helenowski, I.B.; Yount, S.E.; Sturgeon, C. Financial burden and quality of life among thyroid cancer survivors. Surgery 2020, 167, 631–637. [Google Scholar] [CrossRef]
- Rezig, L.; Servadio, A.; Torregrossa, L.; Miccoli, P.; Basolo, F.; Shintu, L.; Caldarelli, S. Diagnosis of post-surgical fine-needle aspiration biopsies of thyroid lesions with indeterminate cytology using HRMAS NMR-based metabolomics. Metabolomics 2018, 14, 141. [Google Scholar] [CrossRef]
- Toraih, E.A.; Ibrahiem, A.T.; Fawzy, M.S.; Hussein, M.H.; Al-Qahtani, S.A.M.; Shaalan, A.A.M. MicroRNA-34a: A Key Regulator in the Hallmarks of Renal Cell Carcinoma. Oxid. Med. Cell. Longev. 2017, 2017, 269379. [Google Scholar] [CrossRef] [Green Version]
- Toraih, E.A.; Aly, N.M.; Abdallah, H.Y.; Al-Qahtani, S.A.; Shaalan, A.A.; Hussein, M.H.; Fawzy, M.S. MicroRNA—Target cross-talks: Key players in glioblastoma multiforme. Tumor Biol. 2017, 39, 1010428317726842. [Google Scholar] [CrossRef] [Green Version]
- Heneghan, H.; Miller, N.; Kerin, M. MiRNAs as biomarkers and therapeutic targets in cancer. Curr. Opin. Pharmacol. 2010, 10, 543–550. [Google Scholar] [CrossRef]
- Cuellar, T.L.; McManus, M.T. MicroRNAs and endocrine biology. J. Endocrinol. 2005, 187, 327–332. [Google Scholar] [CrossRef] [Green Version]
- Lima, C.R.; Geraldo, M.V.; Fuziwara, C.S.; Kimura, E.T.; Santos, M.F. MiRNA-146b-5p upregulates migration and invasion of different Papillary Thyroid Carcinoma cells. BMC Cancer 2016, 16, 108. [Google Scholar] [CrossRef] [Green Version]
- Swierniak, M.; Wojcicka, A.; Czetwertynska, M.; Stachlewska, E.; Maciag, M.; Wiechno, W.; Gornicka, B.; Bogdanska, M.; Koperski, L.; De La Chapelle, A.; et al. In-Depth Characterization of the MicroRNA Transcriptome in Normal Thyroid and Papillary Thyroid Carcinoma. J. Clin. Endocrinol. Metab. 2013, 98, E1401–E1409. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Nishida, N.; Calin, G.; Pantel, K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 2014, 11, 145–156. [Google Scholar] [CrossRef]
- Cheng, L.; Sun, X.; Scicluna, B.J.; Coleman, B.M.; Hill, A. Characterization and deep sequencing analysis of exosomal and non-exosomal miRNA in human urine. Kidney Int. 2014, 86, 433–444. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Geng, H.; Liu, X.; Cao, M.; Zhang, X. A meta-analysis of circulating microRNAs in the diagnosis of papillary thyroid carcinoma. PLoS ONE 2021, 16, e0251676. [Google Scholar] [CrossRef]
- Shao, N.; Xue, L.; Wang, R.; Luo, K.; Zhi, F.; Lan, Q. miR-454-3p Is an Exosomal Biomarker and Functions as a Tumor Suppressor in Glioma. Mol. Cancer Ther. 2018, 18, 459–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hannafon, B.N.; Trigoso, Y.D.; Calloway, C.L.; Zhao, Y.D.; Lum, D.H.; Welm, A.L.; Zhao, Z.J.; Blick, K.E.; Dooley, W.C.; Ding, W.Q. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016, 18, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, K.; Li, G.; Chen, W.; Song, L.; Wei, T.; Li, Z.; Gong, R.; Lei, J.; Shi, H.; Zhu, J. Plasma Exosomal miR-146b-5p and miR-222-3p are Potential Biomarkers for Lymph Node Metastasis in Papillary Thyroid Carcinomas. OncoTargets Ther. 2020, 13, 1311–1319. [Google Scholar] [CrossRef] [Green Version]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Elshazli, R.M.; Toraih, E.A.; Elgaml, A.; El-Mowafy, M.; El-Mesery, M.; Amin, M.; Hussein, M.H.; Killackey, M.; Fawzy, M.S.; Kandil, E. Diagnostic and prognostic value of hematological and immunological markers in COVID-19 infection: A meta-analysis of 6320 patients. PLoS ONE 2020, 15, e0238160. [Google Scholar] [CrossRef]
- Witwer, K.W.; Soekmadji, C.; Hill, A.F.; Wauben, M.H.; Buzás, E.I.; Di Vizio, D.; Falcon-Perez, J.M.; Gardiner, C.; Hochberg, F.; Kurochkin, I.V.; et al. Updating the MISEV minimal requirements for extracellular vesicle studies: Building bridges to reproducibility. J. Extracell. Vesicles 2017, 6, 1396823. [Google Scholar] [CrossRef] [Green Version]
- Samsonov, R.; Burdakov, V.; Shtam, T.; Radzhabova, Z.; Vasilyev, D.; Tsyrlina, E.; Titov, S.; Иванов, М.; Berstein, L.; Filatov, M.; et al. Plasma exosomal miR-21 and miR-181a differentiates follicular from papillary thyroid cancer. Tumor Biol. 2016, 37, 12011–12021. [Google Scholar] [CrossRef]
- Yin, G.; Kong, W.; Zheng, S.; Shan, Y.; Zhang, J.; Ying, R.; Wu, H. Exosomal miR-130a-3p promotes the progression of differentiated thyroid cancer by targeting insulin-like growth factor 1. Oncol. Lett. 2021, 21, 283. [Google Scholar] [CrossRef]
- Xin, Y.; Meng, K.; Guo, H.; Chen, B.; Zheng, C.; Yu, K. Exosomal hsa-miR-129-2 and hsa-miR-889 from a 6-microRNA signature might be a potential biomarker for predicting prognosis of papillary thyroid carcinoma. Comb. Chem. High Throughput Screen. 2021, 24, 1. [Google Scholar] [CrossRef]
- Wen, Q.; Wang, Y.; Li, X.; Jin, X.; Wang, G. Decreased serum exosomal miR-29a expression and its clinical significance in papillary thyroid carcinoma. J. Clin. Lab. Anal. 2021, 35, e23560. [Google Scholar] [CrossRef]
- Li, S.; Zhang, S.; Yang, W.; Li, F.; Long, H. Diagnostic Value of Exosomal miR-148a-3p in the Serum of Patients with Differentiated Thyroid Cancer. Clin. Lab. 2021, 67, 290–297. [Google Scholar] [CrossRef]
- Zou, X.; Gao, F.; Wang, Z.-Y.; Zhang, H.; Liu, Q.-X.; Jiang, L.; Zhou, X.; Zhu, W. A three-microRNA panel in serum as novel biomarker for papillary thyroid carcinoma diagnosis. Chin. Med. J. 2020, 133, 2543–2551. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Zhao, J.; Li, M.; Liu, X.; Xu, Y.; Li, W.; Wu, S.; Su, Z. Exosomal miRNAs are potential diagnostic biomarkers between malignant and benign thyroid nodules based on next-generation sequencing. Carcinogenesis 2019, 41, 18–24. [Google Scholar] [CrossRef]
- Liang, M.; Yu, S.; Tang, S.; Bai, L.; Cheng, J.; Gu, Y.; Li, S.; Zheng, X.; Duan, L.; Wang, L.; et al. A Panel of Plasma Exosomal miRNAs as Potential Biomarkers for Differential Diagnosis of Thyroid Nodules. Front. Genet. 2020, 11, 449. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.; Tan, Y.; Guo, L.; Tang, A.; Zhao, Y. Identification of exosomal miRNA biomarkers for diagnosis of papillary thyroid cancer by small RNA sequencing. Eur. J. Endocrinol. 2020, 182, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Deng, X.; Fan, Y. Exosomal miRNA423-5p mediated oncogene activity in papillary thyroid carcinoma: A potential diagnostic and biological target for cancer therapy. Neoplasma 2019, 66, 516–523. [Google Scholar] [CrossRef]
- Wang, Z.; Lv, J.; Zou, X.; Huang, Z.; Zhang, H.; Liu, Q.; Jiang, L.; Zhou, X.; Zhu, W. A three plasma microRNA signature for papillary thyroid carcinoma diagnosis in Chinese patients. Gene 2019, 693, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Heberle, H.; Meirelles, G.V.; Da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef]
- Russell, P.S.S.; Chikkala, S.M.; Earnest, R.; Viswanathan, S.A.; Russell, S.; Mammen, P.M. Diagnostic accuracy and clinical utility of non-English versions of Edinburgh Post-Natal Depression Scale for screening post-natal depression in India: A meta-analysis. World J. Psychiatry 2020, 10, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Zamora, J.; Abraira, V.; Muriel, A.; Khan, K.; Coomarasamy, A. Meta-DiSc: A software for meta-analysis of test accuracy data. BMC Med. Res. Methodol. 2006, 6, 31. [Google Scholar] [CrossRef]
- Yang, Q.; Diamond, M.P.; Al-Hendy, A.; Yang, Q. The emerging role of extracellular vesicle-derived miRNAs: Implication in cancer progression and stem cell related diseases. J. Clin. Epigenet. 2016, 2, 13. [Google Scholar] [PubMed]
- Volinia, S.; Calin, G.; Liu, C.-G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [Green Version]
- Elnaggar, G.N.; El-Hifnawi, N.M.; Ismail, A.; Yahia, M.; Elshimy, R.A. Micro RNA-148a Targets Bcl-2 in Patients with Non-Small Cell Lung Cancer. Asian Pac. J. Cancer Prev. 2021, 22, 1949–1955. [Google Scholar] [CrossRef]
- Wan, K.; Tu, Z.; Liu, Z.; Cai, Y.; Chen, Y.; Ling, C. Upregulated osteoprotegerin expression promotes lung cancer cell invasion by increasing miR-20a expression. Exp. Ther. Med. 2021, 22, 846. [Google Scholar] [CrossRef]
- Xu, F.; Jiang, L.; Zhao, Q.; Zhang, Z.; Liu, Y.; Yang, S.; Yu, M.; Chen, H.; Zhang, J.; Zhang, J. Whole-transcriptome and proteome analyses identify key differentially expressed mRNAs, miRNAs, lncRNAs and circRNAs associated with HCC. Oncogene 2021, 40, 4820–4831. [Google Scholar] [CrossRef]
- Lee, J.C.; Zhao, J.-T.; Gundara, J.; Serpell, J.; Bach, L.A.; Sidhu, S. Papillary thyroid cancer–derived exosomes contain miRNA-146b and miRNA-222. J. Surg. Res. 2015, 196, 39–48. [Google Scholar] [CrossRef]
- Wojtas, B.; Ferraz, C.; Stokowy, T.; Hauptmann, S.; Lange, D.; Dralle, H.; Musholt, T.; Jarzab, B.; Paschke, R.; Eszlinger, M. Differential miRNA expression defines migration and reduced apoptosis in follicular thyroid carcinomas. Mol. Cell. Endocrinol. 2014, 388, 1–9. [Google Scholar] [CrossRef]
- He, H.; Jazdzewski, K.; Li, W.; Liyanarachchi, S.; Nagy, R.; Volinia, S.; Calin, G.; Liu, C.-G.; Franssila, K.; Suster, S.; et al. The role of microRNA genes in papillary thyroid carcinoma. Proc. Natl. Acad. Sci. USA 2005, 102, 19075–19080. [Google Scholar] [CrossRef] [Green Version]
- Tetzlaff, M.T.; Liu, A.; Xu, X.; Master, S.R.; Baldwin, D.A.; Tobias, J.W.; Livolsi, V.A.; Baloch, Z.W. Differential Expression of miRNAs in Papillary Thyroid Carcinoma Compared to Multinodular Goiter Using Formalin Fixed Paraffin Embedded Tissues. Endocr. Pathol. 2007, 18, 163–173. [Google Scholar] [CrossRef]
- Zhou, G.; Xiao, M.; Zhao, L.; Tang, J.; Zhang, L. MicroRNAs as novel biomarkers for the differentiation of malignant versus benign thyroid lesions: A meta-analysis. Genet. Mol. Res. 2015, 14, 7279–7289. [Google Scholar] [CrossRef]
- Stokowy, T.; Wojtas, B.; Fujarewicz, K.; Jarząb, B.; Eszlinger, M.; Paschke, R. miRNAs with the Potential to Distinguish Follicular Thyroid Carcinomas from Benign Follicular Thyroid Tumors: Results of a Meta-analysis. Horm. Metab. Res. 2014, 46, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Laukiene, R.; Jakubkevicius, V.; Ambrozaityte, L.; Cimbalistiene, L.; Utkus, A. Dysregulation of microRNAs as the risk factor of lymph node metastasis in papillary thyroid carcinoma: Systematic review. Endokrynol. Pol. 2021, 72, 145–152. [Google Scholar] [CrossRef]
- Ito, Y.; Higashiyama, T.; Takamura, Y.; Miya, A.; Kobayashi, K.; Matsuzuka, F.; Kuma, K.; Miyauchi, A. Risk Factors for Recurrence to the Lymph Node in Papillary Thyroid Carcinoma Patients without Preoperatively Detectable Lateral Node Metastasis: Validity of Prophylactic Modified Radical Neck Dissection. World J. Surg. 2007, 31, 2085–2091. [Google Scholar] [CrossRef] [PubMed]
- Su, D.; Ji, Z.; Xue, P.; Guo, S.; Jia, Q.; Sun, H. Long-Noncoding RNA FGD5-AS1 Enhances the Viability, Migration, and Invasion of Glioblastoma Cells by Regulating the miR-103a-3p/TPD52 Axis. Cancer Manag. Res. 2020, 12, 6317–6329. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Chen, J.; Yue, C.; Zhang, H.; Tong, J.; Li, J.; Chen, T. The role of miR-182-5p in hepatocarcinogenesis of trichloroethylene in mice. Toxicol. Sci. 2016, 156, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Sierra-Ramirez, J.; Seseña-Mendez, E.; Godinez-Victoria, M.; Hernandez-Caballero, M. An insight into the promoter methylation of PHF20L1 and the gene association with metastasis in breast cancer. Adv. Clin. Exp. Med. 2021, 30, 507–515. [Google Scholar] [CrossRef]
- Akyay, O.Z.; Gov, E.; Kenar, H.; Arga, K.Y.; Selek, A.; Tarkun, I.; Canturk, Z.; Cetinarslan, B.; Gurbuz, Y.; Sahin, B. Mapping the Molecular Basis and Markers of Papillary Thyroid Carcinoma Progression and Metastasis Using Global Transcriptome and microRNA Profiling. OMICS J. Integr. Biol. 2020, 24, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Cibas, E.S.; Ali, S.Z. The 2017 Bethesda System for Reporting Thyroid Cytopathology. Thyroid 2017, 27, 1341–1346. [Google Scholar] [CrossRef]
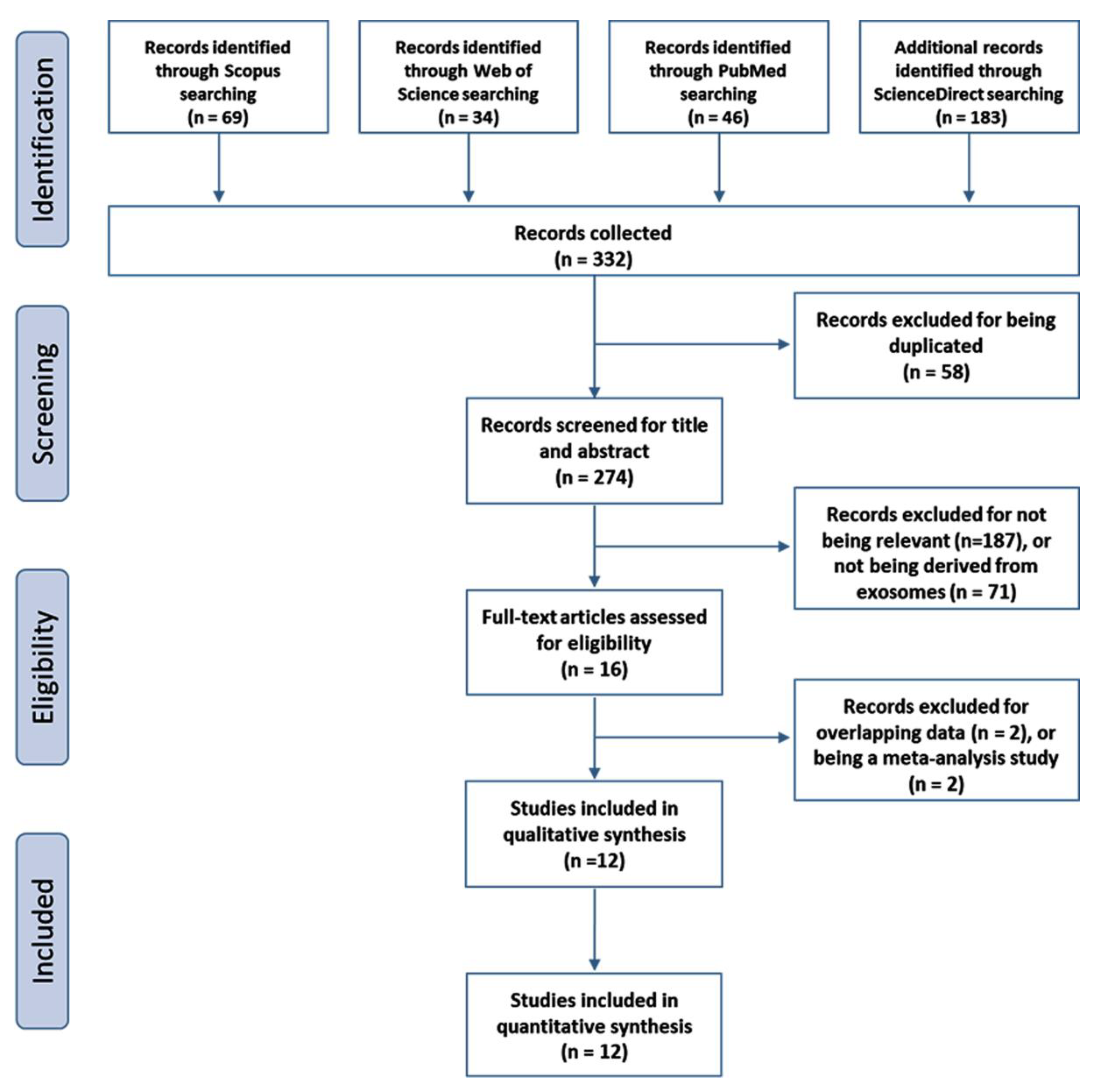
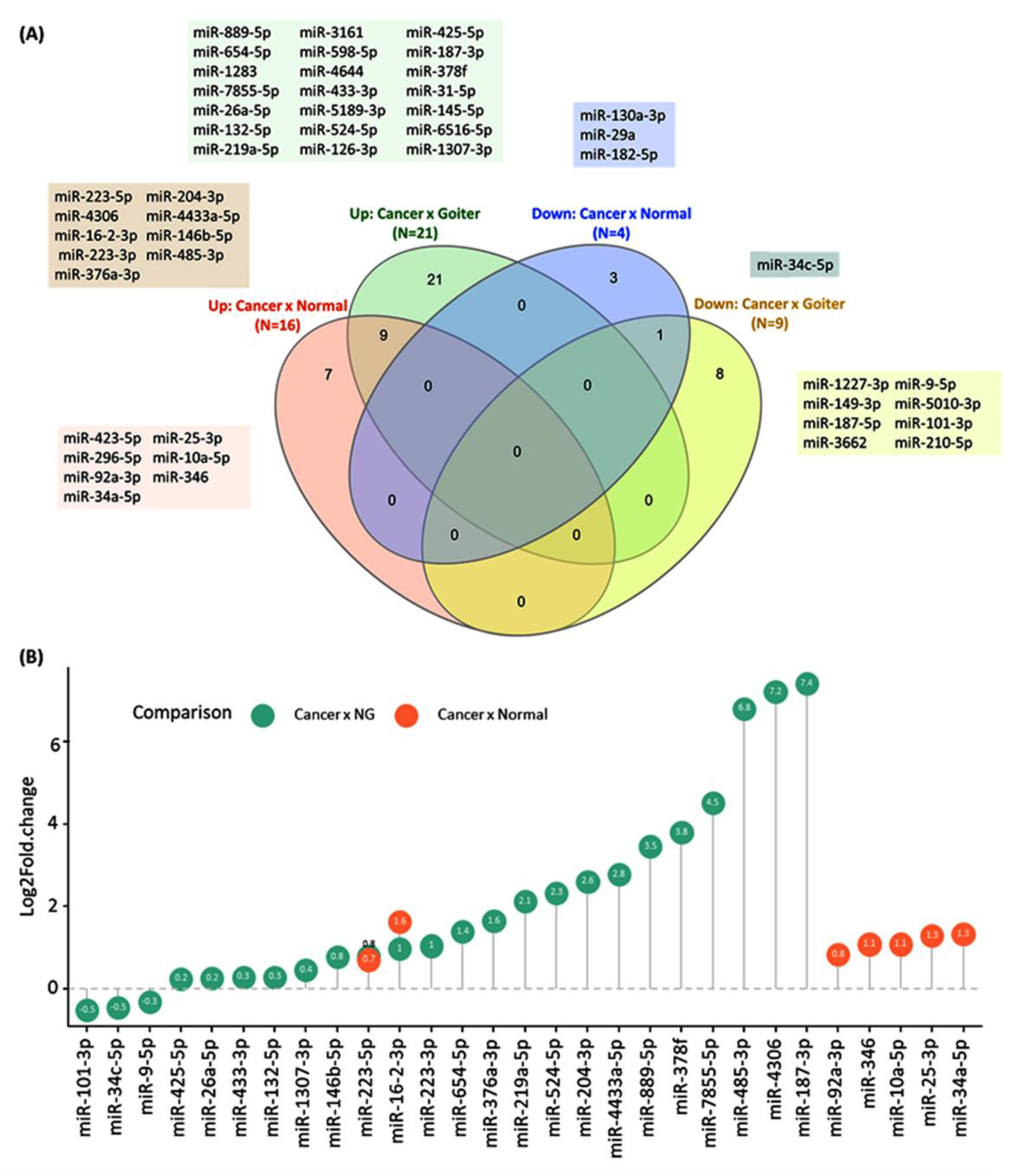



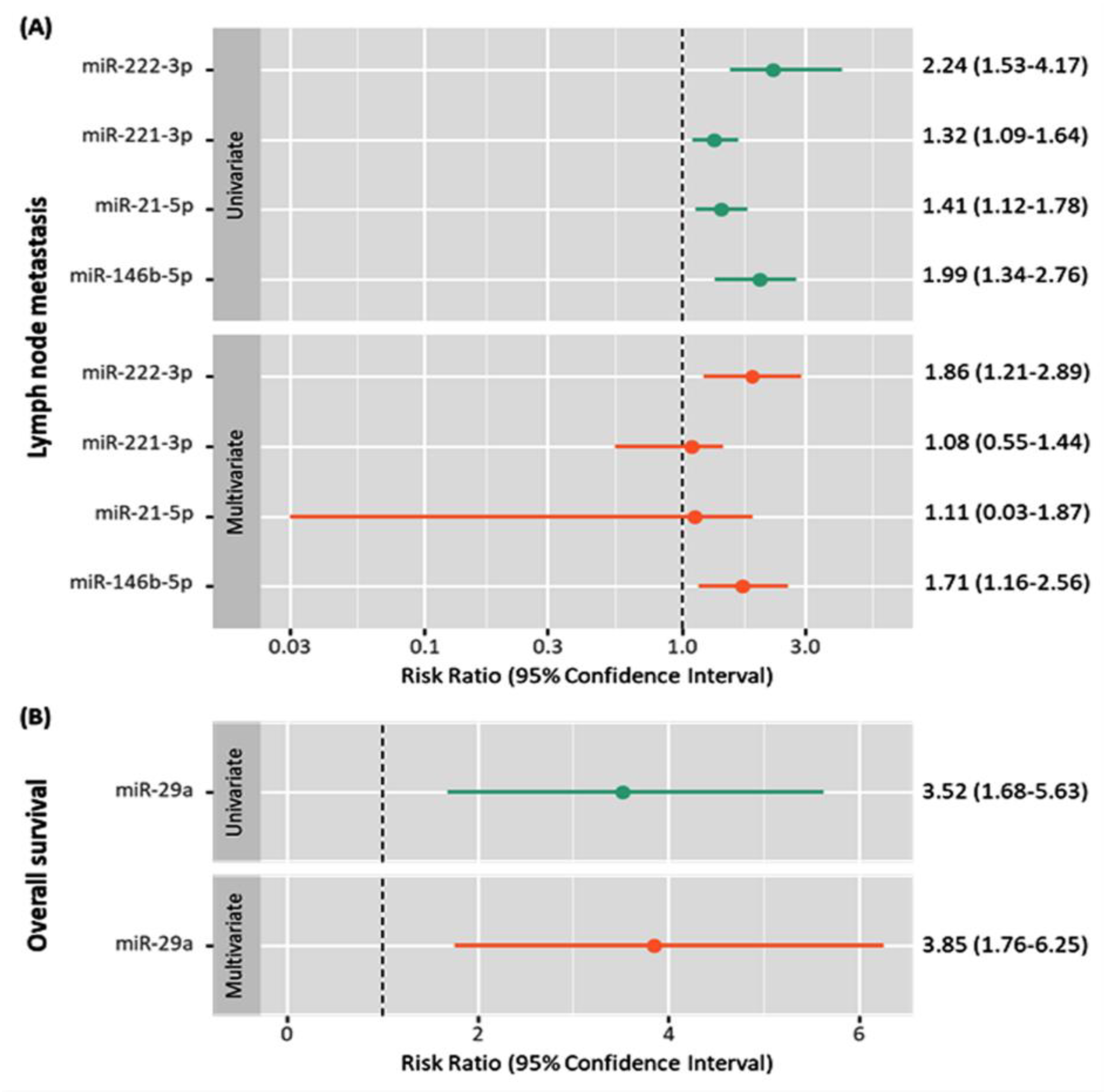

| First Author | Year | Country | No of Patients | No of Controls | Test Method | Name of PCR Kit | Method of Exosomes Isolation | MISEV | Target Exosome miRNA | Tumor Subtype | Mean Age, Y | Female (%) | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yin | 2021 | China | 40 | 40 | qRT-PCR (SYBR Green) | SYBR Green Super mix (Bio-Rad Laboratories, Inc.) | Invitrogen™ Exosome Isolation Kit (Thermo Fisher Scientific, Inc.) | 7 | miR-130a-3p | DTC | 64.9 | 27.5% | [26] |
| Xin | 2021 | China | 491 | -- | Not estimated | -- | -- | miR-129-2 miR-889 | PTC | -- | -- | [27] | |
| Wen | 2021 | China | 119 | 100 | qRT-PCR | TaqMan MicroRNA RT Kit (Applied Biosystems) | ExoQuick Exosome Precipitation Solution (System Biosciences) | 3 | miR-29a | PTC | -- | 52.1% | [28] |
| Li | 2021 | China | -- | qRT-PCR | Not estimated | -- | -- | miR-148a-3p | DTC | -- | -- | [29] | |
| Zou | 2020 | China | 100 | 96 | qRT-PCR | SYBR Green (SYBR® Premix Ex TaqTM II, TaKaRa, Dalian, China). | ExoQuick Exosome Precipitation Solution (System Biosciences, Mountain View, CA, USA). | 6 | miR-25-3p miR-296-5p miR-92a-3p | PTC | -- | -- | [30] |
| Pan | 2020 | China | 13 | 7 | Small RNA sequencing | TruSeq SR Cluster Kit v3-cBot-HS (Illumina, San Diego, CA, USA) | Exosomes were isolated from the plasma through ultracentrifugation method. | 7 | miR-5189-3p miR-5010-3p miR-598-5p miR-3161 miR-6516-5p miR-4644 miR-1283 miR-1227-3p miR-149-3p miR-210-5p miR-3662 miR-187-5p | PTC | -- | 100% | [31] |
| Liang | 2020 | China | 51 | 69 | qRT-PCR | SYBR Green PCR Kit (QIAGEN) | Exosome Precipitation Solution (EXOQ20A-1, SBI, Mountain View, CA, USA) | 8 | miR-16-2-3p miR-223-5p miR-34c-5p miR-182-5p miR-223-3p miR-146b-5p miR-16-2-3p miR-223-5p | PTC | 44.0 | 51.4% | [32] |
| Jiang | 2020 | China | 64 | qRT-PCR | Not determined | Not determined | -- | miR-146b-5p miR-221-3p miR-222-3p miR-21-5p miR-204-5p | PTC | 41.2 | 78.1% | [21] | |
| Dai | 2020 | China | 96 | 30 | qRT-PCR | MiR-X miRNA qRTPCR SYBR Kit (Takara) and miDETECT A Track™ miRNA RT-qPCR Primers (Ribobio). | Exosomes were isolated with a combination of centrifugation and ultracentrifugation. | 5 | miR-485-3p miR-4433a-5p miR-4306 miR-376a-3p miR-204-3p | PTC | 56.6 | -- | [33] |
| Ye | 2019 | China | 60 | 30 | qRT-PCR | miScript SYBR Green PCR Kit (Qiagen, Germany). | Exosomes were isolated with ultracentrifugation. | 5 | miRNA423-5p | PTC | -- | -- | [34] |
| Wang | 2019 | China | 120 | 160 | qRT-PCR | The expression levels of miRNAs in plasma and exosomes were measured using SYBR Green dye | Exosomes of peripheral plasma were isolated by using ExoQuick™ (System Biosciences, Mountain View, CL, USA) | 6 | miR-346 miR-10a-5p miR-34a-5p | PTC | -- | -- | [35] |
| Samsonov | 2016 | Russia | 10 | 8 | qRT-PCR | qPCR was performed using Cancer Focus microRNA PCR Panels and ExiLENT SYBR Green master mix (both from Exiqon, Denmark) on CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, USA). | Exosomes were isolated with centrifugation method. | 5 | miR-21 miR-181a | PTC | 54.5 | 80% | [25] |
| miRNA | Expression | LNM | TNM Stage | Tumor Size | ETE | BRAF Mutation | Short Survival | Recurrence | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| miR-130a-3p | Low | (+) | (+) | (+) | [26] | ||||
| miR-29a | Low | (+) | (+) | (+) | (+) | (+) | [28] | ||
| miR-148a-3p | Low | (+) | (+) | [29] | |||||
| miR-146b-5p | High | (+) | (+) | [21] | |||||
| miR-222-3p | High | (+) | (+) | [21] | |||||
| miR-423-5p | High | (+) | [34] | ||||||
| miR-204-3p | High | (+) | [33] | ||||||
| miR-4306 | Low | (+) | [33] | ||||||
| miR-4433a-5p | High | (+) | (+) | (+) | (+) | [33] | |||
| miR-485-3p | High | (+) | (+) | (+) | (+) | (+) | [33] | ||
| miR-21-5p | High | (+) | [21] | ||||||
| miR-204-5p | High | (+) | [21] | ||||||
| miR-221-3p | High | (+) | [21] | ||||||
| miR-182-5p | High | (+) | [32] | ||||||
| miR-26b-5p | High | (+) | [32] | ||||||
| miR-126-3p | High | (+) | [32] | ||||||
| miR-542-3p | High | (+) | [32] | ||||||
| miR-32-5p | High | (+) | [32] | ||||||
| miR-363-3p | High | (+) | [32] | ||||||
| miR-1912 | Low | (+) | [32] | ||||||
| miR-323a-5p | Low | (+) | [32] | ||||||
| miR-543 | Low | (+) | [32] | ||||||
| miR-381-3p | Low | (+) | [32] | ||||||
| miR-128-3p | Low | (+) | [32] | ||||||
| miR-139-5p | Low | (+) | [32] | ||||||
| miR-885-3p | Low | (+) | [32] | ||||||
| miR-409-5p | Low | (+) | [32] | ||||||
| miR-28-5p | Low | (+) | [32] | ||||||
| miR-151a-5p | Low | (+) | [32] | ||||||
| miR-490-3p | Low | (+) | [32] |
| miRNAs | Cases | Controls | Expression | AUC | Lower | Upper | Ref. | |
|---|---|---|---|---|---|---|---|---|
| Tumor size ≥ 1 cm vs. <1 cm | miR-204-3p | 56 | 40 | High | 0.798 | 0.71 | 0.88 | [33] |
| ETE vs. none | miR-485-3p | 59 | 37 | High | 0.726 | 0.62 | 0.83 | [33] |
| BRAF mutation vs. wild type | miR-485-3p | 65 | 31 | High | 0.890 | 0.83 | 0.96 | [33] |
| Late stage vs. stage I/II | miR-485-3p | 33 | 63 | High | 0.753 | 0.65 | 0.86 | [33] |
| miR-29a | 41 | 78 | Low | 0.758 | 0.70 | 0.81 | [28] | |
| Recurrence vs. none | miR-29a | 30 | 89 | Low | 0.753 | 0.68 | 0.80 | [28] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toraih, E.A.; Elshazli, R.M.; Trinh, L.N.; Hussein, M.H.; Attia, A.A.; Ruiz, E.M.L.; Zerfaoui, M.; Fawzy, M.S.; Kandil, E. Diagnostic and Prognostic Performance of Liquid Biopsy-Derived Exosomal MicroRNAs in Thyroid Cancer Patients: A Systematic Review and Meta-Analysis. Cancers 2021, 13, 4295. https://doi.org/10.3390/cancers13174295
Toraih EA, Elshazli RM, Trinh LN, Hussein MH, Attia AA, Ruiz EML, Zerfaoui M, Fawzy MS, Kandil E. Diagnostic and Prognostic Performance of Liquid Biopsy-Derived Exosomal MicroRNAs in Thyroid Cancer Patients: A Systematic Review and Meta-Analysis. Cancers. 2021; 13(17):4295. https://doi.org/10.3390/cancers13174295
Chicago/Turabian StyleToraih, Eman A., Rami M. Elshazli, Lily N. Trinh, Mohammad H. Hussein, Abdallah A. Attia, Emmanuelle M. L. Ruiz, Mourad Zerfaoui, Manal S. Fawzy, and Emad Kandil. 2021. "Diagnostic and Prognostic Performance of Liquid Biopsy-Derived Exosomal MicroRNAs in Thyroid Cancer Patients: A Systematic Review and Meta-Analysis" Cancers 13, no. 17: 4295. https://doi.org/10.3390/cancers13174295
APA StyleToraih, E. A., Elshazli, R. M., Trinh, L. N., Hussein, M. H., Attia, A. A., Ruiz, E. M. L., Zerfaoui, M., Fawzy, M. S., & Kandil, E. (2021). Diagnostic and Prognostic Performance of Liquid Biopsy-Derived Exosomal MicroRNAs in Thyroid Cancer Patients: A Systematic Review and Meta-Analysis. Cancers, 13(17), 4295. https://doi.org/10.3390/cancers13174295









