Androgen Receptor-Mediated Nuclear Transport of NRDP1 in Prostate Cancer Cells Is Associated with Worse Patient Outcomes
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Characteristics
2.2. Reagents
2.3. Cell Lines and Culture
2.4. Immunofluorescence and Confocal Microscopy
2.5. Subcellular Fractionation
2.6. Transfections and Plasmids
2.7. Protein Extraction and Western Blot
2.8. Immunoprecipitation
2.9. Statistical Analyses
3. Results
3.1. NRDP1 Protein Is Expressed in the Nucleus as Well as the Cytosol in Prostate Cancer Cell Lines
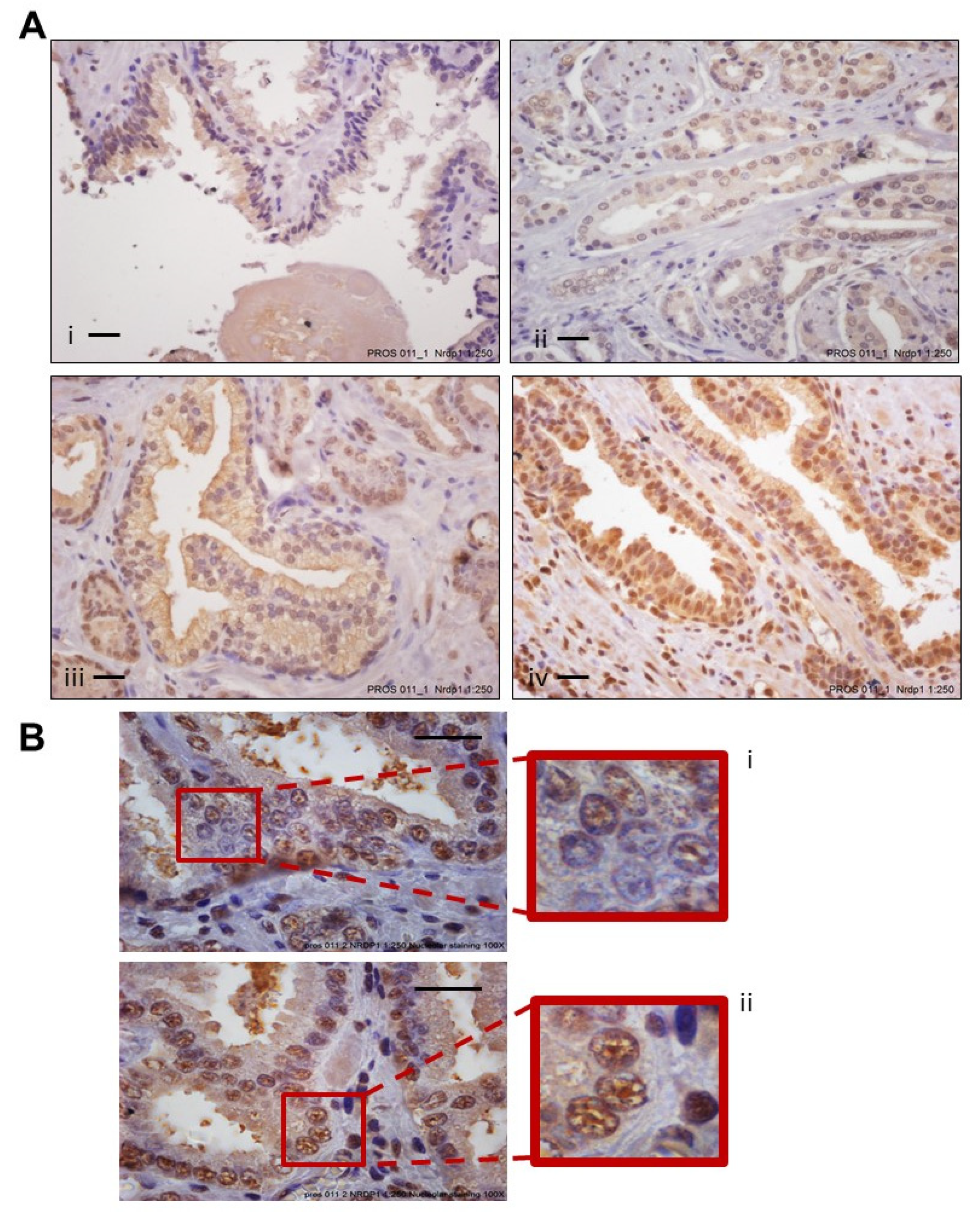
3.2. Increased Levels of Nuclear NRDP1 in Patient Tumors from Prostatectomy Samples Are Associated with Worse Patient Outcome
3.3. Strong Association between Levels of Androgen Receptor and NRDP1 in Prostate Cancer Patient Tumor Samples
3.4. Transport of NRDP1 into the Nucleus Is Associated with Nuclear Transport of the Androgen Receptor
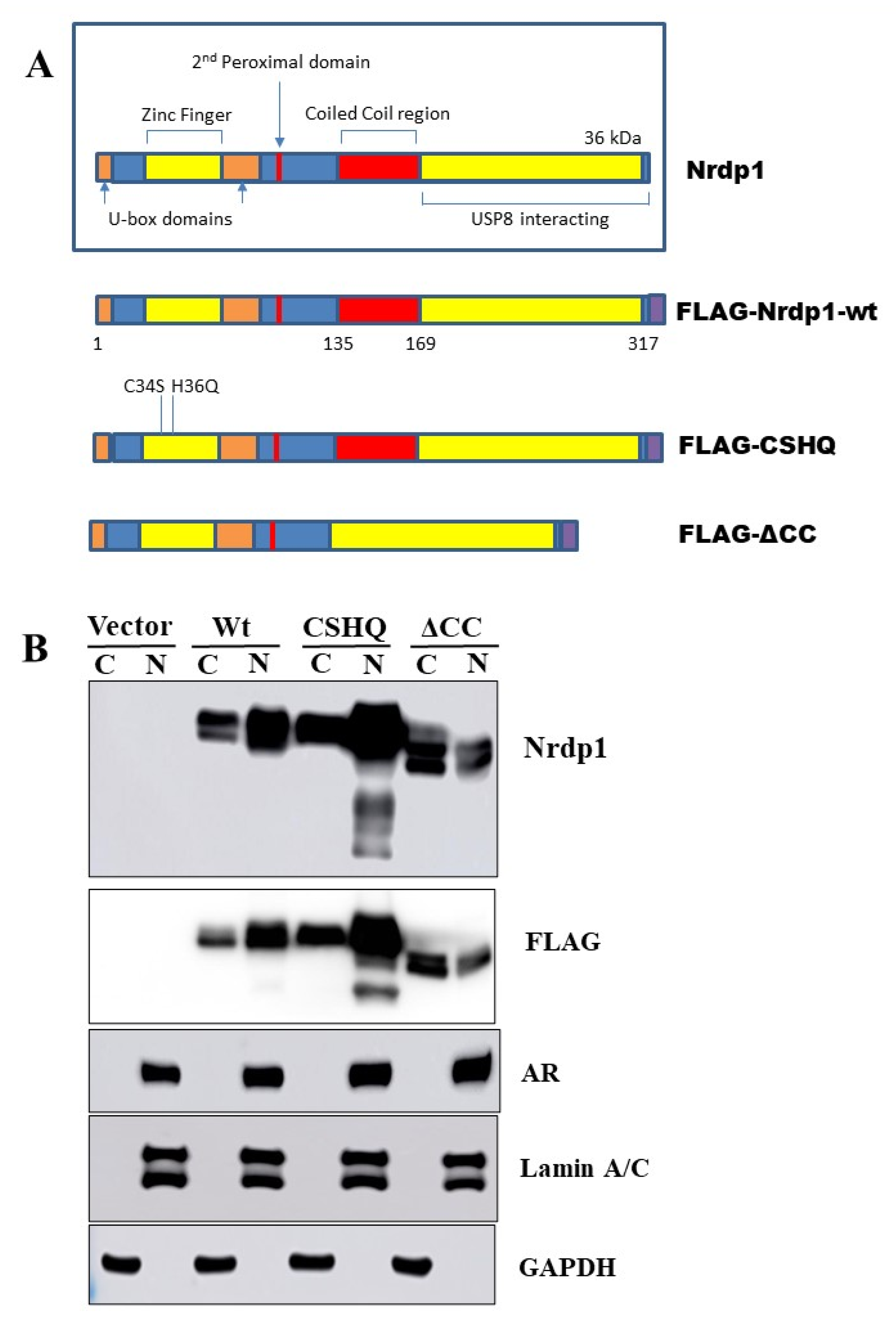
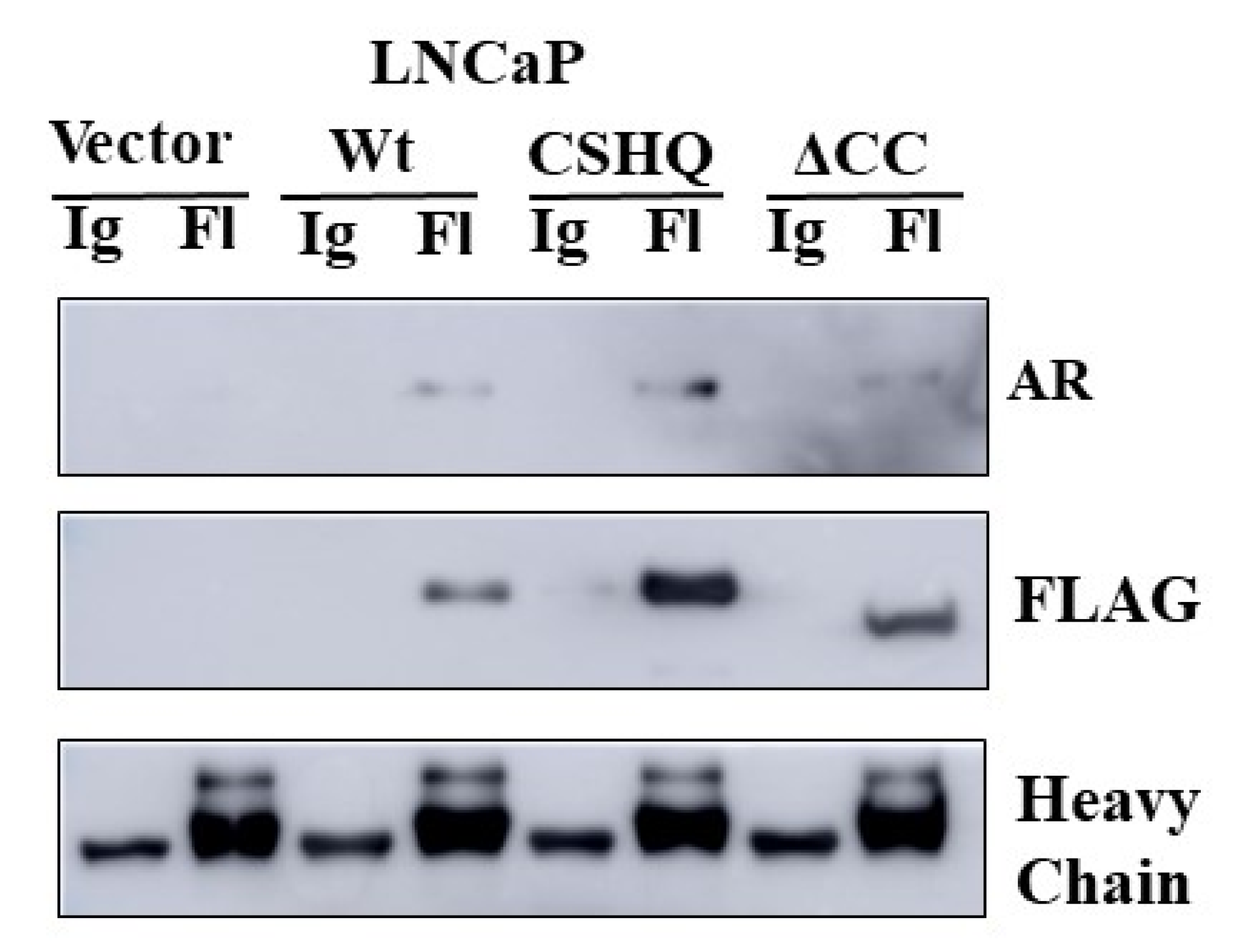
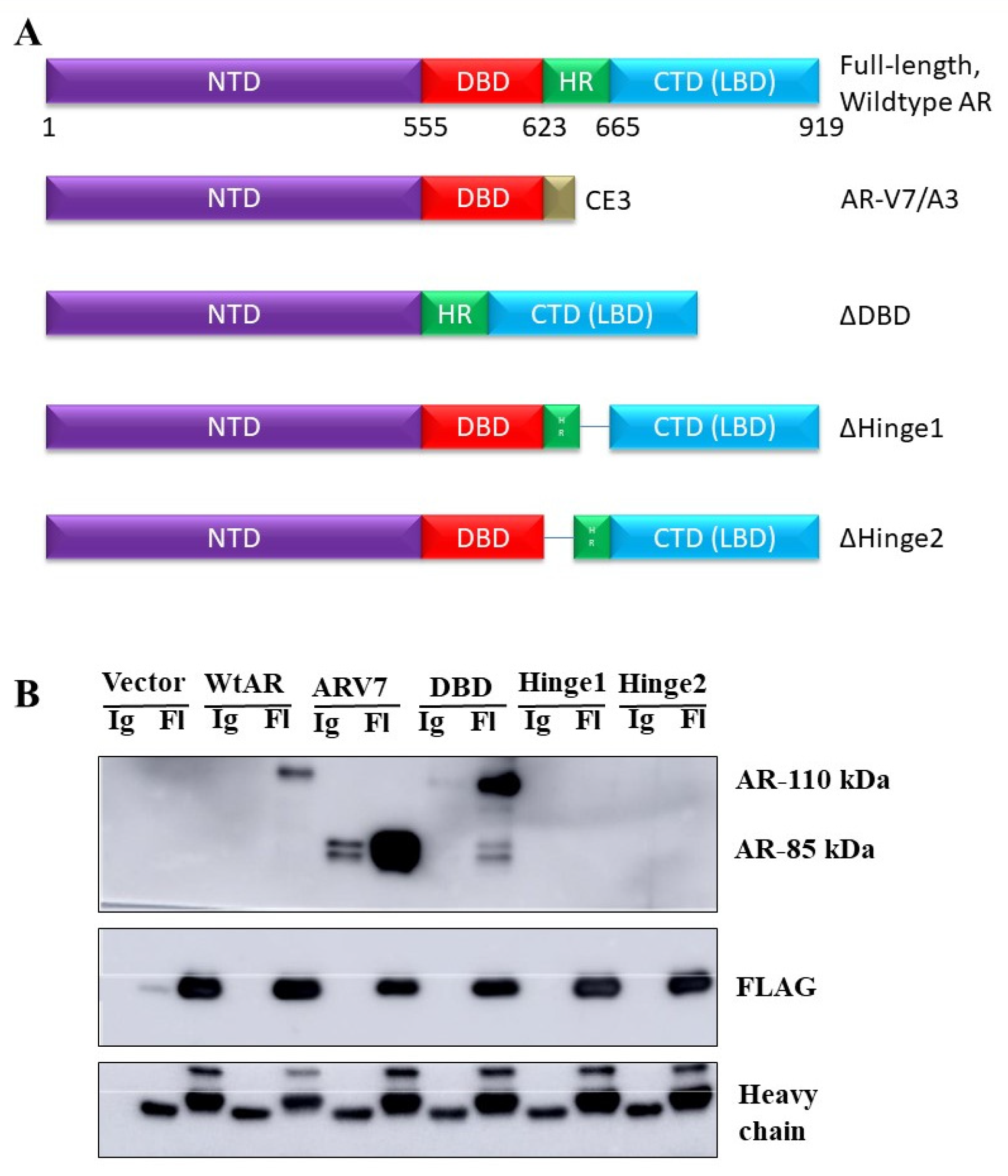
3.5. The Interaction between NRDP1 and AR Is Dependent on Presence of the AR Hinge Region but Does Not Require the NRDP1 Coiled Coil Domain
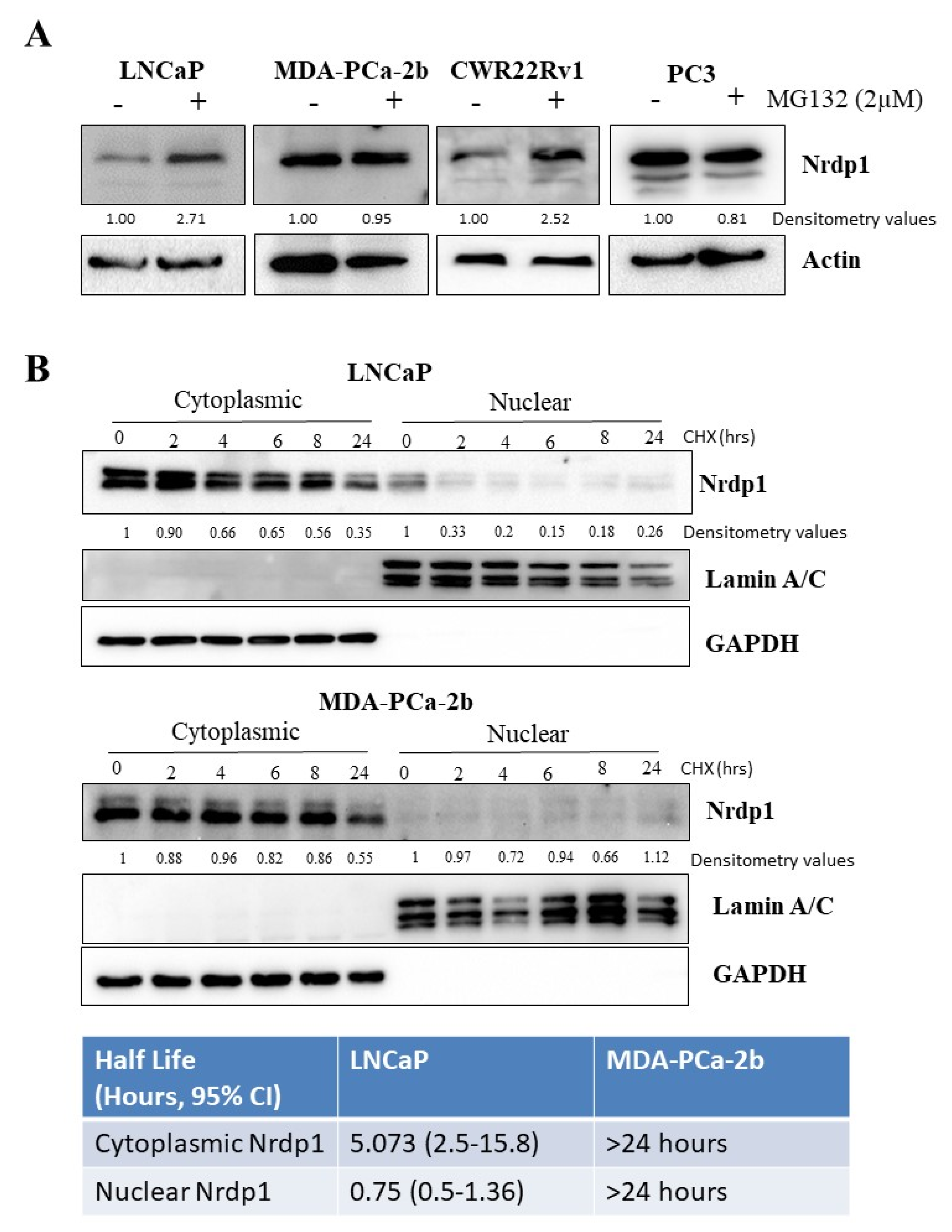
3.6. NRDP1 Can Act as a Ubiquitin Ligase in Both the Cytosol and the Nucleus of CaP Cells
3.7. Nuclear NRDP1 Levels Are Regulated by Proteasomal Degradation in Some but Not All Prostate Cancer Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, Y. E3 ubiquitin ligases as cancer targets and biomarkers. Neoplasia 2006, 8, 645–654. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Chen, J.; Huang, X.; Zhang, E.; He, J.; Cai, Z. Novel Insights into E3 ubiquitin ligase in cancer chemoresistance. Am. J. Med. Sci. 2018, 355, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Meng, T.; Chen, L.; Wei, W.; Wang, P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduct. Target. Ther. 2020, 5, 1–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Liu, S.; Tao, Y. Regulating tumor suppressor genes: Post-translational modifications. Signal Transduct. Target. Ther. 2020, 5, 1–25. [Google Scholar] [CrossRef]
- Shao, X.; Lu, Q.; Wang, G.; Huang, W.; Yang, L.; Chen, Z. Reduced expression of Nrdp1 predicts a poor prognosis in human hepatocellular carcinoma. OncoTargets Ther. 2018, 11, 4955–4963. [Google Scholar] [CrossRef] [Green Version]
- Luhtala, S.; Staff, S.; Kallioniemi, A.; Tanner, M.; Isola, J. Clinicopathological and prognostic correlations of HER3 expression and its degradation regulators, NEDD4-1 and NRDP1, in primary breast cancer. BMC Cancer 2018, 18, 1045. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Siddiqui, S.; Bose, S.; Mooso, B.; Asuncion, A.; Bedolla, R.G.; Vinall, R.; Tepper, C.G.; Gandour-Edwards, R.; Shi, X.; et al. Nrdp1-mediated regulation of ErbB3 expression by the androgen receptor in androgen-dependent but not castrate-resistant prostate cancer cells. Cancer Res. 2010, 70, 5994–6003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Wang, L.; Bao, H.; Zou, S.; Fu, C.; Gong, H.; Gao, Y.; Tang, Y.; Yu, R.; Shi, H. Nrdp1S, short variant of Nrdp1, inhibits human glioma progression by increasing Nrdp1-mediated ErbB3 ubiquitination and degradation. J. Cell Mol. Med. 2016, 20, 422–429. [Google Scholar] [CrossRef]
- Mujoo, K.; Choi, B.K.; Huang, Z.; Zhang, N.; An, Z. Regulation of ERBB3/HER3 signaling in cancer. Oncotarget 2014, 5, 10222–10236. [Google Scholar] [CrossRef] [Green Version]
- Ingalla, E.Q.; Miller, J.K.; Wald, J.H.; Workman, H.C.; Kaur, R.P.; Yen, L.; Fry, W.H.; Borowsky, A.D.; Young, L.J.; Sweeney, C.; et al. Post-transcriptional mechanisms contribute to the suppression of the ErbB3 negative regulator protein Nrdp1 in mammary tumors. J. Biol. Chem. 2010, 285, 28691–28697. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.; Min, S.; Hu, B.; Liu, Q.; Wan, Y. Nrdp1 is involved in hippocampus apoptosis in cardiopulmonary bypass-induced cognitive dysfunction via the regulation of ErbB3 protein levels. Int. J. Mol. Med. 2019, 43, 1747–1757. [Google Scholar] [CrossRef] [Green Version]
- Mo, X.; Liu, D.; Li, W.; Hu, Z.; Hu, Y.; Li, J.; Guo, J.; Tang, B.; Zhang, Z.; Bai, Y.; et al. Genetic screening for mutations in the Nrdp1 gene in Parkinson disease patients in a Chinese population. Park. Relat. Disord. 2010, 16, 222–224. [Google Scholar] [CrossRef]
- Shen, J.; Song, Y.; Shen, J.; Lin, Y.; Wu, X.; Yan, Y.; Niu, M.; Zhou, L.; Huang, Y.; Gao, Y.; et al. Nrdp1 is Associated with Neuronal Apoptosis in Lipopolysaccharide-Induced Neuroinflammation. Neurochem. Res. 2015, 40, 971–979. [Google Scholar] [CrossRef]
- Zhou, C.; Liu, Q.; Zhao, W.; Yang, L.; Huang, Z.; Yang, Z. Nrdp1 increases neuron apoptosis via downregulation of Bruce following intracerebral haemorrhage. J. Inflamm. 2019, 16, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Savoy, R.M.; Chen, L.; Siddiqui, S.; Melgoza, F.U.; Durbin-Johnson, B.; Drake, C.; Jathal, M.K.; Bose, S.; Steele, T.M.; Mooso, B.A.; et al. Transcription of Nrdp1 by the androgen receptor is regulated by nuclear filamin A in prostate cancer. Endocr. Relat. Cancer 2015, 22, 369–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bedolla, R.G.; Wang, Y.; Asuncion, A.; Chamie, K.; Siddiqui, S.; Mudryj, M.M.; Prihoda, T.J.; Siddiqui, J.; Chinnaiyan, A.M.; Mehra, R.; et al. Nuclear versus cytoplasmic localization of filamin A in prostate cancer: Immunohistochemical correlation with metastases. Clin. Cancer Res. 2009, 15, 788–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mooso, B.A.; Vinall, R.L.; Tepper, C.G.; Savoy, R.M.; Cheung, J.P.; Singh, S.; Siddiqui, S.; Wang, Y.; Bedolla, R.G.; Martinez, A.; et al. Enhancing the effectiveness of androgen deprivation in prostate cancer by inducing Filamin A nuclear localization. Endocr. Relat. Cancer 2012, 19, 759–777. [Google Scholar] [CrossRef] [Green Version]
- Vinall, R.L.; Tepper, C.G.; Shi, X.-B.; Xue, L.A.; Gandour-Edwards, R.; White, R.W.D.V. The R273H p53 mutation can facilitate the androgen-independent growth of LNCaP by a mechanism that involves H2 relaxin and its cognate receptor LGR7. Oncogene 2006, 25, 2082–2093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navone, N.M.; Olive, M.; Ozen, M.; Davis, R.; Troncoso, P.; Tu, S.M.; Johnston, D.; Pollack, A.; Pathak, S.; von Eschenbach, A.C.; et al. Establishment of two human prostate cancer cell lines derived from a single bone metastasis. Clin. Cancer Res. 1997, 3, 2493–2500. [Google Scholar]
- Chan, S.C.; Dehm, S.M. Constitutive activity of the androgen receptor. Adv. Pharmacol. 2014, 70, 327–366. [Google Scholar] [CrossRef] [Green Version]
- Mulholland, D.J.; Cheng, H.; Reid, K.; Rennie, P.S.; Nelson, C.C. The androgen receptor can promote beta-catenin nuclear translocation independently of adenomatous polyposis coli. J. Biol. Chem. 2002, 277, 17933–17943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Josefsson, A.; Wikstrom, P.; Granfors, T.; Egevad, L.; Karlberg, L.; Stattin, P.; Bergh, A. Tumor size, vascular density and proliferation as prognostic markers in GS 6 and GS 7 prostate tumors in patients with long follow-up and non-curative treatment. Eur. Urol. 2005, 48, 577–583. [Google Scholar] [CrossRef]
- Josefsson, A.; Wikstrom, P.; Egevad, L.; Granfors, T.; Karlberg, L.; Stattin, P.; Bergh, A. Low endoglin vascular density and Ki67 index in Gleason score 6 tumours may identify prostate cancer patients suitable for surveillance. Scand. J. Urol. Nephrol. 2012, 46, 247–257. [Google Scholar] [CrossRef]
- Bubendorf, L.; Tapia, C.; Gasser, T.C.; Casella, R.; Grunder, B.; Moch, H.; Mihatsch, M.J.; Sauter, G. Ki67 labeling index in core needle biopsies independently predicts tumor-specific survival in prostate cancer. Hum. Pathol. 1998, 29, 949–954. [Google Scholar] [CrossRef]
- Berney, D.M.; Gopalan, A.; Kudahetti, S.; Fisher, G.; Ambroisine, L.; Foster, C.S.; Reuter, V.; Eastham, J.; Moller, H.; Kattan, M.W.; et al. Ki-67 and outcome in clinically localised prostate cancer: Analysis of conservatively treated prostate cancer patients from the Trans-Atlantic Prostate Group study. Br. J. Cancer 2009, 100, 888–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisher, G.; Yang, Z.H.; Kudahetti, S.; Moller, H.; Scardino, P.; Cuzick, J.; Berney, D.M.; Transatlantic Prostate, G. Prognostic value of Ki-67 for prostate cancer death in a conservatively managed cohort. Br. J. Cancer 2013, 108, 271–277. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Lu, W.; Chen, Z. Regulation of Androgen Receptor by E3 Ubiquitin Ligases: For More or Less. Recept. Clin. Investig. 2014, 1. [Google Scholar] [CrossRef]
- Wen, S.; Niu, Y.; Huang, H. Posttranslational regulation of androgen dependent and independent androgen receptor activities in prostate cancer. Asian J. Urol. 2020, 7, 203–218. [Google Scholar] [CrossRef]
- Messner, E.A.; Steele, T.M.; Tsamouri, M.M.; Hejazi, N.; Gao, A.C.; Mudryj, M.; Ghosh, P.M. The androgen receptor in prostate cancer: Effect of structure, ligands and spliced variants on therapy. Biomedicines 2020, 8, 422. [Google Scholar] [CrossRef]
- Azad, A.A.; Zoubeidi, A.; Gleave, M.E.; Chi, K.N. Targeting heat shock proteins in metastatic castration-resistant prostate cancer. Nat. Rev. Urol. 2015, 12, 26–36. [Google Scholar] [CrossRef]
- Lv, S.; Song, Q.; Chen, G.; Cheng, E.; Chen, W.; Cole, R.; Wu, Z.; Pascal, L.E.; Wang, K.; Wipf, P.; et al. Regulation and targeting of androgen receptor nuclear localization in castration-resistant prostate cancer. J. Clin. Investig. 2021, 131. [Google Scholar] [CrossRef]
- Wu, X.; Yen, L.; Irwin, L.; Sweeney, C.; Carraway, K.L., III. Stabilization of the E3 ubiquitin ligase Nrdp1 by the deubiquitinating enzyme USP8. Mol. Cell. Biol. 2004, 24, 7748–7757. [Google Scholar] [CrossRef] [Green Version]
- Qiu, X.B.; Goldberg, A.L. Nrdp1/FLRF is a ubiquitin ligase promoting ubiquitination and degradation of the epidermal growth factor receptor family member, ErbB3. Proc. Natl. Acad. Sci. USA 2002, 99, 14843–14848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Printsev, I.; Yen, L.; Sweeney, C.; Carraway, K.L., III. Oligomerization of the Nrdp1 E3 ubiquitin ligase is necessary for efficient autoubiquitination but not ErbB3 ubiquitination. J. Biol. Chem. 2014, 289, 8570–8578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef] [Green Version]
- Schneider, J.A.; Logan, S.K. Revisiting the role of Wnt/beta-catenin signaling in prostate cancer. Mol. Cell. Endocrinol. 2018, 462, 3–8. [Google Scholar] [CrossRef]
- Song, L.N.; Herrell, R.; Byers, S.; Shah, S.; Wilson, E.M.; Gelmann, E.P. Beta-catenin binds to the activation function 2 region of the androgen receptor and modulates the effects of the N-terminal domain and TIF2 on ligand-dependent transcription. Mol. Cell. Biol. 2003, 23, 1674–1687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Georget, V.; Terouanne, B.; Nicolas, J.C.; Sultan, C. Mechanism of antiandrogen action: Key role of hsp90 in conformational change and transcriptional activity of the androgen receptor. Biochemistry 2002, 41, 11824–11831. [Google Scholar] [CrossRef]
- Haelens, A.; Tanner, T.; Denayer, S.; Callewaert, L.; Claessens, F. The hinge region regulates DNA binding, nuclear translocation, and transactivation of the androgen receptor. Cancer Res. 2007, 67, 4514–4523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, M.H.; Li, J.; Xu, H.E.; Melcher, K.; Yong, E.L. Androgen receptor: Structure, role in prostate cancer and drug discovery. Acta Pharmacol. Sin. 2015, 36, 3–23. [Google Scholar] [CrossRef] [Green Version]
- Senapati, D.; Kumari, S.; Heemers, H.V. Androgen receptor co-regulation in prostate cancer. Asian J. Urol. 2020, 7, 219–232. [Google Scholar] [CrossRef]
- Feng, Q.; He, B. Androgen Receptor Signaling in the Development of Castration-Resistant Prostate Cancer. Front. Oncol. 2019, 9, 858. [Google Scholar] [CrossRef] [Green Version]
- Rice, M.A.; Malhotra, S.V.; Stoyanova, T. Second-generation antiandrogens: From discovery to standard of care in castration resistant prostate cancer. Front. Oncol. 2019, 9, 801. [Google Scholar] [CrossRef] [PubMed]
- von Mikecz, A. The nuclear ubiquitin-proteasome system. J. Cell Sci. 2006, 119, 1977–1984. [Google Scholar] [CrossRef] [Green Version]
- Yu, Z.K.; Geyer, R.K.; Maki, C.G. MDM2-dependent ubiquitination of nuclear and cytoplasmic P53. Oncogene 2000, 19, 5892–5897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trotman, L.C.; Wang, X.; Alimonti, A.; Chen, Z.; Teruya-Feldstein, J.; Yang, H.; Pavletich, N.P.; Carver, B.S.; Cordon-Cardo, C.; Erdjument-Bromage, H.; et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell 2007, 128, 141–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Islam, M.T.; Chen, F.; Chen, H. The oncogenic role of ubiquitin specific peptidase (USP8) and its signaling pathways targeting for cancer therapeutics. Arch. Biochem. Biophys. 2021, 701, 108811. [Google Scholar] [CrossRef] [PubMed]
- Rong, Z.; Zhu, Z.; Cai, S.; Zhang, B. Knockdown of USP8 inhibits the growth of lung cancer cells. Cancer Manag. Res. 2020, 12, 12415–12422. [Google Scholar] [CrossRef]
- Luk, S.U.; Xue, H.; Cheng, H.; Lin, D.; Gout, P.W.; Fazli, L.; Collins, C.C.; Gleave, M.E.; Wang, Y. The BIRC6 gene as a novel target for therapy of prostate cancer: Dual targeting of inhibitors of apoptosis. Oncotarget 2014, 5, 6896–6908. [Google Scholar] [CrossRef] [Green Version]
- Ebner, P.; Poetsch, I.; Deszcz, L.; Hoffmann, T.; Zuber, J.; Ikeda, F. The IAP family member BRUCE regulates autophagosome-lysosome fusion. Nat. Commun. 2018, 9, 599. [Google Scholar] [CrossRef] [PubMed]
- Che, L.; Yang, X.; Ge, C.; El-Amouri, S.S.; Wang, Q.E.; Pan, D.; Herzog, T.J.; Du, C. Loss of BRUCE reduces cellular energy level and induces autophagy by driving activation of the AMPK-ULK1 autophagic initiating axis. PLoS ONE 2019, 14, e0216553. [Google Scholar] [CrossRef] [PubMed]



| Characteristics | Number |
|---|---|
| Age | |
| N | 162 |
| Mean (SD) | 69.7 (8.2) |
| Median (Range) | 69 (49–91) |
| Weight (lbs) | |
| N | 160 |
| Mean (SD) | 181.6 (47.5) |
| Median (Range) | 185.9 (64.9–284) |
| Race | |
| White | 102 (63%) |
| Black/African-American | 31 (19.1%) |
| American Indian/AK Native | 3 (1.9%) |
| Asian | 1 (0.6%) |
| Unknown | 25 (15.4%) |
| Ethnicity | |
| Not Hispanic or Latino | 138 (85.2%) |
| Hispanic or Latino | 4 (2.5%) |
| Unknown | 20 (12.3%) |
| Smoking | |
| Current/Former Smoker | 95 (58.6%) |
| Never Smoker | 67 (41.4%) |
| Marker | Odds Ratio (95% CI) | p-Value |
|---|---|---|
| AR.Cancer.N | 0.935 (0.845, 1.034) | 0.193 |
| AR.Cancer.C | 1.153 (0.708, 1.876) | 0.568 |
| Ki67.Cancer | 0.998 (0.997, 1) | 0.0477 |
| NRDP1.Cancer.N | 1.238 (1.063, 1.443) | 0.00684 |
| NRDP1.Cancer.C | 1 (0.81, 1.235) | 1 |
| Marker | 5-Year Recurrence-Free Percentage, High Expression | 5-Year Recurrence-Free Percentage, Low Expression | Difference (95% CI) | p-Value |
|---|---|---|---|---|
| AR N | 74.50% | 87.10% | −12.7% (−31.8%, 6.5%) | 0.196 |
| AR C | 95.50% | 79.10% | 16.4% (7.5%, 25.2%) | <0.001 |
| Ki67 | 77.40% | 94.40% | −17% (−27.1%, −6.9%) | 0.001 |
| NRDP1 N | 73.50% | 92% | −18.5% (−30.9%, −6.1%) | 0.003 |
| NRDP1 C | 85.10% | 87.40% | −2.3% (−12.3%, 7.7%) | 0.650 |
| Marker 1 | Marker 2 | Spearman Correlation | p-Value |
|---|---|---|---|
| AR N | AR C | 0.32 | <0.001 |
| AR N | Ki67 | 0.22 | 0.012 |
| AR N | NRDP1 N | −0.11 | 0.216 |
| AR C | Ki67 | 0.06 | 0.525 |
| AR C | NRDP1 N | −0.46 | <0.001 |
| Ki67 | NRDP1 N | −0.06 | 0.476 |
| Marker 1 | Marker 2 | Spearman Correlation | p-Value |
|---|---|---|---|
| AR N | AR C | 0.03 | 0.790 |
| AR N | Ki67 | 0.51 | <0.001 |
| AR N | NRDP1 N | 0.07 | 0.564 |
| AR C | Ki67 | 0 | 0.974 |
| AR C | NRDP1 N | −0.35 | 0.003 |
| Ki67 | NRDP1 N | 0.18 | 0.128 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Steele, T.; Sam, A.; Evans, S.; Browning, E.; Krig, S.; Macias, K.; Konda, A.; Siddiqui, S.; Durbin-Johnson, B.; Ghosh, P.; et al. Androgen Receptor-Mediated Nuclear Transport of NRDP1 in Prostate Cancer Cells Is Associated with Worse Patient Outcomes. Cancers 2021, 13, 4425. https://doi.org/10.3390/cancers13174425
Steele T, Sam A, Evans S, Browning E, Krig S, Macias K, Konda A, Siddiqui S, Durbin-Johnson B, Ghosh P, et al. Androgen Receptor-Mediated Nuclear Transport of NRDP1 in Prostate Cancer Cells Is Associated with Worse Patient Outcomes. Cancers. 2021; 13(17):4425. https://doi.org/10.3390/cancers13174425
Chicago/Turabian StyleSteele, Thomas, Anhao Sam, Shawna Evans, Elizabeth Browning, Sheryl Krig, Katelyn Macias, Adarsh Konda, Salma Siddiqui, Blythe Durbin-Johnson, Paramita Ghosh, and et al. 2021. "Androgen Receptor-Mediated Nuclear Transport of NRDP1 in Prostate Cancer Cells Is Associated with Worse Patient Outcomes" Cancers 13, no. 17: 4425. https://doi.org/10.3390/cancers13174425
APA StyleSteele, T., Sam, A., Evans, S., Browning, E., Krig, S., Macias, K., Konda, A., Siddiqui, S., Durbin-Johnson, B., Ghosh, P., & Vinall, R. (2021). Androgen Receptor-Mediated Nuclear Transport of NRDP1 in Prostate Cancer Cells Is Associated with Worse Patient Outcomes. Cancers, 13(17), 4425. https://doi.org/10.3390/cancers13174425







