Exosomes in Lung Cancer: Actors and Heralds of Tumor Development
Abstract
:Simple Summary
Abstract
1. Lung Cancer and Exosomes
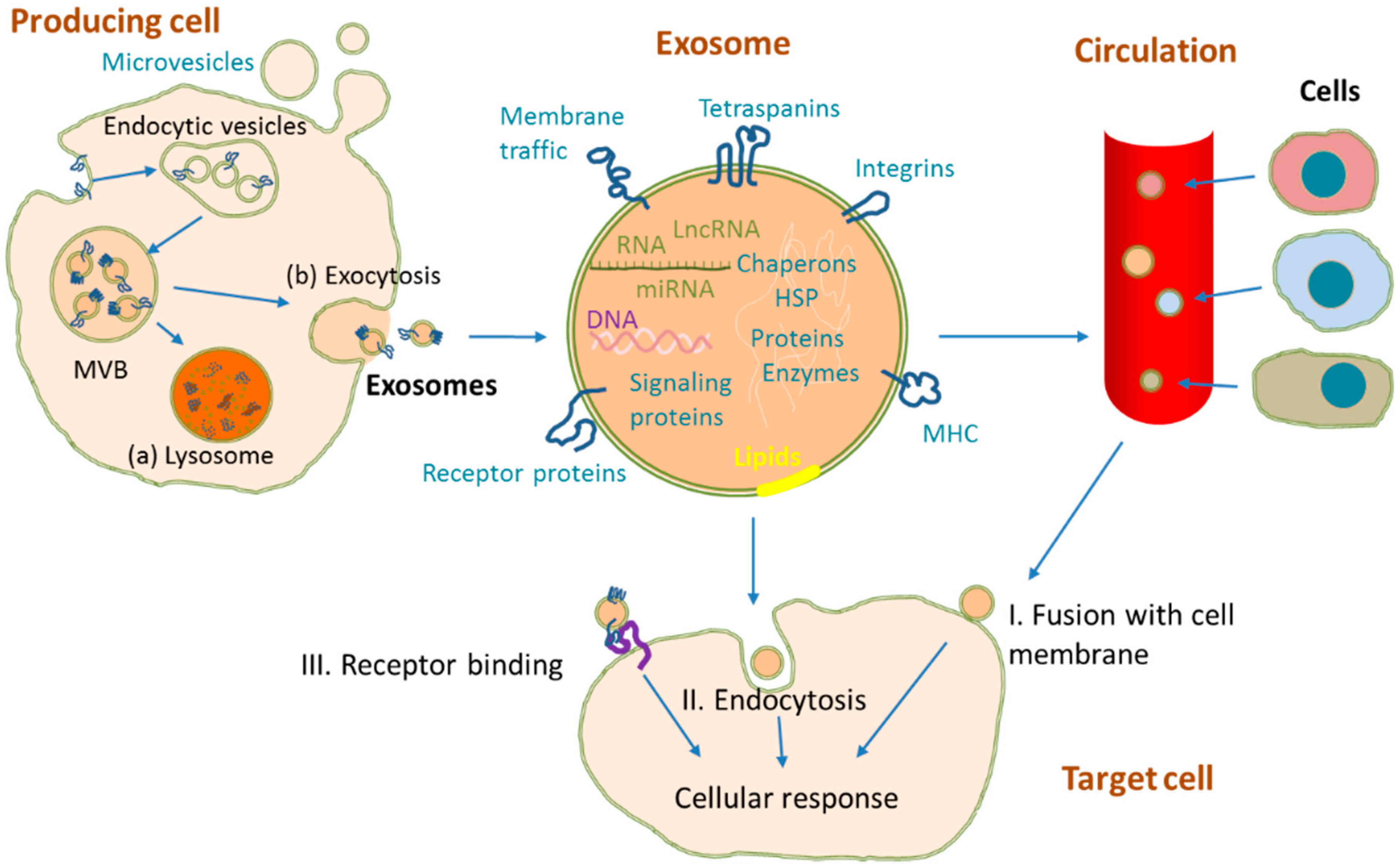
2. Exosome Biogenesis and Structure
3. Exosome Isolation and Identification
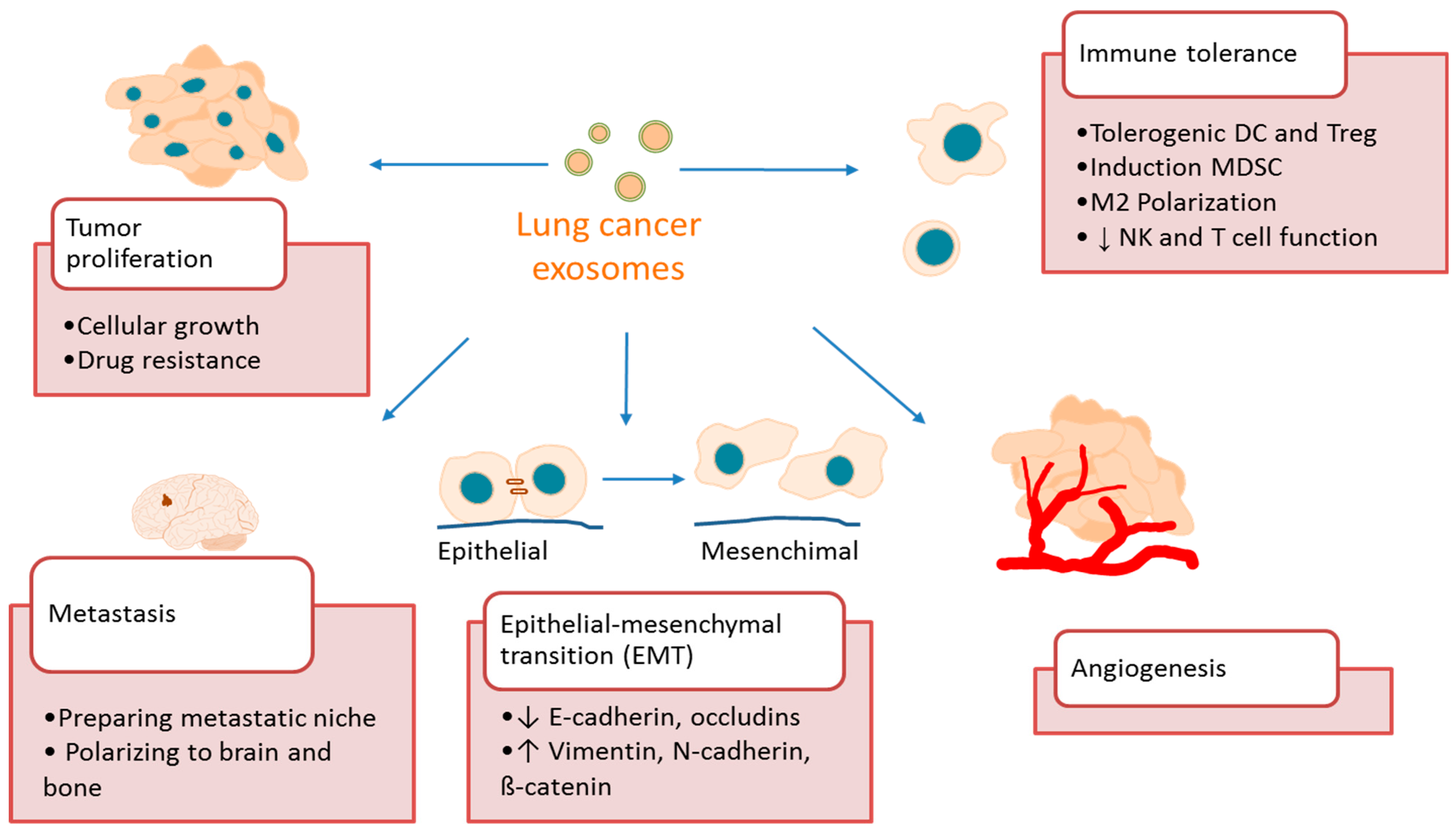
4. Exosome Function in Lung Cancer
4.1. Exosomes Promote Lung Cancer Growth and Metastasis
4.2. Exosomes Promote Lung Cancer Angiogenesis
4.3. Exosomes Promote Lung Cancer Immune Tolerance
5. Exosomes as Biomarkers in Lung Cancer
5.1. Exosomal Proteins
5.2. Exosomal miRNAs
5.3. Other Nucleic Acids
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2021. CA A Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Herbst, R.S.; Morgensztern, D.; Boshoff, C. The biology and management of non-small cell lung cancer. Nature 2018, 553, 446–454. [Google Scholar] [CrossRef]
- Tsao, M.; Sakurada, A.; Cutz, J.-C.; Zhu, C.; Kamel-Reid, S.; Squire, J.; Lorimer, I.; Zhang, T.; Liu, N.; Daneshmand, M.; et al. Erlotinib in lung cancer—Molecular and clinical predictors of outcome. N. Engl. J. Med. 2005, 353, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Gettinger, S.; Horn, L.; Jackman, D.; Spigel, D.; Antonia, S.; Hellmann, M.; Powderly, J.; Heist, R.; Sequist, L.V.; Smith, D.C.; et al. Five-year follow-up of nivolumab in previously treated advanced non–small-cell lung cancer: Results from the CA209-003 study. J. Clin. Oncol. 2018, 36, 1675–1684. [Google Scholar] [CrossRef]
- Leighl, N.B.; Rekhtman, N.; Biermann, W.A.; Huang, J.; Mino-Kenudson, M.; Ramalingam, S.S.; West, H.; Whitlock, S.; Somerfield, M.R. Molecular testing for selection of patients with lung cancer for epidermal growth factor receptor and anaplastic lymphoma kinase tyrosine kinase inhibitors: American Society of Clinical Oncology Endorsement of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology Guideline. J. Clin. Oncol. 2014, 32, 3673–3679. [Google Scholar] [CrossRef] [PubMed]
- Izumchenko, E.; Chang, X.; Brait, M.; Fertig, E.; Kagohara, L.T.; Bedi, A.; Marchionni, L.; Agrawal, N.; Ravi, R.; Jones, S.; et al. Targeted sequencing reveals clonal genetic changes in the progression of early lung neoplasms and paired circulating DNA. Nat. Commun. 2015, 6, 8258. [Google Scholar] [CrossRef] [PubMed]
- Macías, M.; Alegre, E.; Díaz-Lagares, A.; Patiño, A.; Pérez-Gracia, J.L.; Sanmamed, M.; López-López, R.; Varo, N.; González, A. Liquid biopsy: From basic research to clinical practice. Adv. Clin. Chem. 2018, 83, 73–119. [Google Scholar] [CrossRef]
- Freitas, C.; Sousa, C.; Machado, F.; Serino, M.; Santos, V.; Cruz-Martins, N.; Teixeira, A.; Cunha, A.; Pereira, T.; Oliveira, H.P.; et al. The role of liquid biopsy in early diagnosis of lung cancer. Front. Oncol. 2021, 11, 634316. [Google Scholar] [CrossRef]
- Garrido, P.; Conde, E.; De Castro, J.; Gómez-Román, J.J.; Felip, E.; Pijuan, L.; Isla, D.; Sanz, J.; Paz-Ares, L.; López-Ríos, F. Updated guidelines for predictive biomarker testing in advanced non-small-cell lung cancer: A National Consensus of the Spanish Society of Pathology and the Spanish Society of Medical Oncology. Clin. Transl. Oncol. 2019, 22, 989–1003. [Google Scholar] [CrossRef] [Green Version]
- Shin, D.H.; Shim, H.S.; Kim, T.J.; Park, H.S.; La Choi, Y.; Kim, W.S.; Kim, L.; Chang, S.H.; Song, J.S.; Kim, H.J.; et al. Provisional guideline recommendation for EGFR gene mutation testing in liquid samples of lung cancer patients: A proposal by the korean cardiopulmonary pathology study group. J. Pathol. Transl. Med. 2019, 53, 153–158. [Google Scholar] [CrossRef] [Green Version]
- Corcoran, R.B.; Chabner, B.A. Application of Cell-free DNA Analysis to Cancer Treatment. N. Engl. J. Med. 2018, 379, 1754–1765. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Xu, X.; Qian, Z.; Zhang, C.; Niu, Y.; Wang, Z.; Sun, J.; Zhang, X.; Yu, Y. The biological functions and clinical applications of exosomes in lung cancer. Cell. Mol. Life Sci. 2019, 76, 4613–4633. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Hurley, J.; Roberts, D.; Chakrabortty, S.; Enderle, D.; Noerholm, M.; Breakefield, X.; Skog, J. Exosome-based liquid biopsies in cancer: Opportunities and challenges. Ann. Oncol. 2021, 32, 466–477. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Guo, M.; Lin, D.; Liang, D.; Zhao, L.; Zhao, R.; Wang, Y. Docetaxel-loaded exosomes for targeting non-small cell lung cancer: Preparation and evaluation in vitro and in vivo. Drug Deliv. 2021, 28, 1510–1523. [Google Scholar] [CrossRef]
- Harding, C.; Heuser, J.; Stahl, P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 1983, 97, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.T.; Teng, K.; Wu, C.; Adam, M.; Johnstone, R.M. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J. Cell Biol. 1985, 101, 942–948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colombo, M.; Moita, C.F.; van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Théry, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013, 126, 5553–5565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balaj, L.; Lessard, R.; Dai, L.; Cho, Y.-J.; Pomeroy, S.L.; Breakefield, X.O.; Skog, J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2011, 2, 180. [Google Scholar] [CrossRef] [PubMed]
- Ramteke, A.; Ting, H.; Agarwal, C.; Mateen, S.; Somasagara, R.; Hussain, A.; Graner, M.; Frederick, B.; Agarwal, R.; Deep, G. Exosomes secreted under hypoxia enhance invasiveness and stemness of prostate cancer cells by targeting adherens junction molecules. Mol. Carcinog. 2013, 54, 554–565. [Google Scholar] [CrossRef] [Green Version]
- Szczepanski, M.J.; Szajnik, M.; Welsh, A.; Whiteside, T.L.; Boyiadzis, M. Blast-derived microvesicles in sera from patients with acute myeloid leukemia suppress natural killer cell function via membrane-associated transforming growth factor-1. Haematologica 2011, 96, 1302–1309. [Google Scholar] [CrossRef]
- Lai, C.P.; Mardini, O.; Ericsson, M.; Prabhakar, S.; Maguire, C.A.; Chen, J.W.; Tannous, B.A.; Breakefield, X.O. Dynamic biodistribution of extracellular vesicles in vivo using a multimodal imaging reporter. ACS Nano 2013, 8, 483–494. [Google Scholar] [CrossRef] [Green Version]
- Rabinowits, G.; Gerçel-Taylor, C.; Day, J.M.; Taylor, D.D.; Kloecker, G.H. Exosomal MicroRNA: A diagnostic marker for lung cancer. Clin. Lung Cancer 2009, 10, 42–46. [Google Scholar] [CrossRef]
- Skotland, T.; Sandvig, K.; Llorente, A. Lipids in exosomes: Current knowledge and the way forward. Prog. Lipid Res. 2017, 66, 30–41. [Google Scholar] [CrossRef]
- Lai, A.; Elfeky, O.; Rice, G.E.; Salomon, C. Optimized specific isolation of placenta-derived exosomes from maternal circulation. Preeclampsia 2017, 1710, 131–138. [Google Scholar] [CrossRef]
- Escola, J.-M.; Kleijmeer, M.J.; Stoorvogel, W.; Griffith, J.M.; Yoshie, O.; Geuze, H.J. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 1998, 273, 20121–20127. [Google Scholar] [CrossRef] [Green Version]
- Theodoraki, M.-N.; Hoffmann, T.K.; Whiteside, T.L. Separation of plasma-derived exosomes into CD3(+) and CD3(−) fractions allows for association of immune cell and tumour cell markers with disease activity in HNSCC patients. Clin. Exp. Immunol. 2018, 192, 271–283. [Google Scholar] [CrossRef] [Green Version]
- Baranyai, T.; Herczeg, K.; Onódi, Z.; Voszka, I.; Módos, K.; Marton, N.; Nagy, G.; Mäger, I.; Wood, M.J.; El Andaloussi, S.; et al. Isolation of exosomes from blood plasma: Qualitative and quantitative comparison of ultracentrifugation and size exclusion chromatography methods. PLoS ONE 2015, 10, e0145686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alegre, E.; Rebmann, V.; LeMaoult, J.; Rodriguez, C.; Horn, P.A.; Diaz-Lagares, A.; Echeveste, J.I.; González, A. In vivo identification of an HLA-G complex as ubiquitinated protein circulating in exosomes. Eur. J. Immunol. 2013, 43, 1933–1939. [Google Scholar] [CrossRef] [Green Version]
- Barreiro, K.; Dwivedi, O.P.; Leparc, G.; Rolser, M.; Delic, D.; Forsblom, C.; Groop, P.; Groop, L.; Huber, T.B.; Puhka, M.; et al. Comparison of urinary extracellular vesicle isolation methods for transcriptomic biomarker research in diabetic kidney disease. J. Extracell. Vesicles 2020, 10, e12038. [Google Scholar] [CrossRef] [PubMed]
- Campoy, I.; Lanau, L.; Altadill, T.; Sequeiros, T.; Cabrera, S.; Cubo-Abert, M.; Pérez-Benavente, A.; Garcia, A.; Borrós, S.; Santamaria, A.; et al. Exosome-like vesicles in uterine aspirates: A comparison of ultracentrifugation-based isolation protocols. J. Transl. Med. 2016, 14, 180. [Google Scholar] [CrossRef] [Green Version]
- Tauro, B.J.; Greening, D.; Mathias, R.; Ji, H.; Mathivanan, S.; Scott, A.; Simpson, R.J. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods 2012, 56, 293–304. [Google Scholar] [CrossRef]
- Greening, D.W.; Xu, R.; Ji, H.; Tauro, B.J.; Simpson, R.J. A protocol for exosome isolation and characterization: Evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol. Biol. 2015, 1295, 179–209. [Google Scholar] [CrossRef] [PubMed]
- Macías, M.; Rebmann, V.; Mateos, B.; Varo, N.; Perez-Gracia, J.L.; Alegre, E.; González, A. Comparison of six commercial serum exosome isolation methods suitable for clinical laboratories. Effect in cytokine analysis. Clin. Chem. Lab. Med. 2019, 57, 1539–1545. [Google Scholar] [CrossRef] [PubMed]
- Gámez-Valero, A.; Monguió-Tortajada, M.; Carreras-Planella, L.; Franquesa, M.; Beyer, K.; Borràs, F.E. Size-exclusion chromatography-based isolation minimally alters extracellular Vesicles’ characteristics compared to precipitating agents. Sci. Rep. 2016, 6, srep33641. [Google Scholar] [CrossRef] [Green Version]
- Van Deun, J.; Mestdagh, P.; Sormunen, R.; Cocquyt, V.; Vermaelen, K.; Vandesompele, J.; Bracke, M.; De Wever, O.; Hendrix, A. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J. Extracell. Vesicles 2014, 3. [Google Scholar] [CrossRef] [Green Version]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3–22. [Google Scholar] [CrossRef]
- Livshits, M.A.; Khomyakova, E.; Evtushenko, E.; Lazarev, V.N.; Kulemin, N.; Semina, S.E.; Generozov, E.; Govorun, V.M. Isolation of exosomes by differential centrifugation: Theoretical analysis of a commonly used protocol. Sci. Rep. 2015, 5, 17319. [Google Scholar] [CrossRef]
- Cvjetkovic, A.; Lötvall, J.; Lässer, C. The influence of rotor type and centrifugation time on the yield and purity of extracellular vesicles. J. Extracell. Vesicles 2014, 3, 23111. [Google Scholar] [CrossRef]
- Witwer, K.W.; Buzás, E.I.; Bemis, L.; Bora, A.; Lässer, C.; Lötvall, J.; Hoen, E.N.N.; Piper, M.G.; Sivaraman, S.; Skog, J.; et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles 2013, 2, 20360. [Google Scholar] [CrossRef]
- Jeppesen, D.; Hvam, M.L.; Primdahl-Bengtson, B.; Boysen, A.T.; Whitehead, B.; Dyrskjøt, L.; Ørntoft, T.F.; Howard, K.A.; Ostenfeld, M.S. Comparative analysis of discrete exosome fractions obtained by differential centrifugation. J. Extracell. Vesicles 2014, 3, 25011. [Google Scholar] [CrossRef] [Green Version]
- Momen-Heravi, F.; Balaj, L.; Alian, S.; Trachtenberg, A.J.; Hochberg, F.H.; Skog, J.; Kuo, W.P. Impact of biofluid viscosity on size and sedimentation efficiency of the isolated microvesicles. Front. Physiol. 2012, 3, 162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Wang, C.; Li, T.; Liu, Z.; Li, L. Comparison of ultracentrifugation and density gradient separation methods for isolating Tca8113 human tongue cancer cell line-derived exosomes. Oncol. Lett. 2014, 8, 1701–1706. [Google Scholar] [CrossRef] [Green Version]
- Lobb, R.; Becker, M.; Wen, S.W.; Wong, C.S.F.; Wiegmans, A.P.; Leimgruber, A.; Möller, A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 2015, 4, 27031. [Google Scholar] [CrossRef] [PubMed]
- Nordin, J.; Lee, Y.; Vader, P.; Mäger, I.; Johansson, H.; Heusermann, W.; Wiklander, O.P.; Hällbrink, M.; Seow, Y.; Bultema, J.J.; et al. Ultrafiltration with size-exclusion liquid chromatography for high yield isolation of extracellular vesicles preserving intact biophysical and functional properties. Nanomed. Nanotechnol. Biol. Med. 2015, 11, 879–883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarovni, N.; Corrado, A.; Guazzi, P.; Zocco, D.; Lari, E.; Radano, G.; Muhhina, J.; Fondelli, C.; Gavrilova, J.; Chiesi, A. Integrated isolation and quantitative analysis of exosome shuttled proteins and nucleic acids using immunocapture approaches. Methods 2015, 87, 46–58. [Google Scholar] [CrossRef]
- Wubbolts, R.; Leckie, R.S.; Veenhuizen, P.T.M.; Schwarzmann, G.; Möbius, W.; Hoernschemeyer, J.; Slot, J.-W.; Geuze, H.J.; Stoorvogel, W. Proteomic and biochemical analyses of human B cell-derived exosomes. J. Biol. Chem. 2003, 278, 10963–10972. [Google Scholar] [CrossRef] [Green Version]
- Popović, M.; de Marco, A. Canonical and selective approaches in exosome purification and their implications for diagnostic accuracy. Transl. Cancer Res. 2017, 7, S209–S225. [Google Scholar] [CrossRef]
- Serrano-Pertierra, E.; Oliveira-Rodríguez, M.; Rivas, M.; Oliva, P.; Villafani, J.; Navarro, A.; Blanco-López, M.C.; Cernuda-Morollón, E. Characterization of plasma-derived extracellular vesicles isolated by different methods: A comparison study. Bioengineering 2019, 6, 8. [Google Scholar] [CrossRef] [Green Version]
- Brownlee, Z.; Lynn, K.D.; Thorpe, P.E.; Schroit, A.J. A novel “salting-out” procedure for the isolation of tumor-derived exosomes. J. Immunol. Methods 2014, 407, 120–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.B.; Yang, J.S.; Bin Lee, G.; Moon, M.H. Evaluation of exosome separation from human serum by frit-inlet asymmetrical flow field-flow fractionation and multiangle light scattering. Anal. Chim. Acta 2020, 1124, 137–145. [Google Scholar] [CrossRef]
- Alvarez, P.V.; Blazquez, R.; Sánchez, M.F.; DelaRosa, O.; Jorge, I.; Tapia-Araya, A.; Casado, J. Comparative study of isolated human mesenchymal stem cell derived exosomes for clinical use. Acta Bioquím. Clín. Latinoam. 2015, 49, 311–320. [Google Scholar]
- Diaz, G.; Bridges, C.; Lucas, M.; Cheng, Y.; Schorey, J.S.; Dobos, K.M.; Kruh-Garcia, N.A. Protein digestion, ultrafiltration, and size exclusion chromatography to optimize the isolation of exosomes from human blood plasma and serum. J. Vis. Exp. 2018, e57467. [Google Scholar] [CrossRef] [PubMed]
- Kooijmans, S.A.A.; Aleza, C.G.; Roffler, S.R.; Van Solinge, W.; Vader, P.; Schiffelers, R.M. Display of GPI-anchored anti-EGFR nanobodies on extracellular vesicles promotes tumour cell targeting. J. Extracell. Vesicles 2016, 5, 31053. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, M.; Ilmer, M.; Silva, L.P.; Hawke, D.; Recio, A.; Vorontsova, M.A.; Alt, E.; Vykoukal, J. Benchtop isolation and characterization of functional exosomes by sequential filtration. J. Chromatogr. A 2014, 1371, 125–135. [Google Scholar] [CrossRef]
- Heinemann, M.L.; Vykoukal, J. Sequential filtration: A gentle method for the isolation of functional extracellular vesicles. In Extracellular Vesicles: Methods and Protocols; Kuo, W.P., Jia, S., Eds.; Springer: New York, NY, USA, 2017; pp. 33–41. [Google Scholar]
- Liu, C.; Guoqing, H.; Tian, F.; Yang, N.; Yanping, D.; Ding, Y.; Wei, J.; Hu, G.; Nie, G.; Sun, J. Field-free isolation of exosomes from extracellular vesicles by microfluidic viscoelastic flows. ACS Nano 2017, 11, 6968–6976. [Google Scholar] [CrossRef] [Green Version]
- Musante, L.; Tataruch, D.; Gu, D.; Martin, A.B.; Calzaferri, G.; Aherne, S.; Holthofer, H. A simplified method to recover urinary vesicles for clinical applications and sample banking. Sci. Rep. 2014, 4, 7532. [Google Scholar] [CrossRef]
- Xu, Y.; Qin, S.; An, T.; Tang, Y.; Huang, Y.; Zheng, L. MiR-145 detection in urinary extracellular vesicles increase diagnostic efficiency of prostate cancer based on hydrostatic filtration dialysis method. Prostate 2017, 77, 1167–1175. [Google Scholar] [CrossRef]
- Vogel, R.; Coumans, F.A.W.; Maltesen, R.; Böing, A.N.; Bonnington, K.E.; Broekman, M.L.; Broom, M.F.; Buzás, E.I.; Christiansen, G.; Hajji, N.; et al. A standardized method to determine the concentration of extracellular vesicles using tunable resistive pulse sensing. J. Extracell. Vesicles 2016, 5, 31242. [Google Scholar] [CrossRef]
- Böing, A.N.; van der Pol, E.; Grootemaat, A.E.; Coumans, F.A.W.; Sturk, A.; Nieuwland, R. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J. Extracell. Vesicles 2014, 3, 23430. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Ramos, I.; Bancu, I.; Oliveira-Tercero, A.; Armengol, M.P.; Menezes-Neto, A.; Del Portillo, H.A.; Lauzurica-Valdemoros, R.; Borràs, F.E. Size-exclusion chromatography-based enrichment of extracellular vesicles from urine samples. J. Extracell. Vesicles 2015, 4, 27369. [Google Scholar] [CrossRef] [Green Version]
- Giddings, J.C.; Yang, F.J.; Myers, M.N. Flow-field-flow fractionation: A versatile new separation method. Science 1976, 193, 1244–1245. [Google Scholar] [CrossRef]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Mark, M.T.; Molina, H.; Martin, A.B.; Bojmar, L.; et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nature 2018, 20, 332–343. [Google Scholar] [CrossRef]
- Clayton, A.; Court, J.; Navabi, H.; Adams, M.; Mason, M.D.; Hobot, J.A.; Newman, G.R.; Jasani, B. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. J. Immunol. Methods 2001, 247, 163–174. [Google Scholar] [CrossRef]
- Deregibus, M.C.; Figliolini, F.; D′Antico, S.; Manzini, P.M.; Pasquino, C.; De Lena, M.; Tetta, C.; Brizzi, M.F.; Camussi, G. Charge-based precipitation of extracellular vesicles. Int. J. Mol. Med. 2016, 38, 1359–1366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, M.; Crow, J.; Roth, M.; Zeng, Y.; Godwin, A.K. Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab Chip 2014, 14, 3773–3780. [Google Scholar] [CrossRef] [Green Version]
- Kanwar, S.S.; Dunlay, C.J.; Simeone, D.M.; Nagrath, S. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip 2014, 14, 1891–1900. [Google Scholar] [CrossRef]
- Santana, S.M.; Antonyak, M.A.; Cerione, R.A.; Kirby, B.J. Microfluidic isolation of cancer-cell-derived microvesicles from hetergeneous extracellular shed vesicle populations. Biomed. Microdevices 2014, 16, 869–877. [Google Scholar] [CrossRef] [Green Version]
- Dudani, J.; Gossett, D.R.; Tse, H.T.K.; Lamm, R.J.; Kulkarni, R.P.; Di Carlo, D. Rapid inertial solution exchange for enrichment and flow cytometric detection of microvesicles. Biomicrofluidics 2015, 9, 014112. [Google Scholar] [CrossRef] [Green Version]
- Davies, R.T.; Kim, J.; Jang, S.C.; Choi, E.-J.; Gho, Y.S.; Park, J. Microfluidic filtration system to isolate extracellular vesicles from blood. Lab Chip 2012, 12, 5202–5210. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Yang, Y.; Zeng, Y.; He, M. A microfluidic ExoSearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis. Lab Chip 2015, 16, 489–496. [Google Scholar] [CrossRef] [Green Version]
- Royo, F.; Théry, C.; Falcón-Pérez, J.M.; Nieuwland, R.; Witwer, K.W. Methods for separation and characterization of extracellular vesicles: Results of a worldwide survey performed by the ISEV rigor and standardization subcommittee. Cells 2020, 9, 1955. [Google Scholar] [CrossRef]
- Zaborowski, M.P.; Lee, K.; Na, Y.J.; Sammarco, A.; Zhang, X.; Iwanicki, M.; Cheah, P.S.; Lin, H.-Y.; Zinter, M.; Chou, C.-Y.; et al. Methods for systematic identification of membrane proteins for specific capture of cancer-derived extracellular vesicles. Cell Rep. 2019, 27, 255–268.e6. [Google Scholar] [CrossRef] [Green Version]
- Choi, E.-S.; Faruque, H.; Kim, J.-H.; Kim, K.; Choi, J.; Kim, B.; Kim, B.; Kim, Y.; Woo, M.; Park, J.; et al. CD5L as an extracellular vesicle-derived biomarker for liquid biopsy of lung cancer. Diagnostics 2021, 11, 620. [Google Scholar] [CrossRef] [PubMed]
- Yuana, Y.; Koning, R.; Kuil, M.E.; Rensen, P.C.N.; Koster, A.; Bertina, R.M.; Osanto, S. Cryo-electron microscopy of extracellular vesicles in fresh plasma. J. Extracell. Vesicles 2013, 2, 21494. [Google Scholar] [CrossRef]
- Ashby, J.; Flack, K.; Jimenez, L.A.; Duan, Y.; Khatib, A.-K.; Somlo, G.; Wang, S.E.; Cui, X.; Zhong, W. Distribution profiling of circulating MicroRNAs in serum. Anal. Chem. 2014, 86, 9343–9349. [Google Scholar] [CrossRef] [Green Version]
- Gerratana, L.; Basile, D.; Toffoletto, B.; Bulfoni, M.; Zago, S.; Magini, A.; Lera, M.; Pelizzari, G.; Parisse, P.; Casalis, L.; et al. Biologically driven cut-off definition of lymphocyte ratios in metastatic breast cancer and association with exosomal subpopulations and prognosis. Sci. Rep. 2020, 10, 7010. [Google Scholar] [CrossRef]
- Pan, D.; Chen, J.; Feng, C.; Wu, W.; Wang, Y.; Tong, J.; Zhou, D. Preferential localization of MUC1 glycoprotein in exosomes secreted by non-small cell lung carcinoma cells. Int. J. Mol. Sci. 2019, 20, 323. [Google Scholar] [CrossRef] [Green Version]
- Yoh, K.E.; Lowe, C.J.; Mahajan, S.; Suttmann, R.; Nguy, T.; Reichelt, M.; Yang, J.; Melendez, R.; Li, Y.; Molinero, L.; et al. Enrichment of circulating tumor-derived extracellular vesicles from human plasma. J. Immunol. Methods 2020, 490, 112936. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- Lötvall, J.; Hill, A.; Hochberg, F.; Buzás, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2014, 3, 26913. [Google Scholar] [CrossRef]
- Alegre, E.; Zubiri, L.; Perez-Gracia, J.L.; González-Cao, M.; Soria, L.; Martín-Algarra, S.; González, A. Circulating melanoma exosomes as diagnostic and prognosis biomarkers. Clin. Chim. Acta 2016, 454, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Helwa, I.; Cai, J.; Drewry, M.D.; Zimmerman, A.; Dinkins, M.B.; Khaled, M.L.; Seremwe, M.; Dismuke, W.M.; Bieberich, E.; Stamer, W.D.; et al. A comparative study of serum exosome isolation using differential ultracentrifugation and three commercial reagents. PLoS ONE 2017, 12, e0170628. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, K.R.; Paulsen, B.S.; Bæk, R.; Varming, K.; Sorensen, B.; Jørgensen, M.M. Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma. J. Extracell. Vesicles 2015, 4, 26659. [Google Scholar] [CrossRef] [PubMed]
- Poupardin, R.; Wolf, M.; Strunk, D. Adherence to minimal experimental requirements for defining extracellular vesicles and their functions. Adv. Drug Deliv. Rev. 2021, 176, 113872. [Google Scholar] [CrossRef]
- Mulcahy, L.; Pink, R.; Carter, D.R.F. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3. [Google Scholar] [CrossRef] [Green Version]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Gaudio, E.; Santhanam, R.; Lovat, F.; Fadda, P.; Mao, C.; Nuovo, G.J.; et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. USA 2012, 109, E2110–E2116. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Yan, Y.; Yang, Y.; Hong, X.; Wang, M.; Yang, Z.; Liu, B.; Ye, L. MiR-210 in exosomes derived from CAFs promotes non-small cell lung cancer migration and invasion through PTEN/PI3K/AKT pathway. Cell. Signal. 2020, 73, 109675. [Google Scholar] [CrossRef]
- Zhang, R.; Xia, Y.; Wang, Z.; Zheng, J.; Chen, Y.; Li, X.; Wang, Y.; Ming, H. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2017, 490, 406–414. [Google Scholar] [CrossRef]
- Taverna, S.; Pucci, M.; Giallombardo, M.; Di Bella, M.A.; Santarpia, M.; Reclusa, P.; Gil-Bazo, I.; Rolfo, C.; Alessandro, R. Amphiregulin contained in NSCLC-exosomes induces osteoclast differentiation through the activation of EGFR pathway. Sci. Rep. 2017, 7, 3170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chalmin, F.; Ladoire, S.; Mignot, G.; Vincent, J.; Bruchard, M.; Remy-Martin, J.-P.; Boireau, W.; Rouleau, A.; Simon, B.; Lanneau, D.; et al. Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J. Clin. Investig. 2010, 120, 457–471. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, T.L. Tumor-derived exosomes and their role in cancer progression. J. Clin. Investig. 2016, 74, 103–141. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Luo, F.; Wang, B.; Li, H.; Xu, Y.; Liu, X.; Shi, L.; Lu, X.; Xu, W.; Lu, L.; et al. STAT3-regulated exosomal miR-21 promotes angiogenesis and is involved in neoplastic processes of transformed human bronchial epithelial cells. Cancer Lett. 2016, 370, 125–135. [Google Scholar] [CrossRef]
- Wu, H.; Zhou, J.; Mei, S.; Wu, D.; Mu, Z.; Chen, B.; Xie, Y.; Ye, Y.; Liu, J. Circulating exosomal microRNA-96 promotes cell proliferation, migration and drug resistance by targeting LMO7. J. Cell. Mol. Med. 2016, 21, 1228–1236. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, M.; Hu, C. Exosomal transfer of miR-214 mediates gefitinib resistance in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2018, 507, 457–464. [Google Scholar] [CrossRef]
- Wu, S.; Luo, M.; To, K.K.W.; Zhang, J.; Su, C.; Zhang, H.; An, S.; Wang, F.; Chen, D.; Fu, L. Intercellular transfer of exosomal wild type EGFR triggers osimertinib resistance in non-small cell lung cancer. Mol. Cancer 2021, 20, 1–17. [Google Scholar] [CrossRef]
- Brabletz, T.; Kalluri, R.; Nieto, M.A.; Weinberg, R.A. EMT in cancer. Nat. Rev. Cancer 2018, 18, 128–134. [Google Scholar] [CrossRef]
- Rahman, M.A.; Barger, J.F.; Lovat, F.; Gao, M.; Otterson, G.A.; Nana-Sinkam, P. Lung cancer exosomes as drivers of epithelial mesenchymal transition. Oncotarget 2016, 7, 54852–54866. [Google Scholar] [CrossRef]
- He, S.; Li, Z.; Yu, Y.; Zeng, Q.; Cheng, Y.; Ji, W.; Xia, W.; Lu, S. Exosomal miR-499a-5p promotes cell proliferation, migration and EMT via mTOR signaling pathway in lung adenocarcinoma. Exp. Cell Res. 2019, 379, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, T.Y.; Lee, M.S.; Mun, J.Y.; Ihm, C.; Kim, S.A. Exosome cargo reflects TGF-β1-mediated epithelial-to-mesenchymal transition (EMT) status in A549 human lung adenocarcinoma cells. Biochem. Biophys. Res. Commun. 2016, 478, 643–648. [Google Scholar] [CrossRef]
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Mark, M.T.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, C.; Duan, X.; Yao, X.; Wan, J.; Cheng, Y.; Wang, Y.; Yan, Y.; Zhang, L.; Zhu, L.; Ni, C.; et al. Tumour-derived exosomal miR-3473b promotes lung tumour cell intrapulmonary colonization by activating the nuclear factor-κB of local fibroblasts. J. Cell. Mol. Med. 2020, 24, 7802–7813. [Google Scholar] [CrossRef]
- Gan, D.-X.; Wang, Y.-B.; He, M.-Y.; Chen, Z.-Y.; Qin, X.-X.; Miao, Z.-W.; Chen, Y.-H.; Li, B. Lung cancer cells-controlled Dkk-1 production in brain metastatic cascade drive microglia to acquire a pro-tumorigenic phenotype. Front. Cell Dev. Biol. 2020, 8, 1594. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.-L.; Hung, J.-Y.; Chang, W.-A.; Lin, Y.-S.; Pan, Y.-C.; Tsai, P.-H.; Wu, C.-Y.; Kuo, P.-L. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene 2017, 36, 4929–4942. [Google Scholar] [CrossRef]
- Li, Z.; Zeng, C.; Nong, Q.; Long, F.; Liu, J.; Mu, Z.; Chen, B.; Wu, D.; Wu, H. Exosomal leucine-rich-alpha2-glycoprotein 1 Derived from Non-Small-Cell Lung Cancer Cells Promotes angiogenesis via TGF-β signal pathway. Mol. Ther.-Oncolytics 2019, 14, 313–322. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.-H.; Li, Y.; Zhang, J.; Rong, J.; Ye, S. Epidermal growth factor receptor-containing exosomes induce tumor-specific regulatory T cells. Cancer Investig. 2013, 31, 330–335. [Google Scholar] [CrossRef]
- Berchem, G.; Noman, M.Z.; Bosseler, M.; Paggetti, J.; Baconnais, S.; Le Cam, E.; Nanbakhsh, A.; Moussay, E.; Mami-Chouaib, F.; Janji, B.; et al. Hypoxic tumor-derived microvesicles negatively regulate NK cell function by a mechanism involving TGF-β and miR23a transfer. OncoImmunology 2015, 5, e1062968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, G.; Huang, A.C.; Zhang, W.; Zhang, G.; Wu, M.; Xu, W.; Yu, Z.; Yang, J.; Wang, B.; Sun, H.; et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 2018, 560, 382–386. [Google Scholar] [CrossRef]
- Wu, J.; Shen, Z. Exosomal miRNAs as biomarkers for diagnostic and prognostic in lung cancer. Cancer Med. 2020, 9, 6909–6922. [Google Scholar] [CrossRef]
- Sandfeld-Paulsen, B.; Jakobsen, K.R.; Bæk, R.; Folkersen, B.H.; Rasmussen, T.R.; Meldgaard, P.; Varming, K.; Jørgensen, M.M.; Sorensen, B. Exosomal proteins as diagnostic biomarkers in lung cancer. J. Thorac. Oncol. 2016, 11, 1701–1710. [Google Scholar] [CrossRef] [Green Version]
- Ueda, K.; Ishikawa, N.; Tatsuguchi, A.; Saichi, N.; Fujii, R.; Nakagawa, H. Antibody-coupled monolithic silica microtips for highthroughput molecular profiling of circulating exosomes. Sci. Rep. 2014, 4, srep06232. [Google Scholar] [CrossRef] [PubMed]
- Niu, L.; Song, X.; Wang, N.; Xue, L.; Song, X.; Xie, L. Tumor-derived exosomal proteins as diagnostic biomarkers in non-small cell lung cancer. Cancer Sci. 2018, 110, 433–442. [Google Scholar] [CrossRef] [Green Version]
- Vykoukal, J.; Sun, N.; Aguilar-Bonavides, C.; Katayama, H.; Tanaka, I.; Fahrmann, J.F.; Capello, M.; Fujimoto, J.; Aguilar, M.; Wistuba, I.I.; et al. Plasma-derived extracellular vesicle proteins as a source of biomarkers for lung adenocarcinoma. Oncotarget 2017, 8, 95466–95480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Zhang, Y.; Qiu, F.; Qiu, Z. Proteomic identification of exosomal LRG1: A potential urinary biomarker for detecting NSCLC. Electrophoresis 2011, 32, 1976–1983. [Google Scholar] [CrossRef] [PubMed]
- Sandfeld-Paulsen, B.; Aggerholm-Pedersen, N.; Baek, R.; Jakobsen, K.R.; Meldgaard, P.; Folkersen, B.; Rasmussen, T.R.; Varming, K.; Jørgensen, M.M.; Sorensen, B. Exosomal proteins as prognostic biomarkers in non-small cell lung cancer. Mol. Oncol. 2016, 10, 1595–1602. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Chen, M.; Gu, J.; Niu, K.; Zhao, X.; Zheng, L.; Xu, Z.; Yu, Y.; Li, F.; Meng, L.; et al. Novel biomarkers of dynamic blood PD-L1 expression for immune checkpoint inhibitors in advanced non-small-cell lung cancer patients. Front. Immunol. 2021, 12, 665133. [Google Scholar] [CrossRef]
- Chanteloup, G.; Cordonnier, M.; Isambert, N.; Bertaut, A.; Hervieu, A.; Hennequin, A.; Luu, M.; Zanetta, S.; Coudert, B.; Bengrine, L.; et al. Monitoring HSP70 exosomes in cancer patients’ follow up: A clinical prospective pilot study. J. Extracell. Vesicles 2020, 9, 1766192. [Google Scholar] [CrossRef]
- Cazzoli, R.; Buttitta, F.; DI Nicola, M.; Malatesta, S.; Marchetti, A.; Rom, W.; Pass, H.I. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J. Thorac. Oncol. 2013, 8, 1156–1162. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Jiang, X.; Li, J.; Wang, J.; Binang, H.; Shi, S.; Duan, W.; Zhao, Y.; Zhang, Y. Serum exosomal miR-1269a serves as a diagnostic marker and plays an oncogenic role in non-small cell lung cancer. Thorac. Cancer 2020, 11, 3436–3447. [Google Scholar] [CrossRef]
- Zhang, Z.; Tang, Y.; Song, X.; Xie, L.; Zhao, S.; Song, X. Tumor-derived exosomal miRNAs as diagnostic biomarkers in non-small cell lung cancer. Front. Oncol. 2020, 10, 560025. [Google Scholar] [CrossRef]
- Wang, J.; Xue, H.; Zhu, Z.; Gao, J.; Zhao, M.; Ma, Z. Expression of serum exosomal miR-23b-3p in non-small cell lung cancer and its diagnostic efficacy. Oncol. Lett. 2020, 20, 30. [Google Scholar] [CrossRef]
- Wang, N.; Guo, W.; Song, X.; Liu, L.; Niu, L.; Song, X.; Xie, L. Tumor-associated exosomal miRNA biomarkers to differentiate metastatic vs. nonmetastatic non-small cell lung cancer. Clin. Chem. Lab. Med. 2020, 58, 1535–1545. [Google Scholar] [CrossRef]
- Zhang, Z.-J.; Song, X.-G.; Xie, L.; Wang, K.-Y.; Tang, Y.-Y.; Yu, M.; Feng, X.-D.; Song, X.-R. Circulating serum exosomal miR-20b-5p and miR-3187-5p as efficient diagnostic biomarkers for early-stage non-small cell lung cancer. Exp. Biol. Med. 2020, 245, 1428–1436. [Google Scholar] [CrossRef]
- Yang, G.; Wang, T.; Qu, X.; Chen, S.; Han, Z.; Chen, S.; Chen, M.; Lin, J.; Yu, S.; Gao, L.; et al. Exosomal miR-21/Let-7a ratio distinguishes non-small cell lung cancer from benign pulmonary diseases. Asia-Pac. J. Clin. Oncol. 2020, 16, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Chen, Y.; Chen, H.; Fei, S.; Chen, D.; Cai, X.; Liu, L.; Lin, B.; Su, H.; Zhao, L.; et al. Evaluation of tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non–small cell lung cancer using next-generation sequencing. Clin. Cancer Res. 2017, 23, 5311–5319. [Google Scholar] [CrossRef] [Green Version]
- Dejima, H.; Iinuma, H.; Kanaoka, R.; Matsutani, N.; Kawamura, M. Exosomal microRNA in plasma as a non-invasive biomarker for the recurrence of non-small cell lung cancer. Oncol. Lett. 2017, 13, 1256–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, J.; Wang, Y.; Zou, Y.-Q.; Chen, X.; Huang, B.; Liu, J.; Xu, Y.-M.; Zhang, J.; Yang, W.-M.; Wei-Ming, Y.; et al. Differential miRNA expression in pleural effusions derived from extracellular vesicles of patients with lung cancer, pulmonary tuberculosis, or pneumonia. Tumor Biol. 2016, 37, 15835–15845. [Google Scholar] [CrossRef]
- Halvorsen, A.R.; Bjaanæs, M.; Leblanc, M.; Holm, A.M.; Bolstad, N.; Rubio, L.; Peñalver, J.C.; Cervera, J.; Mojarrieta, J.C.; López-Guerrero, J.A.; et al. A unique set of 6 circulating microRNAs for early detection of non-small cell lung cancer. Oncotarget 2016, 7, 37250–37259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamiya, H.; Mitani, A.; Saito, A.; Ishimori, T.; Saito, M.; Isago, H.; Jo, T.; Yamauchi, Y.; Tanaka, G.; Nagase, T. Exosomal microRNA expression profiling in patients with lung adenocarcinoma-associated malignant pleural effusion. Anticancer Res. 2018, 38, 6707–6714. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Lu, F.; Wang, J.; Wang, K.; Liu, B.; Li, N.; Tang, B. A portable point-of-care testing system to diagnose lung cancer through the detection of exosomal miRNA in urine and saliva. Chem. Commun. 2020, 56, 8968–8971. [Google Scholar] [CrossRef]
- Yang, X.-R.; Pi, C.; Yu, R.; Fan, X.-J.; Peng, X.-X.; Zhang, X.-C.; Chen, Z.-H.; Wu, X.; Shao, Y.; Wu, Y.-L.; et al. Correlation of exosomal microRNA clusters with bone metastasis in non-small cell lung cancer. Clin. Exp. Metastasis 2020, 38, 109–117. [Google Scholar] [CrossRef]
- Yuwen, D.-L.; Sheng, B.-B.; Liu, J.; Wenyu, W.; Shu, Y.-Q. MiR-146a-5p level in serum exosomes predicts therapeutic effect of cisplatin in non-small cell lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 2650–2658. [Google Scholar]
- Zheng, Q.; Ding, H.; Wang, L.; Yan, Y.; Wan, Y.; Yi, Y.; Tao, L.; Zhu, C. Circulating exosomal miR-96 as a novel biomarker for radioresistant non-small-cell lung cancer. J. Oncol. 2021, 2021, 5893981. [Google Scholar] [CrossRef]
- Peng, X.X.; Yu, R.; Wu, X.; Wu, S.Y.; Pi, C.; Chen, Z.H.; Zhang, X.C.; Gao, C.Y.; Shao, Y.W.; Liu, L.; et al. Correlation of plasma exosomal microRNAs with the efficacy of immunotherapy in EGFR/ALK wild-type advanced non-small cell lung cancer. J. Immunother. Cancer 2020, 8, e000376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, B.; Wang, P.; Gu, L.; Liu, J. Use of four genes in exosomes as biomarkers for the identification of lung adenocarcinoma and lung squamous cell carcinoma. Oncol. Lett. 2021, 21, 249. [Google Scholar] [CrossRef]
- Dong, Q.; Dong, L.; Liu, S.; Kong, Y.; Zhang, M.; Wang, X. Tumor-derived exosomal eIF4E as a biomarker for survival prediction in patients with non-small cell lung cancer. Med. Sci. Monit. 2020, 26, e923210. [Google Scholar] [CrossRef] [PubMed]
- Del Re, M.; Cucchiara, F.; Rofi, E.; Fontanelli, L.; Petrini, I.; Gri, N.; Pasquini, G.; Rizzo, M.; Gabelloni, M.; Belluomini, L.; et al. A multiparametric approach to improve the prediction of response to immunotherapy in patients with metastatic NSCLC. Cancer Immunol. Immunother. 2020, 70, 1667–1678. [Google Scholar] [CrossRef] [PubMed]
- Xian, J.; Zeng, Y.; Chen, S.; Lu, L.; Liu, L.; Chen, J.; Rao, B.; Zhao, Z.; Liu, J.; Xie, C.; et al. Discovery of a novel linc01125 isoform in serum exosomes as a promising biomarker for NSCLC diagnosis and survival assessment. Carcinogenesis 2021, 42, 831–841. [Google Scholar] [CrossRef]
- Li, L.; Li, W.; Chen, N.; Zhao, H.; Xu, G.; Zhao, Y.; Pan, X.; Zhang, X.; Zhou, L.; Yu, D.; et al. FLI1 exonic circular RNAs as a novel oncogenic driver to promote tumor metastasis in small cell lung cancer. Clin. Cancer Res. 2018, 25, 1302–1317. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhang, H.; Wang, J.; Li, B.; Wang, X. Circular RNA expression profile of lung squamous cell carcinoma: Identification of potential biomarkers and therapeutic targets. Biosci. Rep. 2020, 40, BSR20194512. [Google Scholar] [CrossRef] [Green Version]
- He, F.; Zhong, X.; Lin, Z.; Lin, J.; Qiu, M.; Li, X.; Hu, Z. Plasma exo-hsa_circRNA_0056616: A potential biomarker for lymph node metastasis in lung adenocarcinoma. J. Cancer 2020, 11, 4037–4046. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Nan, A.; Chen, L.; Li, X.; Jia, Y.; Qiu, M.; Dai, X.; Zhou, H.; Zhu, J.; Zhang, H.; et al. Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol. Cancer 2020, 19, 101. [Google Scholar] [CrossRef]
- Xian, J.; Su, W.; Liu, L.; Rao, B.; Lin, M.; Feng, Y.; Qiu, F.; Chen, J.; Zhou, Q.; Zhao, Z.; et al. Identification of three circular RNA cargoes in serum exosomes as diagnostic biomarkers of non–small-cell lung cancer in the chinese population. J. Mol. Diagn. 2020, 22, 1096–1108. [Google Scholar] [CrossRef]
- Jørgensen, M.M.; Baek, R.; Pedersen, S.; Sondergaard, E.K.L.; Kristensen, S.R.; Varming, K. Extracellular Vesicle (EV) Array: Microarray capturing of exosomes and other extracellular vesicles for multiplexed phenotyping. J. Extracell. Vesicles 2013, 2, 20920. [Google Scholar] [CrossRef] [PubMed]
- Erdbrügger, U.; Blijdorp, C.J.; Bijnsdorp, I.V.; Borràs, F.E.; Burger, D.; Bussolati, B.; Byrd, J.B.; Clayton, A.; Dear, J.W.; Falcón-Pérez, J.M.; et al. Urinary extracellular vesicles: A position paper by the Urine Task Force of the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2021, 10, e12093. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, K.; Kawamoto, K.; Eguchi, H.; Tanemura, M.; Tanida, T.; Tomimaru, Y.; Akita, H.; Hama, N.; Wada, H.; Kobayashi, S.; et al. Clinicopathological significance of leucine-rich α2-glycoprotein-1 in sera of patients with pancreatic cancer. Pancreas 2015, 44, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yin, H.; Zhu, J.; Buckanovich, R.J.; Thorpe, J.D.; Dai, J.; Urban, N.; Lubman, D.M. Validation of LRG1 as a potential biomarker for detection of epithelial ovarian cancer by a blinded study. PLoS ONE 2015, 10, e0121112. [Google Scholar] [CrossRef] [PubMed]
- Alegre, E.; Fusco, J.P.; Restituto, P.; Salas, D.; Rodriguez-Ruiz, E.M.; Andueza, M.-P.; Pajares, M.J.; Patiño-García, A.; Pio, R.; Lozano, M.D.; et al. Total and mutated EGFR quantification in cell-free DNA from non-small cell lung cancer patients detects tumor heterogeneity and presents prognostic value. Tumor Biol. 2016, 37, 13687–13694. [Google Scholar] [CrossRef]
- Fortunato, O.; Gasparini, P.; Boeri, M.; Sozzi, G. Exo-miRNAs as a new tool for liquid biopsy in lung cancer. Cancers 2019, 11, 888. [Google Scholar] [CrossRef] [Green Version]
- Kuchuk, M.; Addison, C.L.; Clemons, M.; Kuchuk, I.; Wheatley-Price, P. Incidence and consequences of bone metastases in lung cancer patients. J. Bone Oncol. 2013, 2, 22–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brinkmann, K.; Enderle, D.; Koestler, T.; Bentink, S.; Emenegger, J.; Spiel, A.; Mueller, R.; O′Neill, V.; Skog, J.; Noerholm, M. Abstract 545: Plasma-based diagnostics for detection of EML4-ALK fusion transcripts in NSCLC patients. Cancer Res. 2015, 75, 545. [Google Scholar] [CrossRef]
- Karimzadeh, M.R.; Seyedtaghia, M.R.; Soudyab, M.; Nezamnia, M.; Kidde, J.; Sahebkar, A. Exosomal long noncoding RNAs: Insights into emerging diagnostic and therapeutic applications in lung cancer. J. Oncol. 2020, 2020, 7630197. [Google Scholar] [CrossRef]
- Wu, Z.; Xu, Z.; Yu, B.; Zhang, J.; Yu, B. The Potential Diagnostic Value of Exosomal Long Noncoding RNAs in Solid Tumors: A Meta-Analysis and Systematic Review. BioMed Res. Int. 2020, 2020, 6786875. [Google Scholar] [CrossRef] [PubMed]
- Macías, M.; Alegre, E.; Alkorta-Aranburu, G.; Patiño-García, A.; Mateos, B.; Andueza, M.P.; Gúrpide, A.; Lopez-Picazo, J.M.; Gil-Bazo, I.; Perez-Gracia, J.L.; et al. The dynamic use of EGFR mutation analysis in cell-free DNA as a follow-up biomarker during different treatment lines in non-small-cell lung cancer patients. Dis. Markers 2019, 2019, 7954921. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Method | Isolation Principle | Assessment Parameters | Advantages | Disadvantages | Examples of Available Commercial Kits | References | ||
|---|---|---|---|---|---|---|---|---|
| Time | Purity | Recovery | ||||||
| Ultracentrifugation | Density by centrifugations at increasing speeds | +++ | + | + | Isolation of large volumes, no addition of chemicals, no pretreatment needed, most used method | Time consuming, expensive equipment, low purity, low reproducibility, damage of vesicles | [27,29,30,31,32,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49] | |
| Density gradient ultracentrifugation | Density by centrifugations in a density gradient | +++ | ++ | + | Effective in separation of EV from protein aggregates, high purity, no addition of chemicals | Time consuming, complex, low yields, fails to separate large vesicles with similar sedimentation rates | OptiPrep | [30,31,32,35,42,43,46,47] |
| Ultrafiltration | Size and molecular weight. Membranes with defined pore diameter or molecular weight cut offs | ++ | + | ++ | Simple and fast procedure, no special instrumentation, scalable | Clogging and trapping of vesicles on the filter, low yield, deformation of vesicles and lysis of exosomes, low purity | Amicon Ultra Centrifugal filters Vivaspin Centrifugal Concentrators | [30,43,44,50,51,52,53,54,55] |
| Hydrostatic filtration dialysis | Size. Diffusion of particles across a porous membrane at concentration gradient | +++ | + | ++ | Simple, inexpensive, scalable, appropriate for diluted samples as urine | Selectivity of separation dependent on the cut-off, low purity | [29,56,57,58] | |
| Size exclusion chromatography | Size. Small particles penetrate a porous stationary phase and elute at different rates | ++ | ++ | ++ | Preserves vesicles integrity and biological activity, high recovery and reproducibility | Low yield, might require concentration, difficulty in scaling | Exo-spin qEV Extracellular Vesicle Isolation | [27,33,34,39,43,44,47,52,53,59,60,61] |
| Asymmetric flow field-flow fractionation (AF4) | Size. Separation of particles in a channel with parabolic longitudinal flow combined with an external gradient | ++ | ++ | ++ | Possible EV subpopulation separation, possibility to couple to multidetection systems | Time consuming procedure, requires special equipment | [47,50,62,63] | |
| Immunoaffinity | Specific binding between antigens expressed on the exosome surface and corresponding antibodies | ++ | +++ | + | High purity and specificity, high selectivity, preservation of the activity of exosomal proteins, no protein contamination | Low yield, expensive, no scaling-up, EV cannot be readily eluted off the complexes with antibodies, antigenic epitopes might be blocked or masked | Dynabeads ExoFlow96 and 32 Exosome IP Kits ExoRNeasy Serum/Plasma Maxi Kit | [31,32,33,39,45,46,47,64] |
| Precipitation with polymers | Change in either the solubility, aggregate formation or both, after reagent addition | ++ | + | +++ | High recovery, simple and fast procedure, no expensive equipment requirement, scalable | Low purity | ExoQuick Invitrogen Total Exosome Isolation Kit | [33,34,35,39,43,45,47,48,49,65] |
| Microfluidics technology | Separation according to size, external markers or innovative sorting mechanisms such as acoustic, electrophoretic or electromagnetic fields | ++ | +++ | +++ | High purity and recovery, efficiency, minimal sample volume and reagent consumption, fast, reduce cross-contamination | Cost, additional equipment and complexity of devices | [39,66,67,68,69,70,71] | |
| Molecule | Sample | Number of Subjects | Isolation Methods | Characterization Methods | Utility | Comments | Authors |
|---|---|---|---|---|---|---|---|
| CD151, CD171, and tetraspanin 8 | Plasma | 336 LC + 126 C | EV array | - | Diagnosis | AUC calculated between LC and controls and when subdividing in AC, SCC and SCLC. NYESO1, HER2, EGFRvIII, SFTPD, Florilin1, CD142 and Mucin 16 also analyzed | Sandfeld-Paulsen et al. [111] |
| CD91 (+CEA) | Serum | Screening set: 10 C, 10 IP, 14 AC, 12 SCC Validation set: 54 C, 19 IP, 105 AC, 34 SCC | Immune-affinity for screening set ELISA with anti-CD9 in validation set | - | Diagnosis | Screening set: isolation by immune-affinity with anti-CD9 tips and proteomic study to identify CD9 Validation Set: ELISA with anti CD9 as capture antibody and anti-CD91 as detection antibody | Ueda et al. [112] |
| AHSG and ECM1 | Serum | 125 NSCLC + 46 C | Ultracentrifugation | TEM/NTA/WB | Diagnosis (including early stage) | Differentially expressed proteins identified by mass spectrometry | Niu et al. [113] |
| Panel of 30 proteins | Plasma | 109 advanced NSCLC + 110 C | EV Array | - | Diagnosis | Array for 37 proteins | Jakobsen et al. [84] |
| SRGN, TPM3, THBS1 and HUWE1 | Plasma | 13 AC + 15 C | Density gradient | TEM/NTA/WB | Diagnosis | 108 differentially expressed proteins identified by mass spectrometry | Vykoukal et al. [114] |
| CD5L, CLEC3B, ITIH4, SERFINF1, SAA4, SERFINC1, and C20ORF3 | Serum | 20 AC + 20 SCC + 20 SCLC + 20 C | Polyethylene glycol -based precipitation and immunoaffinity separation using antibodies against CD9, CD63, CD81, and EpCAM | TEM/NTA/DLS/WB | Diagnosis | Differentially expressed proteins identified by mass spectrometry; 55 confirmed by Western blot. CD5L highest AUC | Choi et al. [74] |
| LRG1 | Urine | 8 NSCLC + 10 C | Ultracentrifugation | TEM | Diagnosis | Differentially expressed proteins identified by mass spectrometry | Li et al. [115] |
| CD171 (1), NY-ESO-1 (2) | EDTA Plasma | 276 NSCLC | EV array | - | Prognosis: (1) OS, (2) HR | Array for 49 proteins | Sandfeld-Paulsen et al. [116] |
| PD-L1 | Plasma | 33 NSCLC | Precipitation | TEM/NTA/WB | Prognosis: OS and PFS | Quantification with Simoa Bead Technology | Yang et al. [117] |
| HSP70 | EDTA Plasma | 20NSCLC+ 14 C + 10 BC | Ultracentrifugation | NTA/TEM | Diagnosis, prognosis (metastasis detection), monitoring | HSP70 barely detected in plasma. Exosomal HSP70 correlates with tissue analysis | Chanteloup et al. [118] |
| Molecule | Sample | Number of Subjects | Isolation Methods | Characterization Methods | Utility | Comments | Authors |
|---|---|---|---|---|---|---|---|
| (1) miR-378a, miR-379, miR-139-5p, and miR-200b-5p (2) miR-151a-5p, miR-30a-3p, miR-200b-5p, miR-629, miR-100, and miR-154-3p | Plasma | Screening set: 10 AC+ 10 LG + 10 C Validation set: 50 AC+ 30 LG + 25 C | Precipitation | - | (1) Diagnosis AC+ LG vs. C (2) Diagnosis AC vs. LG | Wide-range miRNAs analysis (742 microRNAs) | Cazzoli et al. [119] |
| miR-9-3p, miR-205-5p, miR-210-5p and miR-1269a | Serum | Training set: 74 NSCLC + 74 C Validation set: 73 NSCLC + 75 C | Precipitation | TEM/NTA/WB | Diagnosis | 10 miRNAs to be analyzed were selected previously from TCGA database | Wang et al. [120] |
| miR-5684 (1) and miR-125b-5p (1, 2, 3) +CEA | Serum | 330 NSCLC + 312 C | Ultracentrifugation | TEM/tunable resistive pulse sensing/WB | (1) Diagnosis, (2) Prognosis: Metastasis detection and survival, (3) therapy monitoring | 22 miRNAs profiled by microarrays and verified by quantitative PCR | Zhang et al. [121] |
| miR-23b-3p + CEA + CYFRA21-1 | Serum | 80 NSCLC + 60 P + 30 C | Precipitation | TEM/NTA | Diagnosis Prognosis: tumor size, depth of invasion, liver metastasis and TNM stage | Quantification by RT-PCR. miRNA-39 was used as the external reference gene | Wang et al. [122] |
| let-7f-5p (1) miR-320a, miR-622 and let-7f-5p (2) + CEA and CYFRA21-1 | Plasma | 80 NSCLC + 30 C | Membrane affinity spin columns | - | (1) Diagnosis (2) Metastasis detection | miRNA array | Wang et al. [123] |
| miR-20b-5p and miR-3187-5p | Serum | 276 NSCLC (104 stage I) + 282 C | Ultracentrifugation | TEM/NTA/WB | Diagnosis (including early stage) | miRNAs profiled by microarrays and verified by quantitative PCR | Zhang et al. [124] |
| miR-21/Let-7a ratio | Serum | 75 NSCLC + 23 BPN + 18 PID +24 C | Precipitation | - | Diagnosis (including versus benign and inflammatory lung diseases) | Quantification by RT-PCR | Yang et al. [125] |
| let-7, miR-21, miR-24, and miR-486 (1) miR-181-5p, miR-30a-3p, miR-30e-3p, and miR-361-5p (2) miR-10b-5p, miR-15b-5p, and miR-320b (3) | Plasma | Testing set: stage I (16 AC + 10 SCC) + 12 C Validation set: stage I (10 AC + 10 SCC) + 30 C Symptomatic set 60 | Ultracentrifugation + immune-affinity with anti-EpCAM beads | NTA/WB | (1) Diagnosis at early stage (2) Histological classification: AC (3) Histological classification: SCC | Small RNA profile with RNA NGS and subsequent confirmation with RT-PCR. Normalization with cel-miR-39 | Jin et al. [126] |
| miR-4257 and miR-21 | EDTA Plasma | Screening set: 6 NSCLC Validation set: 129 stage I + 34 stage II +32 stage III + 30 C | Ultracentrifugation | TEM | Histological classification Prognosis: TNM stage, tumor size, lymphatic invasion, disease-free survival | miRNA selected with an array in 6 NSCLC patients (3 with and 3 without recurrence) | Dejima et al. [127] |
| miR-205-5p and miR-200b | Pleural effusion | 9 LC + 9 P + 9 T | Ultracentrifugation | TEM/NTA/WB | Diagnosis | Small RNA sequencing and subsequent confirmation with RT-PCR in 8 randomly chosen miRNAs | Lin et al. [128] |
| miR-429, miR-205, miR-200b, miR-203, miR-125b and miR-34b | Serum | Discovery set: 38 NSCLC + 16 COPD + 16 C Technical validation set: 16 NSCLC + 8 COPD + 6 C External validation set: 100 NSCLC + 58 C | Precipitation | - | Diagnosis (including early stage) | 754 microRNAs screened with TaqMan Low Density Arrays. In the 10 miRNAs upregulated a technical validation was performed by RT-PCR. Global normalization was performed | Halvorsen et al. [129] |
| miR-182 and miR-210 | Pleural effusion | 41 AC + 15 BPE | Precipitation | - | Diagnosis | miR-21, miR-31, miR-182, and miR-210 analyzed by RT-PCR. Normalization with miR-16 | Tamiya et al. [130] |
| miRNA-205 | Urine and saliva | 5 LC+ 5 C | Fe3O4@SiO2-aptamer nanoparticles | WB | Diagnosis | Development of a POCT device | Zhou et al. [131] |
| miR-574-5p and miR-328-3p and miR-423-3p | Plasma | 30 NSCLC (16 with and 14 without bone metastasis) + 14 C | Ultracentrifugation | WB | Bone metastasis detection | Small RNA sequencing | Yang et al. [132] |
| miR-146a-5p | Serum | 100 NSCLC with cisplatin-based chemotherapy | Precipitation | TEM/NTA/WB | Chemotherapy resistance Prognosis | Absolute miRNA levels quantify with RT-PCR with standard curves. Relative levels related to exosomal protein content | Yuwen et al. [133] |
| miR-1246 (1) and miR-96 (1,2,3) | Heparin Plasma | 52 NSCLC (27 Radioresistant + 25 radiosensitive) + 45 C | Lipid nanoprobe | TEM/NTA/WB | (1) Diagnosis (2) Radioresistance detection (3) Prognosis: OS | miR-21, miR-1246, let-7g, miR-210, miR-214, and miR-96 analyzed by RT-PCR. Normalization with cel-miR-39 | Zheng et al. [134] |
| hsa-miR-320d, hsa-miR-320c, and hsa-miR-320b | Plasma | 5 NSCLC with partial response to PD-1/PD-L1 inhibitors + 4 with progression + 7 C | Ultracentrifugation | TEM | Response to PD-1/PD-L1 inhibitors | Small RNA profile with RNA NGS; 155 miRNAs differentially expressed versus controls | Peng et al. [135] |
| Molecule | Sample | Number of Subjects | Isolation Methods | Characterization Methods | Utility | Comments | Authors |
|---|---|---|---|---|---|---|---|
| TP63, KRT5, CEACAM6 and SFTPB mRNAs | Serum | 54 AC + 16 SCC | Ultracentrifugation | TEM/NTA/WB | Histological classification | 17 miRNAs to be analyzed were selected previously from TCGA database as differentially expressed between AC and SCC. ACTB and SLC25A6 were used as internal references | Cao et al. [136] |
| eIF4E RNA | Serum | 99 NSCLC + 40 C | Precipitation | TEM/NTA/WB | Diagnosis Prognosis: stage, distant metastases, OS and PFS | eIF4E data extracted from TCGA database | Dong et al. [137] |
| PD-L1 (1) and IFN-γ (1,2) mRNA | EDTA Plasma | 38 NSCLC | Membrane affinity spin columns | - | (1) Response to treatment (2) PFS | Quantification by ddPCR with ACTB as internal control | Del Re et al. [138] |
| MALAT-1 | Serum | 77 NSCLC + 30 C | Precipitation | TEM/NTA/WB | Diagnosis Prognosis (Lymph node metastasis, TNM stage) | Quantification by RT-PCR. GAPDH was used for normalization | Zhang et al. [90] |
| linc01125 | Serum | 277 NSCLC + 187 C + 5 P + 59 T + 58 COPD | Precipitation | - | Diagnosis Prognosis (stage, OS) | RNA-Seq for lncRNA profile and subsequent quantification of linc01125 by RT-PCR with spiked in controls | Xian et al. [139] |
| FECR | Serum | 35 with limited SCLC and 26 with extensive SCLC +55 C | Affinity Chromatography | TEM/WB | Diagnosis Prognosis (survival) Response to chemotherapy | RT-PCR with β-actin as control | Li et al. [140] |
| circ_0014235 and circ_0025580 | Plasma | 30 SCC + 30 C | Precipitation | - | Diagnosis Prognosis (TNM stage and tumor size) | circRNA sequencing and confirmation with RT-PCR with GAPDH as internal control | Wang et al. [141] |
| circRNA_0056616 | EDTA plasma | 90 AC (42 with lymph node metastasis and 48 without) | Precipitation | TEM/WB | Lymph node metastasis predictor | RT-PCR. Normalization as Wang’s methods | He et al. [142] |
| circSATB2 | Serum | 83 NSCLC + 95 C | Ultracentrifugation | TEM/NTA/WB | Diagnosis Prognosis (metastasis detection) | RT-PCR. GAPDH and U6 were used as internal references and cel-miR-39 as an external reference | Zhang et al. [143] |
| circ_0047921, and circ_0007761 (1) circ_0056285 (1,2) | Serum | Screening set: 30 NSCLC + 45 C Training set: 120 NSCLC + 165 C Validation set 1: 62 NSCLC + 95 C Validation set 2: 63 NSCL + 58 COPD + 46 T | Precipitation | TEM/NTA/WB/FC | (1) Diagnosis (including early stage) (2) Prognosis: state of progression and lymph-node metastases | 1701 circRNAs initially identified by RNA-seq, 17 of them were differentially expressed and 8 of them were validated by RT-PCR with GAPDH and ACTB as spiked-in controls | Xian et al. [144] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sandúa, A.; Alegre, E.; González, Á. Exosomes in Lung Cancer: Actors and Heralds of Tumor Development. Cancers 2021, 13, 4330. https://doi.org/10.3390/cancers13174330
Sandúa A, Alegre E, González Á. Exosomes in Lung Cancer: Actors and Heralds of Tumor Development. Cancers. 2021; 13(17):4330. https://doi.org/10.3390/cancers13174330
Chicago/Turabian StyleSandúa, Amaia, Estibaliz Alegre, and Álvaro González. 2021. "Exosomes in Lung Cancer: Actors and Heralds of Tumor Development" Cancers 13, no. 17: 4330. https://doi.org/10.3390/cancers13174330
APA StyleSandúa, A., Alegre, E., & González, Á. (2021). Exosomes in Lung Cancer: Actors and Heralds of Tumor Development. Cancers, 13(17), 4330. https://doi.org/10.3390/cancers13174330







