Tumor-Derived Exosomes: Hidden Players in PD-1/PD-L1 Resistance
Abstract
:Simple Summary
Abstract
1. Introduction
2. Exosomes
2.1. Overview
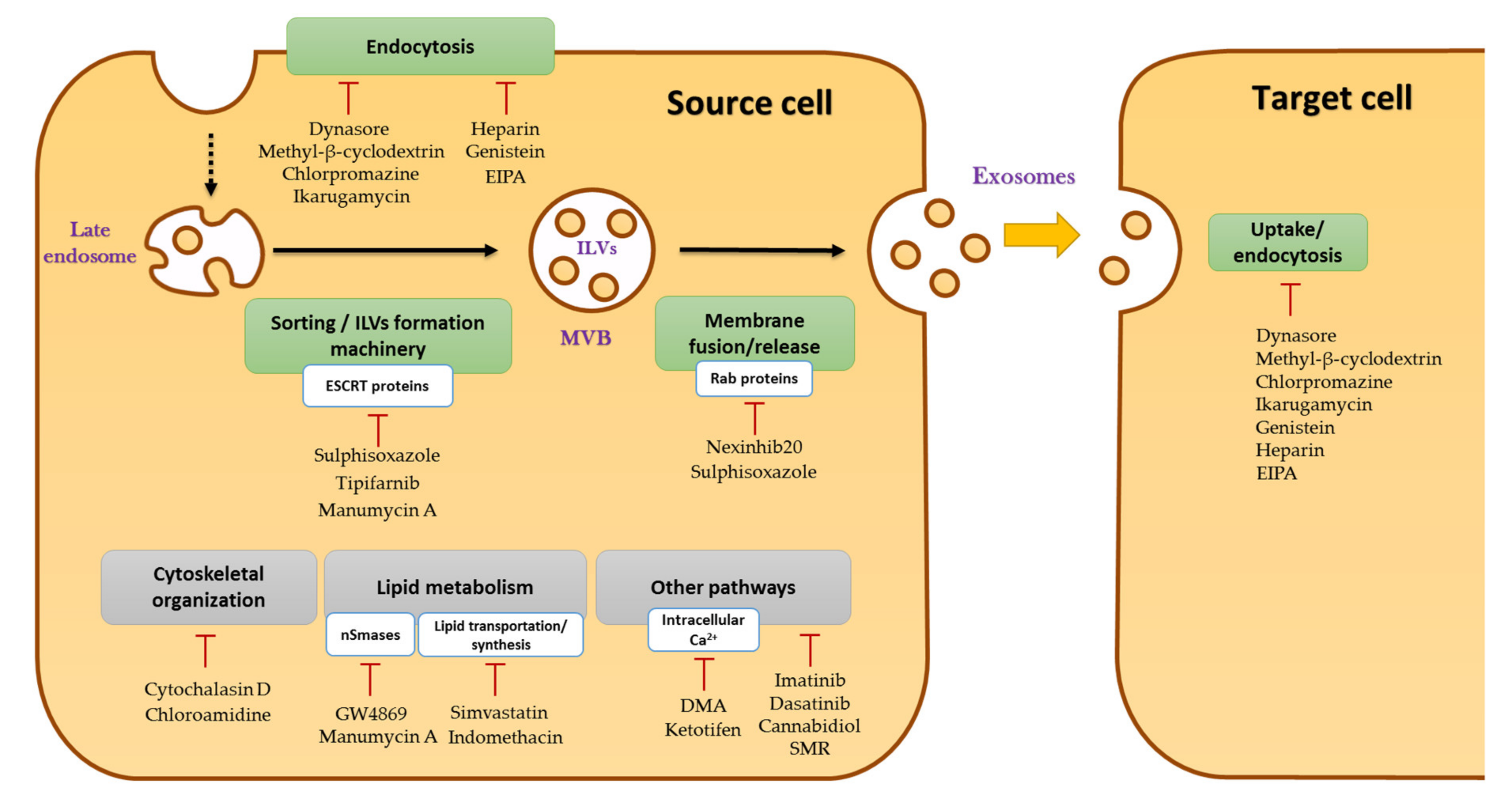
2.2. Biogenesis of Exosomes
2.3. Functions of Exosomes in Cancer Development and Immunity
2.3.1. Exosomes in Cancer Development
2.3.2. TEXs in Immunity
3. Exosomes and Immune Checkpoints
3.1. Exosomes and PD-L1
3.2. Exosomes and TIM-3
3.3. Exosomes and CD73/CD39
3.4. What about CTLA-4, LAG-3, and TIGIT in TEX?
4. Challenges and Future Directions
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hodi, F.S.; O’Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C.; et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. N. Engl. J. Med. 2010, 363, 711–723. [Google Scholar] [CrossRef]
- Iwai, Y.; Ishida, M.; Tanaka, Y.; Okazaki, T.; Honjo, T.; Minato, N. Involvement of PD-L1 on Tumor Cells in the Escape from Host Immune System and Tumor Immunotherapy by PD-L1 Blockade. Proc. Natl. Acad. Sci. USA 2002, 99, 12293–12297. [Google Scholar] [CrossRef] [Green Version]
- Balar, A.V.; Weber, J.S. PD-1 and PD-L1 Antibodies in Cancer: Current Status and Future Directions. Cancer Immunol. Immunother. 2017, 66, 551–564. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, J.S.; Teng, M.W.L.; Smyth, M.J. Cancer Immunoediting and Resistance to T Cell-Based Immunotherapy. Nat. Rev. Clin. Oncol. 2019, 16, 151–167. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.D.; Warren, R.L.; Gibb, E.A.; Martin, S.D.; Spinelli, J.J.; Nelson, B.H.; Holt, R.A. Neo-Antigens Predicted by Tumor Genome Meta-Analysis Correlate with Increased Patient Survival. Genome Res. 2014, 24, 743–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, Q.; Wang, D.; Sun, K.; Wang, L.; Zhang, Y. Resistance Mechanisms of Anti-PD1/PDL1 Therapy in Solid Tumors. Front. Cell Dev. Biol. 2020, 8, 672. [Google Scholar] [CrossRef] [PubMed]
- Limagne, E.; Richard, C.; Thibaudin, M.; Fumet, J.-D.; Truntzer, C.; Lagrange, A.; Favier, L.; Coudert, B.; Ghiringhelli, F. Tim-3/Galectin-9 Pathway and MMDSC Control Primary and Secondary Resistances to PD-1 Blockade in Lung Cancer Patients. Oncoimmunology 2019, 8, e1564505. [Google Scholar] [CrossRef]
- McLane, L.M.; Abdel-Hakeem, M.S.; Wherry, E.J. CD8 T Cell Exhaustion During Chronic Viral Infection and Cancer. Annu. Rev. Immunol. 2019, 37, 457–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Wang, S.; Zhou, Q. The Resistance Mechanisms of Lung Cancer Immunotherapy. Front. Oncol. 2020, 10, 568059. [Google Scholar] [CrossRef]
- Whiteside, T.L. Exosomes and Tumor-Mediated Immune Suppression. J. Clin. Invest. 2021, 126, 1216–1223. [Google Scholar] [CrossRef] [Green Version]
- Trams, E.G.; Lauter, C.J.; Salem, N., Jr.; Heine, U. Exfoliation of Membrane Ecto-Enzymes in the Form of Micro-Vesicles. Biochim. Biophys. Acta BBA-Biomembr. 1981, 645, 63–70. [Google Scholar] [CrossRef]
- Kharaziha, P.; Ceder, S.; Li, Q.; Panaretakis, T. Tumor Cell-Derived Exosomes: A Message in a Bottle. Biochim. Biophys. Acta 2012, 1826, 103–111. [Google Scholar] [CrossRef]
- Wubbolts, R.; Leckie, R.S.; Veenhuizen, P.T.M.; Schwarzmann, G.; Möbius, W.; Hoernschemeyer, J.; Slot, J.-W.; Geuze, H.J.; Stoorvogel, W. Proteomic and Biochemical Analyses of Human B Cell-Derived Exosomes. Potential Implications for Their Function and Multivesicular Body Formation. J. Biol. Chem. 2003, 278, 10963–10972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- Andre, F.; Schartz, N.E.; Movassagh, M.; Flament, C.; Pautier, P.; Morice, P.; Pomel, C.; Lhomme, C.; Escudier, B.; Le Chevalier, T.; et al. Malignant Effusions and Immunogenic Tumour-Derived Exosomes. Lancet 2002, 360, 295–305. [Google Scholar] [CrossRef]
- Boukouris, S.; Mathivanan, S. Exosomes in Bodily Fluids Are a Highly Stable Resource of Disease Biomarkers. Proteomics Clin. Appl. 2015, 9, 358–367. [Google Scholar] [CrossRef] [Green Version]
- Hessvik, N.P.; Llorente, A. Current Knowledge on Exosome Biogenesis and Release. Cell. Mol. Life Sci. CMLS 2018, 75, 193–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKelvey, K.J.; Powell, K.L.; Ashton, A.W.; Morris, J.M.; McCracken, S.A. Exosomes: Mechanisms of Uptake. J. Circ. Biomark. 2015, 4, 7. [Google Scholar] [CrossRef] [Green Version]
- Keller, S.; Sanderson, M.P.; Stoeck, A.; Altevogt, P. Exosomes: From Biogenesis and Secretion to Biological Function. Immunol. Lett. 2006, 107, 102–108. [Google Scholar] [CrossRef]
- Robbins, P.D.; Morelli, A.E. Regulation of Immune Responses by Extracellular Vesicles. Nat. Rev. Immunol. 2014, 14, 195–208. [Google Scholar] [CrossRef] [Green Version]
- Stuffers, S.; Sem Wegner, C.; Stenmark, H.; Brech, A. Multivesicular Endosome Biogenesis in the Absence of ESCRTs. Traffic Cph. Den. 2009, 10, 925–937. [Google Scholar] [CrossRef]
- Fader, C.M.; Sánchez, D.G.; Mestre, M.B.; Colombo, M.I. TI-VAMP/VAMP7 and VAMP3/Cellubrevin: Two v-SNARE Proteins Involved in Specific Steps of the Autophagy/Multivesicular Body Pathways. Biochim. Biophys. Acta BBA-Mol. Cell Res. 2009, 1793, 1901–1916. [Google Scholar] [CrossRef] [Green Version]
- Johnstone, R.M. Exosomes Biological Significance: A Concise Review. Blood Cells. Mol. Dis. 2006, 36, 315–321. [Google Scholar] [CrossRef]
- Borges, F.T.; Reis, L.A.; Schor, N. Extracellular Vesicles: Structure, Function, and Potential Clinical Uses in Renal Diseases. Braz. J. Med. Biol. Res. Rev. Bras. Pesqui. Medicas E Biol. 2013, 46, 824–830. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Yuan, X.; Shi, H.; Wu, L.; Qian, H.; Xu, W. Exosomes in Cancer: Small Particle, Big Player. J. Hematol. Oncol. 2015, 8, 83. [Google Scholar] [CrossRef] [Green Version]
- Bobrie, A.; Théry, C. Exosomes and Communication between Tumours and the Immune System: Are All Exosomes Equal? Biochem. Soc. Trans. 2013, 41, 263–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chowdhury, R.; Webber, J.P.; Gurney, M.; Mason, M.D.; Tabi, Z.; Clayton, A. Cancer Exosomes Trigger Mesenchymal Stem Cell Differentiation into Pro-Angiogenic and pro-Invasive Myofibroblasts. Oncotarget 2015, 6, 715–731. [Google Scholar] [CrossRef] [PubMed]
- Ringuette Goulet, C.; Bernard, G.; Tremblay, S.; Chabaud, S.; Bolduc, S.; Pouliot, F. Exosomes Induce Fibroblast Differentiation into Cancer-Associated Fibroblasts through TGFβ Signaling. Mol. Cancer Res. MCR 2018, 16, 1196–1204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meehan, K.; Vella, L.J. The Contribution of Tumour-Derived Exosomes to the Hallmarks of Cancer. Crit. Rev. Clin. Lab. Sci. 2016, 53, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Ekström, E.J.; Bergenfelz, C.; von Bülow, V.; Serifler, F.; Carlemalm, E.; Jönsson, G.; Andersson, T.; Leandersson, K. WNT5A Induces Release of Exosomes Containing Pro-Angiogenic and Immunosuppressive Factors from Malignant Melanoma Cells. Mol. Cancer 2014, 13, 88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ko, S.Y.; Lee, W.; Kenny, H.A.; Dang, L.H.; Ellis, L.M.; Jonasch, E.; Lengyel, E.; Naora, H. Cancer-Derived Small Extracellular Vesicles Promote Angiogenesis by Heparin-Bound, Bevacizumab-Insensitive VEGF, Independent of Vesicle Uptake. Commun. Biol. 2019, 2, 386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ludwig, N.; Yerneni, S.S.; Razzo, B.M.; Whiteside, T.L. Exosomes from HNSCC Promote Angiogenesis through Reprogramming of Endothelial Cells. Mol. Cancer Res. MCR 2018, 16, 1798–1808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, F.; Li, F.; Lin, X.; Xu, F.; Cui, R.-R.; Zhong, J.-Y.; Zhu, T.; Shan, S.-K.; Liao, X.-B.; Yuan, L.-Q.; et al. Exosomes Increased Angiogenesis in Papillary Thyroid Cancer Microenvironment. Endocr. Relat. Cancer 2019, 26, 525–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, M.; Qin, H.; Poon, T.C.W.; Sze, S.-C.; Ding, X.; Co, N.N.; Ngai, S.-M.; Chan, T.-F.; Wong, N. Hepatocellular Carcinoma-Derived Exosomes Promote Motility of Immortalized Hepatocyte through Transfer of Oncogenic Proteins and RNAs. Carcinogenesis 2015, 36, 1008–1018. [Google Scholar] [CrossRef] [Green Version]
- Hendrix, A.; Maynard, D.; Pauwels, P.; Braems, G.; Denys, H.; Van den Broecke, R.; Lambert, J.; Van Belle, S.; Cocquyt, V.; Gespach, C.; et al. Effect of the Secretory Small GTPase Rab27B on Breast Cancer Growth, Invasion, and Metastasis. J. Natl. Cancer Inst. 2010, 102, 866–880. [Google Scholar] [CrossRef]
- Paolillo, M.; Schinelli, S. Integrins and Exosomes, a Dangerous Liaison in Cancer Progression. Cancers 2017, 9, 95. [Google Scholar] [CrossRef] [Green Version]
- Jeppesen, D.K.; Nawrocki, A.; Jensen, S.G.; Thorsen, K.; Whitehead, B.; Howard, K.A.; Dyrskjøt, L.; Ørntoft, T.F.; Larsen, M.R.; Ostenfeld, M.S. Quantitative Proteomics of Fractionated Membrane and Lumen Exosome Proteins from Isogenic Metastatic and Nonmetastatic Bladder Cancer Cells Reveal Differential Expression of EMT Factors. Proteomics 2014, 14, 699–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kidd, S.; Spaeth, E.; Dembinski, J.L.; Dietrich, M.; Watson, K.; Klopp, A.; Battula, V.L.; Weil, M.; Andreeff, M.; Marini, F.C. Direct Evidence of Mesenchymal Stem Cell Tropism for Tumor and Wounding Microenvironments Using in Vivo Bioluminescent Imaging. Stem Cells Dayt. Ohio 2009, 27, 2614–2623. [Google Scholar] [CrossRef] [Green Version]
- Zitvogel, L.; Regnault, A.; Lozier, A.; Wolfers, J.; Flament, C.; Tenza, D.; Ricciardi-Castagnoli, P.; Raposo, G.; Amigorena, S. Eradication of Established Murine Tumors Using a Novel Cell-Free Vaccine: Dendritic Cell-Derived Exosomes. Nat. Med. 1998, 4, 594–600. [Google Scholar] [CrossRef]
- Raimondo, S.; Pucci, M.; Alessandro, R.; Fontana, S. Extracellular Vesicles and Tumor-Immune Escape: Biological Functions and Clinical Perspectives. Int. J. Mol. Sci. 2020, 21, 2286. [Google Scholar] [CrossRef] [Green Version]
- Andreola, G.; Rivoltini, L.; Castelli, C.; Huber, V.; Perego, P.; Deho, P.; Squarcina, P.; Accornero, P.; Lozupone, F.; Lugini, L.; et al. Induction of Lymphocyte Apoptosis by Tumor Cell Secretion of FasL-Bearing Microvesicles. J. Exp. Med. 2002, 195, 1303–1316. [Google Scholar] [CrossRef]
- Huber, V.; Fais, S.; Iero, M.; Lugini, L.; Canese, P.; Squarcina, P.; Zaccheddu, A.; Colone, M.; Arancia, G.; Gentile, M.; et al. Human Colorectal Cancer Cells Induce T-Cell Death through Release of Proapoptotic Microvesicles: Role in Immune Escape. Gastroenterology 2005, 128, 1796–1804. [Google Scholar] [CrossRef]
- Liu, C.; Yu, S.; Zinn, K.; Wang, J.; Zhang, L.; Jia, Y.; Kappes, J.C.; Barnes, S.; Kimberly, R.P.; Grizzle, W.E.; et al. Murine Mammary Carcinoma Exosomes Promote Tumor Growth by Suppression of NK Cell Function. J. Immunol. Baltim. Md 1950 2006, 176, 1375–1385. [Google Scholar] [CrossRef] [Green Version]
- Navarro-Tableros, V.; Gomez, Y.; Camussi, G.; Brizzi, M.F. Extracellular Vesicles: New Players in Lymphomas. Int. J. Mol. Sci. 2018, 20, 41. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Yang, L.; Cui, Y.; Zhou, Y.; Yin, X.; Guo, J.; Zhang, G.; Wang, T.; He, Q.-Y. Cytoskeleton-Centric Protein Transportation by Exosomes Transforms Tumor-Favorable Macrophages. Oncotarget 2016, 7, 67387–67402. [Google Scholar] [CrossRef] [Green Version]
- Gabrilovich, D.I.; Nagaraj, S. Myeloid-Derived Suppressor Cells as Regulators of the Immune System. Nat. Rev. Immunol. 2009, 9, 162–174. [Google Scholar] [CrossRef]
- Xiang, X.; Poliakov, A.; Liu, C.; Liu, Y.; Deng, Z.; Wang, J.; Cheng, Z.; Shah, S.V.; Wang, G.-J.; Zhang, L.; et al. Induction of Myeloid-Derived Suppressor Cells by Tumor Exosomes. Int. J. Cancer 2009, 124, 2621–2633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Jiang, J.; Xia, W.; Huang, J. Tumor-Related Exosomes Contribute to Tumor-Promoting Microenvironment: An Immunological Perspective. J. Immunol. Res. 2017, 2017, 1073947. [Google Scholar] [CrossRef] [PubMed]
- Benites, B.D.; Alvarez, M.C.; Saad, S.T.O. Small Particles, Big Effects: The Interplay Between Exosomes and Dendritic Cells in Antitumor Immunity and Immunotherapy. Cells 2019, 8, 1648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chalmin, F.; Ladoire, S.; Mignot, G.; Vincent, J.; Bruchard, M.; Remy-Martin, J.-P.; Boireau, W.; Rouleau, A.; Simon, B.; Lanneau, D.; et al. Membrane-Associated Hsp72 from Tumor-Derived Exosomes Mediates STAT3-Dependent Immunosuppressive Function of Mouse and Human Myeloid-Derived Suppressor Cells. J. Clin. Invest. 2010, 120, 457–471. [Google Scholar] [CrossRef]
- Gobbo, J.; Marcion, G.; Cordonnier, M.; Dias, A.M.M.; Pernet, N.; Hammann, A.; Richaud, S.; Mjahed, H.; Isambert, N.; Clausse, V.; et al. Restoring Anticancer Immune Response by Targeting Tumor-Derived Exosomes With a HSP70 Peptide Aptamer. J. Natl. Cancer Inst. 2016, 108, 330. [Google Scholar] [CrossRef] [PubMed]
- Chanteloup, G.; Cordonnier, M.; Isambert, N.; Bertaut, A.; Hervieu, A.; Hennequin, A.; Luu, M.; Zanetta, S.; Coudert, B.; Bengrine, L.; et al. Monitoring HSP70 Exosomes in Cancer Patients’ Follow up: A Clinical Prospective Pilot Study. J. Extracell. Vesicles 2020, 9, 1766192. [Google Scholar] [CrossRef]
- Azambuja, J.H.; Ludwig, N.; Yerneni, S.; Rao, A.; Braganhol, E.; Whiteside, T.L. Molecular Profiles and Immunomodulatory Activities of Glioblastoma-Derived Exosomes. Neuro-Oncol. Adv. 2020, 2, vdaa056. [Google Scholar] [CrossRef]
- Chen, G.; Huang, A.C.; Zhang, W.; Zhang, G.; Wu, M.; Xu, W.; Yu, Z.; Yang, J.; Wang, B.; Sun, H.; et al. Exosomal PD-L1 Contributes to Immunosuppression and Is Associated with Anti-PD-1 Response. Nature 2018, 560, 382–386. [Google Scholar] [CrossRef]
- Cheng, Z.; Wang, L.; Wu, C.; Huang, L.; Ruan, Y.; Xue, W. Tumor-Derived Exosomes Induced M2 Macrophage Polarization and Promoted the Metastasis of Osteosarcoma Cells Through Tim-3. Arch. Med. Res. 2021, 52, 200–210. [Google Scholar] [CrossRef]
- Cordonnier, M.; Nardin, C.; Chanteloup, G.; Derangere, V.; Algros, M.-P.; Arnould, L.; Garrido, C.; Aubin, F.; Gobbo, J. Tracking the Evolution of Circulating Exosomal-PD-L1 to Monitor Melanoma Patients. J. Extracell. Vesicles 2020, 9, 1710899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Zeng, H.; Zhang, H.; Han, Y. The Role of Exosomal PD-L1 in Tumor Immunotherapy. Transl. Oncol. 2021, 14, 101047. [Google Scholar] [CrossRef]
- Theodoraki, M.-N.; Yerneni, S.S.; Hoffmann, T.K.; Gooding, W.E.; Whiteside, T.L. Clinical Significance of PD-L1+ Exosomes in Plasma of Head and Neck Cancer Patients. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 896–905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theodoraki, M.-N.; Hoffmann, T.K.; Jackson, E.K.; Whiteside, T.L. Exosomes in HNSCC Plasma as Surrogate Markers of Tumour Progression and Immune Competence. Clin. Exp. Immunol. 2018, 194, 67–78. [Google Scholar] [CrossRef] [Green Version]
- Theodoraki, M.-N.; Yerneni, S.; Gooding, W.E.; Ohr, J.; Clump, D.A.; Bauman, J.E.; Ferris, R.L.; Whiteside, T.L. Circulating Exosomes Measure Responses to Therapy in Head and Neck Cancer Patients Treated with Cetuximab, Ipilimumab, and IMRT. Oncoimmunology 2019, 8, 1593805. [Google Scholar] [CrossRef] [PubMed]
- Ricklefs, F.L.; Alayo, Q.; Krenzlin, H.; Mahmoud, A.B.; Speranza, M.C.; Nakashima, H.; Hayes, J.L.; Lee, K.; Balaj, L.; Passaro, C.; et al. Immune Evasion Mediated by PD-L1 on Glioblastoma-Derived Extracellular Vesicles. Sci. Adv. 2018, 4, 2766. [Google Scholar] [CrossRef] [Green Version]
- Martinez, V.G.; O’Neill, S.; Salimu, J.; Breslin, S.; Clayton, A.; Crown, J.; O’Driscoll, L. Resistance to HER2-Targeted Anti-Cancer Drugs Is Associated with Immune Evasion in Cancer Cells and Their Derived Extracellular Vesicles. Oncoimmunology 2017, 6, e1362530. [Google Scholar] [CrossRef] [PubMed]
- Monypenny, J.; Milewicz, H.; Flores-Borja, F.; Weitsman, G.; Cheung, A.; Chowdhury, R.; Burgoyne, T.; Arulappu, A.; Lawler, K.; Barber, P.R.; et al. ALIX Regulates Tumor-Mediated Immunosuppression by Controlling EGFR Activity and PD-L1 Presentation. Cell Rep. 2018, 24, 630–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Li, C.-W.; Chan, L.-C.; Wei, Y.; Hsu, J.-M.; Xia, W.; Cha, J.-H.; Hou, J.; Hsu, J.L.; Sun, L.; et al. Exosomal PD-L1 Harbors Active Defense Function to Suppress T Cell Killing of Breast Cancer Cells and Promote Tumor Growth. Cell Res. 2018, 28, 862–864. [Google Scholar] [CrossRef] [Green Version]
- Poggio, M.; Hu, T.; Pai, C.-C.; Chu, B.; Belair, C.D.; Chang, A.; Montabana, E.; Lang, U.E.; Fu, Q.; Fong, L.; et al. Suppression of Exosomal PD-L1 Induces Systemic Anti-Tumor Immunity and Memory. Cell 2019, 177, 414–427. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.H.; Kim, H.; Choi, Y.J.; Kim, S.Y.; Lee, J.-E.; Sung, K.J.; Sung, Y.H.; Pack, C.-G.; Jung, M.-K.; Han, B.; et al. Exosomal PD-L1 Promotes Tumor Growth through Immune Escape in Non-Small Cell Lung Cancer. Exp. Mol. Med. 2019, 51, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Li, C.; Zhi, C.; Liang, W.; Wang, X.; Chen, X.; Lv, T.; Shen, Q.; Song, Y.; Lin, D.; et al. Clinical Significance of PD-L1 Expression in Serum-Derived Exosomes in NSCLC Patients. J. Transl. Med. 2019, 17, 355. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Fan, Y.; Che, X.; Zhang, M.; Li, Z.; Li, C.; Wang, S.; Wen, T.; Hou, K.; Shao, X.; et al. Anti-PD-1 Therapy Response Predicted by the Combination of Exosomal PD-L1 and CD28. Front. Oncol. 2020, 10, 760. [Google Scholar] [CrossRef]
- Fan, Y.; Che, X.; Qu, J.; Hou, K.; Wen, T.; Li, Z.; Li, C.; Wang, S.; Xu, L.; Liu, Y.; et al. Exosomal PD-L1 Retains Immunosuppressive Activity and Is Associated with Gastric Cancer Prognosis. Ann. Surg. Oncol. 2019, 26, 3745–3755. [Google Scholar] [CrossRef]
- Zhang, M.; Fan, Y.; Che, X.; Hou, K.; Zhang, C.; Li, C.; Wen, T.; Wang, S.; Cheng, Y.; Liu, Y.; et al. 5-FU-Induced Upregulation of Exosomal PD-L1 Causes Immunosuppression in Advanced Gastric Cancer Patients. Front. Oncol. 2020, 10, 492. [Google Scholar] [CrossRef] [Green Version]
- Razzo, B.M.; Ludwig, N.; Hong, C.-S.; Sharma, P.; Fabian, K.P.; Fecek, R.J.; Storkus, W.J.; Whiteside, T.L. Tumor-Derived Exosomes Promote Carcinogenesis of Murine Oral Squamous Cell Carcinoma. Carcinogenesis 2020, 41, 625–633. [Google Scholar] [CrossRef]
- Gao, J.; Qiu, X.; Li, X.; Fan, H.; Zhang, F.; Lv, T.; Song, Y. Expression Profiles and Clinical Value of Plasma Exosomal Tim-3 and Galectin-9 in Non-Small Cell Lung Cancer. Biochem. Biophys. Res. Commun. 2018, 498, 409–415. [Google Scholar] [CrossRef]
- Klibi, J.; Niki, T.; Riedel, A.; Pioche-Durieu, C.; Souquere, S.; Rubinstein, E.; Le Moulec, S.; Moulec, S.L.E.; Guigay, J.; Hirashima, M.; et al. Blood Diffusion and Th1-Suppressive Effects of Galectin-9-Containing Exosomes Released by Epstein-Barr Virus-Infected Nasopharyngeal Carcinoma Cells. Blood 2009, 113, 1957–1966. [Google Scholar] [CrossRef] [Green Version]
- Keryer-Bibens, C.; Pioche-Durieu, C.; Villemant, C.; Souquère, S.; Nishi, N.; Hirashima, M.; Middeldorp, J.; Busson, P. Exosomes Released by EBV-Infected Nasopharyngeal Carcinoma Cells Convey the Viral Latent Membrane Protein 1 and the Immunomodulatory Protein Galectin 9. BMC Cancer 2006, 6, 283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Huang, D.; Baloche, V.; Zhang, L.; Xu, J.; Li, B.; Zhao, X.; He, J.; Mai, H.; Chen, Q.; et al. Galectin-9 Promotes a Suppressive Microenvironment in Human Cancer by Enhancing STING Degradation. Oncogenesis 2020, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Clayton, A.; Al-Taei, S.; Webber, J.; Mason, M.D.; Tabi, Z. Cancer Exosomes Express CD39 and CD73, Which Suppress T Cells through Adenosine Production. J. Immunol. Baltim. Md 1950 2011, 187, 676–683. [Google Scholar] [CrossRef] [PubMed]
- Salimu, J.; Webber, J.; Gurney, M.; Al-Taei, S.; Clayton, A.; Tabi, Z. Dominant Immunosuppression of Dendritic Cell Function by Prostate-Cancer-Derived Exosomes. J. Extracell. Vesicles 2017, 6, 1368823. [Google Scholar] [CrossRef] [Green Version]
- Ludwig, N.; Yerneni, S.S.; Azambuja, J.H.; Gillespie, D.G.; Menshikova, E.V.; Jackson, E.K.; Whiteside, T.L. Tumor-Derived Exosomes Promote Angiogenesis via Adenosine A2B Receptor Signaling. Angiogenesis 2020, 23, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Schuler, P.J.; Saze, Z.; Hong, C.-S.; Muller, L.; Gillespie, D.G.; Cheng, D.; Harasymczuk, M.; Mandapathil, M.; Lang, S.; Jackson, E.K.; et al. Human CD4+CD39+ Regulatory T Cells Produce Adenosine upon Co-Expression of Surface CD73 or Contact with CD73+ Exosomes or CD73+ Cells. Clin. Exp. Immunol. 2014, 177, 531–543. [Google Scholar] [CrossRef]
- Freeman, G.J.; Long, A.J.; Iwai, Y.; Bourque, K.; Chernova, T.; Nishimura, H.; Fitz, L.J.; Malenkovich, N.; Okazaki, T.; Byrne, M.C.; et al. Engagement of the PD-1 Immunoinhibitory Receptor by a Novel B7 Family Member Leads to Negative Regulation of Lymphocyte Activation. J. Exp. Med. 2000, 192, 1027–1034. [Google Scholar] [CrossRef] [Green Version]
- Keir, M.E.; Butte, M.J.; Freeman, G.J.; Sharpe, A.H. PD-1 and Its Ligands in Tolerance and Immunity. Annu. Rev. Immunol. 2008, 26, 677–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, X.; Wang, J.; Deng, X.; Xiong, F.; Ge, J.; Xiang, B.; Wu, X.; Ma, J.; Zhou, M.; Li, X.; et al. Role of the Tumor Microenvironment in PD-L1/PD-1-Mediated Tumor Immune Escape. Mol. Cancer 2019, 18, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Francisco, L.M.; Sage, P.T.; Sharpe, A.H. The PD-1 Pathway in Tolerance and Autoimmunity. Immunol. Rev. 2010, 236, 219–242. [Google Scholar] [CrossRef] [PubMed]
- Jalali, S.; Price-Troska, T.; Bothun, C.; Villasboas, J.; Kim, H.-J.; Yang, Z.-Z.; Novak, A.J.; Dong, H.; Ansell, S.M. Reverse Signaling via PD-L1 Supports Malignant Cell Growth and Survival in Classical Hodgkin Lymphoma. Blood Cancer J. 2019, 9, 22. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Chen, W.; Xu, Z.P.; Gu, W. PD-L1 Distribution and Perspective for Cancer Immunotherapy-Blockade, Knockdown, or Inhibition. Front. Immunol. 2019, 10, 2022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blank, C.; Gajewski, T.F.; Mackensen, A. Interaction of PD-L1 on Tumor Cells with PD-1 on Tumor-Specific T Cells as a Mechanism of Immune Evasion: Implications for Tumor Immunotherapy. Cancer Immunol. Immunother. 2005, 54, 307–314. [Google Scholar] [CrossRef]
- Azuma, T.; Yao, S.; Zhu, G.; Flies, A.S.; Flies, S.J.; Chen, L. B7-H1 Is a Ubiquitous Antiapoptotic Receptor on Cancer Cells. Blood 2008, 111, 3635–3643. [Google Scholar] [CrossRef]
- Lux, A.; Kahlert, C.; Grützmann, R.; Pilarsky, C. C-Met and PD-L1 on Circulating Exosomes as Diagnostic and Prognostic Markers for Pancreatic Cancer. Int. J. Mol. Sci. 2019, 20, 3305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Gong, Y.; Lv, Z.; Li, L.; Yuan, Y. Expression of PD1/PDL1 in Gastric Cancer at Different Microsatellite Status and Its Correlation with Infiltrating Immune Cells in the Tumor Microenvironment. J. Cancer 2021, 12, 1698–1707. [Google Scholar] [CrossRef] [PubMed]
- Ayala-Mar, S.; Donoso-Quezada, J.; González-Valdez, J. Clinical Implications of Exosomal PD-L1 in Cancer Immunotherapy. J. Immunol. Res. 2021, 2021, 8839978. [Google Scholar] [CrossRef]
- Yin, Z.; Yu, M.; Ma, T.; Zhang, C.; Huang, S.; Karimzadeh, M.R.; Momtazi-Borojeni, A.A.; Chen, S. Mechanisms Underlying Low-Clinical Responses to PD-1/PD-L1 Blocking Antibodies in Immunotherapy of Cancer: A Key Role of Exosomal PD-L1. J. Immunother. Cancer 2021, 9, e001698. [Google Scholar] [CrossRef]
- Qiu, Y.; Yang, Y.; Yang, R.; Liu, C.; Hsu, J.-M.; Jiang, Z.; Sun, L.; Wei, Y.; Li, C.-W.; Yu, D.; et al. Activated T Cell-Derived Exosomal PD-1 Attenuates PD-L1-Induced Immune Dysfunction in Triple-Negative Breast Cancer. Oncogene 2021, 40, 4992–5001. [Google Scholar] [CrossRef] [PubMed]
- Monney, L.; Sabatos, C.A.; Gaglia, J.L.; Ryu, A.; Waldner, H.; Chernova, T.; Manning, S.; Greenfield, E.A.; Coyle, A.J.; Sobel, R.A.; et al. Th1-Specific Cell Surface Protein Tim-3 Regulates Macrophage Activation and Severity of an Autoimmune Disease. Nature 2002, 415, 536–541. [Google Scholar] [CrossRef] [PubMed]
- Das, M.; Zhu, C.; Kuchroo, V.K. Tim-3 and Its Role in Regulating Anti-Tumor Immunity. Immunol. Rev. 2017, 276, 97–111. [Google Scholar] [CrossRef] [Green Version]
- Wolf, Y.; Anderson, A.C.; Kuchroo, V.K. TIM3 Comes of Age as an Inhibitory Receptor. Nat. Rev. Immunol. 2020, 20, 173–185. [Google Scholar] [CrossRef]
- Clayton, K.L.; Haaland, M.S.; Douglas-Vail, M.B.; Mujib, S.; Chew, G.M.; Ndhlovu, L.C.; Ostrowski, M.A. T Cell Ig and Mucin Domain-Containing Protein 3 Is Recruited to the Immune Synapse, Disrupts Stable Synapse Formation, and Associates with Receptor Phosphatases. J. Immunol. Baltim. Md 1950 2014, 192, 782–791. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Su, E.W.; Zhu, C.; Hainline, S.; Phuah, J.; Moroco, J.A.; Smithgall, T.E.; Kuchroo, V.K.; Kane, L.P. Phosphotyrosine-Dependent Coupling of Tim-3 to T-Cell Receptor Signaling Pathways. Mol. Cell. Biol. 2011, 31, 3963–3974. [Google Scholar] [CrossRef] [Green Version]
- Rangachari, M.; Zhu, C.; Sakuishi, K.; Xiao, S.; Karman, J.; Chen, A.; Angin, M.; Wakeham, A.; Greenfield, E.A.; Sobel, R.A.; et al. Bat3 Promotes T Cell Responses and Autoimmunity by Repressing Tim-3–Mediated Cell Death and Exhaustion. Nat. Med. 2012, 18, 1394–1400. [Google Scholar] [CrossRef] [Green Version]
- Chiba, S.; Baghdadi, M.; Akiba, H.; Yoshiyama, H.; Kinoshita, I.; Dosaka-Akita, H.; Fujioka, Y.; Ohba, Y.; Gorman, J.V.; Colgan, J.D.; et al. Tumor-Infiltrating DCs Suppress Nucleic Acid-Mediated Innate Immune Responses through Interactions between the Receptor TIM-3 and the Alarmin HMGB1. Nat. Immunol. 2012, 13, 832–842. [Google Scholar] [CrossRef]
- Sakuishi, K.; Apetoh, L.; Sullivan, J.M.; Blazar, B.R.; Kuchroo, V.K.; Anderson, A.C. Targeting Tim-3 and PD-1 Pathways to Reverse T Cell Exhaustion and Restore Anti-Tumor Immunity. J. Exp. Med. 2010, 207, 2187–2194. [Google Scholar] [CrossRef] [PubMed]
- Sakuishi, K.; Ngiow, S.F.; Sullivan, J.M.; Teng, M.W.L.; Kuchroo, V.K.; Smyth, M.J.; Anderson, A.C. TIM3+FOXP3+ Regulatory T Cells Are Tissue-Specific Promoters of T-Cell Dysfunction in Cancer. Oncoimmunology 2013, 2, e23849. [Google Scholar] [CrossRef] [Green Version]
- Yanai, H.; Ban, T.; Wang, Z.; Choi, M.K.; Kawamura, T.; Negishi, H.; Nakasato, M.; Lu, Y.; Hangai, S.; Koshiba, R.; et al. HMGB Proteins Function as Universal Sentinels for Nucleic-Acid-Mediated Innate Immune Responses. Nature 2009, 462, 99–103. [Google Scholar] [CrossRef]
- Zhu, C.; Anderson, A.C.; Schubart, A.; Xiong, H.; Imitola, J.; Khoury, S.J.; Zheng, X.X.; Strom, T.B.; Kuchroo, V.K. The Tim-3 Ligand Galectin-9 Negatively Regulates T Helper Type 1 Immunity. Nat. Immunol. 2005, 6, 1245–1252. [Google Scholar] [CrossRef]
- Jiang, J.; Jin, M.-S.; Kong, F.; Cao, D.; Ma, H.-X.; Jia, Z.; Wang, Y.-P.; Suo, J.; Cao, X. Decreased Galectin-9 and Increased Tim-3 Expression Are Related to Poor Prognosis in Gastric Cancer. PLoS ONE 2013, 8, e81799. [Google Scholar] [CrossRef] [PubMed]
- Jia, K.; He, Y.; Dziadziuszko, R.; Zhao, S.; Zhang, X.; Deng, J.; Wang, H.; Hirsch, F.R.; Zhou, C.; Yu, H.; et al. T Cell Immunoglobulin and Mucin-Domain Containing-3 in Non-Small Cell Lung Cancer. Transl. Lung Cancer Res. 2019, 8, 895–906. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Zhang, X.; Xia, X.; Zhang, C.; Liang, X.; Gao, L.; Zhang, X.; Ma, C. Ectopic Expression of TIM-3 in Lung Cancers: A Potential Independent Prognostic Factor for Patients With NSCLC. Am. J. Clin. Pathol. 2012, 137, 978–985. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Zhou, X.; Huang, X.; Li, Q.; Gao, L.; Jiang, L.; Huang, M.; Zhou, J. Tim-3 Expression in Cervical Cancer Promotes Tumor Metastasis. PLoS ONE 2013, 8, e53834. [Google Scholar] [CrossRef]
- Shang, Y.; Li, Z.; Li, H.; Xia, H.; Lin, Z. TIM-3 Expression in Human Osteosarcoma: Correlation with the Expression of Epithelial-Mesenchymal Transition-Specific Biomarkers. Oncol. Lett. 2013, 6, 490–494. [Google Scholar] [CrossRef] [Green Version]
- Komohara, Y.; Morita, T.; Annan, D.A.; Horlad, H.; Ohnishi, K.; Yamada, S.; Nakayama, T.; Kitada, S.; Suzu, S.; Kinoshita, I.; et al. The Coordinated Actions of TIM-3 on Cancer and Myeloid Cells in the Regulation of Tumorigenicity and Clinical Prognosis in Clear Cell Renal Cell Carcinomas. Cancer Immunol. Res. 2015, 3, 999–1007. [Google Scholar] [CrossRef] [Green Version]
- Wiener, Z.; Kohalmi, B.; Pocza, P.; Jeager, J.; Tolgyesi, G.; Toth, S.; Gorbe, E.; Papp, Z.; Falus, A. TIM-3 Is Expressed in Melanoma Cells and Is Upregulated in TGF-Beta Stimulated Mast Cells. J. Invest. Dermatol. 2007, 127, 906–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haubner, S.; Perna, F.; Köhnke, T.; Schmidt, C.; Berman, S.; Augsberger, C.; Schnorfeil, F.M.; Krupka, C.; Lichtenegger, F.S.; Liu, X.; et al. Coexpression Profile of Leukemic Stem Cell Markers for Combinatorial Targeted Therapy in AML. Leukemia 2019, 33, 64–74. [Google Scholar] [CrossRef]
- Jan, M.; Chao, M.P.; Cha, A.C.; Alizadeh, A.A.; Gentles, A.J.; Weissman, I.L.; Majeti, R. Prospective Separation of Normal and Leukemic Stem Cells Based on Differential Expression of TIM3, a Human Acute Myeloid Leukemia Stem Cell Marker. Proc. Natl. Acad. Sci. USA 2011, 108, 5009–5014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enninga, E.A.L.; Nevala, W.K.; Holtan, S.G.; Leontovich, A.A.; Markovic, S.N. Galectin-9 Modulates Immunity by Promoting Th2/M2 Differentiation and Impacts Survival in Patients with Metastatic Melanoma. Melanoma Res. 2016, 26, 429–441. [Google Scholar] [CrossRef]
- Zhou, W.; Zhou, Y.; Chen, X.; Ning, T.; Chen, H.; Guo, Q.; Zhang, Y.; Liu, P.; Zhang, Y.; Li, C.; et al. Pancreatic Cancer-Targeting Exosomes for Enhancing Immunotherapy and Reprogramming Tumor Microenvironment. Biomaterials 2021, 268, 120546. [Google Scholar] [CrossRef] [PubMed]
- Irie, A.; Yamauchi, A.; Kontani, K.; Kihara, M.; Liu, D.; Shirato, Y.; Seki, M.; Nishi, N.; Nakamura, T.; Yokomise, H.; et al. Galectin-9 as a Prognostic Factor with Antimetastatic Potential in Breast Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2005, 11, 2962–2968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamauchi, A.; Kontani, K.; Kihara, M.; Nishi, N.; Yokomise, H.; Hirashima, M. Galectin-9, a Novel Prognostic Factor with Antimetastatic Potential in Breast Cancer. Breast J. 2006, 12, 196–200. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Sun, L.; Jing, D.; Xu, G.; Zhang, J.; Lin, L.; Zhao, J.; Yao, Z.; Lin, H. Galectin-9 Expression Predicts Favorable Clinical Outcome in Solid Tumors: A Systematic Review and Meta-Analysis. Front. Physiol. 2018, 9, 452. [Google Scholar] [CrossRef] [Green Version]
- Figueiró, F.; Muller, L.; Funk, S.; Jackson, E.K.; Battastini, A.M.O.; Whiteside, T.L. Phenotypic and Functional Characteristics of CD39high Human Regulatory B Cells (Breg). Oncoimmunology 2016, 5, e1082703. [Google Scholar] [CrossRef] [Green Version]
- Saze, Z.; Schuler, P.J.; Hong, C.-S.; Cheng, D.; Jackson, E.K.; Whiteside, T.L. Adenosine Production by Human B Cells and B Cell-Mediated Suppression of Activated T Cells. Blood 2013, 122, 9–18. [Google Scholar] [CrossRef] [Green Version]
- Antonioli, L.; Pacher, P.; Vizi, E.S.; Haskó, G. CD39 and CD73 in Immunity and Inflammation. Trends Mol. Med. 2013, 19, 355–367. [Google Scholar] [CrossRef] [Green Version]
- Sitkovsky, M.V.; Ohta, A. The “danger” Sensors That STOP the Immune Response: The A2 Adenosine Receptors? Trends Immunol. 2005, 26, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Sperlágh, B.; Vizi, E.S. The Role of Extracellular Adenosine in Chemical Neurotransmission in the Hippocampus and Basal Ganglia: Pharmacological and Clinical Aspects. Curr. Top. Med. Chem. 2011, 11, 1034–1046. [Google Scholar] [CrossRef] [PubMed]
- Sperlágh, B.; Haskó, G.; Németh, Z.; Vizi, E.S. ATP Released by LPS Increases Nitric Oxide Production in Raw 264.7 Macrophage Cell Line via P2Z/P2X7 Receptors. Neurochem. Int. 1998, 33, 209–215. [Google Scholar] [CrossRef]
- Baghbani, E.; Noorolyai, S.; Shanehbandi, D.; Mokhtarzadeh, A.; Aghebati-Maleki, L.; Shahgoli, V.K.; Brunetti, O.; Rahmani, S.; Shadbad, M.A.; Baghbanzadeh, A.; et al. Regulation of Immune Responses through CD39 and CD73 in Cancer: Novel Checkpoints. Life Sci. 2021, 282, 119826. [Google Scholar] [CrossRef] [PubMed]
- Novitskiy, S.V.; Ryzhov, S.; Zaynagetdinov, R.; Goldstein, A.E.; Huang, Y.; Tikhomirov, O.Y.; Blackburn, M.R.; Biaggioni, I.; Carbone, D.P.; Feoktistov, I.; et al. Adenosine Receptors in Regulation of Dendritic Cell Differentiation and Function. Blood 2008, 112, 1822–1831. [Google Scholar] [CrossRef]
- Ohta, A.; Kini, R.; Ohta, A.; Subramanian, M.; Madasu, M.; Sitkovsky, M. The Development and Immunosuppressive Functions of CD4(+) CD25(+) FoxP3(+) Regulatory T Cells Are under Influence of the Adenosine-A2A Adenosine Receptor Pathway. Front. Immunol. 2012, 3, 190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beavis, P.A.; Stagg, J.; Darcy, P.K.; Smyth, M.J. CD73: A Potent Suppressor of Antitumor Immune Responses. Trends Immunol. 2012, 33, 231–237. [Google Scholar] [CrossRef]
- Gao, Z.-W.; Wang, H.-P.; Lin, F.; Wang, X.; Long, M.; Zhang, H.-Z.; Dong, K. CD73 Promotes Proliferation and Migration of Human Cervical Cancer Cells Independent of Its Enzyme Activity. BMC Cancer 2017, 17, 135. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Jia, S.; Chen, Y.; Wang, W.; Wu, Z.; Yu, W.; Zhang, M.; Ding, G.; Cao, L. The Distinct Role of CD73 in the Progression of Pancreatic Cancer. J. Mol. Med. Berl. Ger. 2019, 97, 803–815. [Google Scholar] [CrossRef] [Green Version]
- Bowser, J.L.; Lee, J.W.; Yuan, X.; Eltzschig, H.K. The Hypoxia-Adenosine Link during Inflammation. J. Appl. Physiol. Bethesda Md 1985 2017, 123, 1303–1320. [Google Scholar] [CrossRef] [Green Version]
- Synnestvedt, K.; Furuta, G.T.; Comerford, K.M.; Louis, N.; Karhausen, J.; Eltzschig, H.K.; Hansen, K.R.; Thompson, L.F.; Colgan, S.P. Ecto-5’-Nucleotidase (CD73) Regulation by Hypoxia-Inducible Factor-1 Mediates Permeability Changes in Intestinal Epithelia. J. Clin. Invest. 2002, 110, 993–1002. [Google Scholar] [CrossRef]
- Mandapathil, M.; Hilldorfer, B.; Szczepanski, M.J.; Czystowska, M.; Szajnik, M.; Ren, J.; Lang, S.; Jackson, E.K.; Gorelik, E.; Whiteside, T.L. Generation and Accumulation of Immunosuppressive Adenosine by Human CD4+CD25highFOXP3+ Regulatory T Cells. J. Biol. Chem. 2010, 285, 7176–7186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dwyer, K.M.; Hanidziar, D.; Putheti, P.; Hill, P.A.; Pommey, S.; McRae, J.L.; Winterhalter, A.; Doherty, G.; Deaglio, S.; Koulmanda, M.; et al. Expression of CD39 by Human Peripheral Blood CD4+ CD25+ T Cells Denotes a Regulatory Memory Phenotype. Am. J. Transplant. Off. J. Am. Soc. Transplant. Am. Soc. Transpl. Surg. 2010, 10, 2410–2420. [Google Scholar] [CrossRef] [Green Version]
- Ghiringhelli, F.; Bruchard, M.; Chalmin, F.; Rébé, C. Production of Adenosine by Ectonucleotidases: A Key Factor in Tumor Immunoescape. J. Biomed. Biotechnol. 2012, 2012, 473712. [Google Scholar] [CrossRef]
- Niechi, I.; Uribe-Ojeda, A.; Erices, J.I.; Torres, Á.; Uribe, D.; Rocha, J.D.; Silva, P.; Richter, H.G.; San Martín, R.; Quezada, C. Adenosine Depletion as A New Strategy to Decrease Glioblastoma Stem-Like Cells Aggressiveness. Cells 2019, 8, 1353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryzhov, S.; Novitskiy, S.V.; Goldstein, A.E.; Biktasova, A.; Blackburn, M.R.; Biaggioni, I.; Dikov, M.M.; Feoktistov, I. Adenosinergic Regulation of the Expansion and Immunosuppressive Activity of CD11b+Gr1+ Cells. J. Immunol. Baltim. Md 1950 2011, 187, 6120–6129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torres, Á.; Erices, J.I.; Sanchez, F.; Ehrenfeld, P.; Turchi, L.; Virolle, T.; Uribe, D.; Niechi, I.; Spichiger, C.; Rocha, J.D.; et al. Extracellular Adenosine Promotes Cell Migration/Invasion of Glioblastoma Stem-like Cells through A3 Adenosine Receptor Activation under Hypoxia. Cancer Lett. 2019, 446, 112–122. [Google Scholar] [CrossRef]
- Mastelic-Gavillet, B.; Navarro Rodrigo, B.; Décombaz, L.; Wang, H.; Ercolano, G.; Ahmed, R.; Lozano, L.E.; Ianaro, A.; Derré, L.; Valerio, M.; et al. Adenosine Mediates Functional and Metabolic Suppression of Peripheral and Tumor-Infiltrating CD8+ T Cells. J. Immunother. Cancer 2019, 7, 257. [Google Scholar] [CrossRef]
- Zarek, P.E.; Huang, C.-T.; Lutz, E.R.; Kowalski, J.; Horton, M.R.; Linden, J.; Drake, C.G.; Powell, J.D. A2A Receptor Signaling Promotes Peripheral Tolerance by Inducing T-Cell Anergy and the Generation of Adaptive Regulatory T Cells. Blood 2008, 111, 251–259. [Google Scholar] [CrossRef]
- Jin, K.; Mao, C.; Chen, L.; Wang, L.; Liu, Y.; Yuan, J. Adenosinergic Pathway: A Hope in the Immunotherapy of Glioblastoma. Cancers 2021, 13, 229. [Google Scholar] [CrossRef]
- Ludwig, N.; Azambuja, J.H.; Rao, A.; Gillespie, D.G.; Jackson, E.K.; Whiteside, T.L. Adenosine Receptors Regulate Exosome Production. Purinergic Signal. 2020, 16, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Theodoraki, M.-N.; Hoffmann, T.K.; Whiteside, T.L. Separation of Plasma-Derived Exosomes into CD3(+) and CD3(-) Fractions Allows for Association of Immune Cell and Tumour Cell Markers with Disease Activity in HNSCC Patients. Clin. Exp. Immunol. 2018, 192, 271–283. [Google Scholar] [CrossRef] [Green Version]
- Whiteside, T.L. Immune Modulation of T-Cell and NK (Natural Killer) Cell Activities by TEXs (Tumour-Derived Exosomes). Biochem. Soc. Trans. 2013, 41, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Andrews, L.P.; Marciscano, A.E.; Drake, C.G.; Vignali, D.A.A. LAG3 (CD223) as a Cancer Immunotherapy Target. Immunol. Rev. 2017, 276, 80–96. [Google Scholar] [CrossRef] [PubMed]
- Balko, J.M.; Johnson, D.B.; Wang, D.Y.; Ericsson-Gonzalez, P.; Nixon, M.; Salgado, R.; Sanchez, V.; Schreeder, D.; Kim, J.Y.; Bordeaux, J.; et al. MHC-II Expression to Drive a Unique Pattern of Adaptive Resistance to Antitumor Immunity through Receptor Checkpoint Engagement. J. Clin. Oncol. 2018, 36, 180. [Google Scholar] [CrossRef]
- Gauvreau, M.-E.; Côté, M.-H.; Bourgeois-Daigneault, M.-C.; Rivard, L.-D.; Xiu, F.; Brunet, A.; Shaw, A.; Steimle, V.; Thibodeau, J. Sorting of MHC Class II Molecules into Exosomes through a Ubiquitin-Independent Pathway. Traffic Cph. Den. 2009, 10, 1518–1527. [Google Scholar] [CrossRef]
- Lee, Y.S.; Kim, S.H.; Cho, J.A.; Kim, C.W. Introduction of the CIITA Gene into Tumor Cells Produces Exosomes with Enhanced Anti-Tumor Effects. Exp. Mol. Med. 2011, 43, 281–290. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Harden, K.; Gonzalez, L.C.; Francesco, M.; Chiang, E.; Irving, B.; Tom, I.; Ivelja, S.; Refino, C.J.; Clark, H.; et al. The Surface Protein TIGIT Suppresses T Cell Activation by Promoting the Generation of Mature Immunoregulatory Dendritic Cells. Nat. Immunol. 2009, 10, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Solomon, B.L.; Garrido-Laguna, I. TIGIT: A Novel Immunotherapy Target Moving from Bench to Bedside. Cancer Immunol. Immunother. CII 2018, 67, 1659–1667. [Google Scholar] [CrossRef]
- Liu, H.; Gao, W.; Yuan, J.; Wu, C.; Yao, K.; Zhang, L.; Ma, L.; Zhu, J.; Zou, Y.; Ge, J. Exosomes Derived from Dendritic Cells Improve Cardiac Function via Activation of CD4(+) T Lymphocytes after Myocardial Infarction. J. Mol. Cell. Cardiol. 2016, 91, 123–133. [Google Scholar] [CrossRef]
- Li, M.; Soder, R.; Abhyankar, S.; Abdelhakim, H.; Braun, M.W.; Trinidad, C.V.; Pathak, H.B.; Pessetto, Z.; Deighan, C.; Ganguly, S.; et al. WJMSC-Derived Small Extracellular Vesicle Enhance T Cell Suppression through PD-L1. J. Extracell. Vesicles 2021, 10, e12067. [Google Scholar] [CrossRef]
- Huang, T.; Li, F.; Cheng, X.; Wang, J.; Zhang, W.; Zhang, B.; Tang, Y.; Li, Q.; Zhou, C.; Tu, S. Wnt Inhibition Sensitizes PD-L1 Blockade Therapy by Overcoming Bone Marrow-Derived Myofibroblasts-Mediated Immune Resistance in Tumors. Front. Immunol. 2021, 12, 619209. [Google Scholar] [CrossRef]
- Ritprajak, P.; Azuma, M. Intrinsic and Extrinsic Control of Expression of the Immunoregulatory Molecule PD-L1 in Epithelial Cells and Squamous Cell Carcinoma. Oral Oncol. 2015, 51, 221–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, A.C.; Joller, N.; Kuchroo, V.K. Lag-3, Tim-3, and TIGIT: Co-Inhibitory Receptors with Specialized Functions in Immune Regulation. Immunity 2016, 44, 989–1004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liangsupree, T.; Multia, E.; Riekkola, M.-L. Modern Isolation and Separation Techniques for Extracellular Vesicles. J. Chromatogr. A 2021, 1636, 461773. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Ludwig, S.; Muller, L.; Hong, C.S.; Kirkwood, J.M.; Ferrone, S.; Whiteside, T.L. Immunoaffinity-Based Isolation of Melanoma Cell-Derived Exosomes from Plasma of Patients with Melanoma. J. Extracell. Vesicles 2018, 7, 1435138. [Google Scholar] [CrossRef]
- Datta, A.; Kim, H.; Lal, M.; McGee, L.; Johnson, A.; Moustafa, A.A.; Jones, J.C.; Mondal, D.; Ferrer, M.; Abdel-Mageed, A.B. Manumycin A Suppresses Exosome Biogenesis and Secretion via Targeted Inhibition of Ras/Raf/ERK1/2 Signaling and HnRNP H1 in Castration-Resistant Prostate Cancer Cells. Cancer Lett. 2017, 408, 73–81. [Google Scholar] [CrossRef]
- Datta, A.; Kim, H.; McGee, L.; Johnson, A.E.; Talwar, S.; Marugan, J.; Southall, N.; Hu, X.; Lal, M.; Mondal, D.; et al. High-Throughput Screening Identified Selective Inhibitors of Exosome Biogenesis and Secretion: A Drug Repurposing Strategy for Advanced Cancer. Sci. Rep. 2018, 8, 8161. [Google Scholar] [CrossRef] [Green Version]
- Bobrie, A.; Krumeich, S.; Reyal, F.; Recchi, C.; Moita, L.F.; Seabra, M.C.; Ostrowski, M.; Théry, C. Rab27a Supports Exosome-Dependent and -Independent Mechanisms That Modify the Tumor Microenvironment and Can Promote Tumor Progression. Cancer Res. 2012, 72, 4920–4930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catalano, M.; O’Driscoll, L. Inhibiting Extracellular Vesicles Formation and Release: A Review of EV Inhibitors. J. Extracell. Vesicles 2020, 9, 1703244. [Google Scholar] [CrossRef] [Green Version]
- Hayatudin, R.; Fong, Z.; Ming, L.C.; Goh, B.-H.; Lee, W.-L.; Kifli, N. Overcoming Chemoresistance via Extracellular Vesicle Inhibition. Front. Mol. Biosci. 2021, 8, 629874. [Google Scholar] [CrossRef]
- Zhang, H.; Lu, J.; Liu, J.; Zhang, G.; Lu, A. Advances in the Discovery of Exosome Inhibitors in Cancer. J. Enzyme Inhib. Med. Chem. 2020, 35, 1322–1330. [Google Scholar] [CrossRef]
- Christianson, H.C.; Svensson, K.J.; van Kuppevelt, T.H.; Li, J.-P.; Belting, M. Cancer Cell Exosomes Depend on Cell-Surface Heparan Sulfate Proteoglycans for Their Internalization and Functional Activity. Proc. Natl. Acad. Sci. USA 2013, 110, 17380–17385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa Verdera, H.; Gitz-Francois, J.J.; Schiffelers, R.M.; Vader, P. Cellular Uptake of Extracellular Vesicles Is Mediated by Clathrin-Independent Endocytosis and Macropinocytosis. J. Control. Release Off. J. Control. Release Soc. 2017, 266, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Donaldson, J.G. Search for Inhibitors of Endocytosis: Intended Specificity and Unintended Consequences. Cell. Logist. 2012, 2, 203–208. [Google Scholar] [CrossRef] [Green Version]
- Elkin, S.R.; Oswald, N.W.; Reed, D.K.; Mettlen, M.; MacMillan, J.B.; Schmid, S.L. Ikarugamycin: A Natural Product Inhibitor of Clathrin-Mediated Endocytosis. Traffic Cph. Den. 2016, 17, 1139–1149. [Google Scholar] [CrossRef] [Green Version]
- Kosgodage, U.S.; Trindade, R.P.; Thompson, P.R.; Inal, J.M.; Lange, S. Chloramidine/Bisindolylmaleimide-I-Mediated Inhibition of Exosome and Microvesicle Release and Enhanced Efficacy of Cancer Chemotherapy. Int. J. Mol. Sci. 2017, 18, 1007. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.-P.; Singla, B.; Ghoshal, P.; Faulkner, J.L.; Cherian-Shaw, M.; O’Connor, P.M.; She, J.-X.; Belin de Chantemele, E.J.; Csányi, G. Identification of Novel Macropinocytosis Inhibitors Using a Rational Screen of Food and Drug Administration-Approved Drugs. Br. J. Pharmacol. 2018, 175, 3640–3655. [Google Scholar] [CrossRef] [Green Version]
- Im, E.-J.; Lee, C.-H.; Moon, P.-G.; Rangaswamy, G.G.; Lee, B.; Lee, J.M.; Lee, J.-C.; Jee, J.-G.; Bae, J.-S.; Kwon, T.-K.; et al. Sulfisoxazole Inhibits the Secretion of Small Extracellular Vesicles by Targeting the Endothelin Receptor A. Nat. Commun. 2019, 10, 1387. [Google Scholar] [CrossRef]
- Hu, Y.; Yan, C.; Mu, L.; Huang, K.; Li, X.; Tao, D.; Wu, Y.; Qin, J. Fibroblast-Derived Exosomes Contribute to Chemoresistance through Priming Cancer Stem Cells in Colorectal Cancer. PLoS ONE 2015, 10, e0125625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, R.; Aung, T.; Vogel, D.; Chapuy, B.; Wenzel, D.; Becker, S.; Sinzig, U.; Venkataramani, V.; von Mach, T.; Jacob, R.; et al. Nuclear Trapping through Inhibition of Exosomal Export by Indomethacin Increases Cytostatic Efficacy of Doxorubicin and Pixantrone. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 395–404. [Google Scholar] [CrossRef] [Green Version]
- Kulshreshtha, A.; Singh, S.; Ahmad, M.; Khanna, K.; Ahmad, T.; Agrawal, A.; Ghosh, B. Simvastatin Mediates Inhibition of Exosome Synthesis, Localization and Secretion via Multicomponent Interventions. Sci. Rep. 2019, 9, 16373. [Google Scholar] [CrossRef] [Green Version]
- Johnson, J.L.; Ramadass, M.; Haimovich, A.; McGeough, M.D.; Zhang, J.; Hoffman, H.M.; Catz, S.D. Increased Neutrophil Secretion Induced by NLRP3 Mutation Links the Inflammasome to Azurophilic Granule Exocytosis. Front. Cell. Infect. Microbiol. 2017, 7, 507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, S.; Jutzy, J.M.S.; Aspe, J.R.; McGregor, D.W.; Neidigh, J.W.; Wall, N.R. Survivin Is Released from Cancer Cells via Exosomes. Apoptosis Int. J. Program. Cell Death 2011, 16, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.-B.; Wu, J.Y.; Lillard, J.; Bond, V.C. SMR Peptide Antagonizes Mortalin Promoted Release of Extracellular Vesicles and Affects Mortalin Protection from Complement-Dependent Cytotoxicity in Breast Cancer Cells and Leukemia Cells. Oncotarget 2019, 10, 5419–5438. [Google Scholar] [CrossRef] [Green Version]
- Kosgodage, U.S.; Mould, R.; Henley, A.B.; Nunn, A.V.; Guy, G.W.; Thomas, E.L.; Inal, J.M.; Bell, J.D.; Lange, S. Cannabidiol (CBD) Is a Novel Inhibitor for Exosome and Microvesicle (EMV) Release in Cancer. Front. Pharmacol. 2018, 9, 889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mineo, M.; Garfield, S.H.; Taverna, S.; Flugy, A.; De Leo, G.; Alessandro, R.; Kohn, E.C. Exosomes Released by K562 Chronic Myeloid Leukemia Cells Promote Angiogenesis in a Src-Dependent Fashion. Angiogenesis 2012, 15, 33–45. [Google Scholar] [CrossRef] [Green Version]
- Khan, F.M.; Saleh, E.; Alawadhi, H.; Harati, R.; Zimmermann, W.-H.; El-Awady, R. Inhibition of Exosome Release by Ketotifen Enhances Sensitivity of Cancer Cells to Doxorubicin. Cancer Biol. Ther. 2018, 19, 25–33. [Google Scholar] [CrossRef] [Green Version]
- Multhoff, G.; Botzler, C.; Wiesnet, M.; Müller, E.; Meier, T.; Wilmanns, W.; Issels, R.D. A Stress-Inducible 72-KDa Heat-Shock Protein (HSP72) Is Expressed on the Surface of Human Tumor Cells, but Not on Normal Cells. Int. J. Cancer 1995, 61, 272–279. [Google Scholar] [CrossRef] [PubMed]


| Exosomal Protein | Type of Cancer | Source of Exosomes | Target Protein | Potential Exosome-Related Functions in Cancer |
|---|---|---|---|---|
| PD-L1 | HNSCC | Plasma [58,59,60] | PD-1 |
|
| Glioblastoma | Plasma; Primary cancer cells lines G34, G35, G44, G157 [61] |
| ||
| Breast | Cancer cell lines MDA-MB-231, HCC1954, SKBR3, EFM-192A [62,63,64] |
| ||
| Prostate | Cancer cell lines DU145, PC3, LnCap, TRAMP-C2 [65] |
| ||
| Melanoma | Plasma; Cancer cell lines SK-Mel-2, SK-Mel-28, Mel624 and WM9 (human) and B16F10 (murine) [54,56,65] |
| ||
| NSCLC | Serum; plasma; Cancer cell lines A549, H460, H1975 [66,67] |
| ||
| Lung Squamous Cell Carcinoma and adenocarcinoma | Plasma [68] |
| ||
| Gastric cancer | Plasma [69,70] |
| ||
| Oral-oesophageal | HNSCC cancer cell lines SCC90 (human) and SCCVII (murine) [71]. |
| ||
| TIM-3 | Osteosarcoma | Cancer cell line MG63 [55] | (Non Available) |
|
| NSCLC | Plasma [72] |
| ||
| Gal-9 | NPC | Plasma (patients) [73] Plasma from C15 and C17 xenografted mice [73] | TIM-3 |
|
| NPC xenografts cell lines (C15 and C17) supernatant [74] |
| |||
| Cancer cell lines TW03 [75] | (Non Available) |
| ||
| CD73/CD39 | Bladder, breast, colorectal | Cancer cell lines HT1376 (bladder), CACO2 (CRC), MCF7 (breast) [76] | Adenosine Receptors |
|
| Prostate | Cancer cell lines DU145, PC3 [76,77] |
| ||
| Mesothelioma | Pleural effusion; custom cancer cell line (Meso) [76] |
| ||
| HNSCC | Plasma; cancer cell line UMSCC47 [78] |
| ||
| Plasma [79] |
| |||
| Plasma [59] |
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vautrot, V.; Bentayeb, H.; Causse, S.; Garrido, C.; Gobbo, J. Tumor-Derived Exosomes: Hidden Players in PD-1/PD-L1 Resistance. Cancers 2021, 13, 4537. https://doi.org/10.3390/cancers13184537
Vautrot V, Bentayeb H, Causse S, Garrido C, Gobbo J. Tumor-Derived Exosomes: Hidden Players in PD-1/PD-L1 Resistance. Cancers. 2021; 13(18):4537. https://doi.org/10.3390/cancers13184537
Chicago/Turabian StyleVautrot, Valentin, Hafidha Bentayeb, Sébastien Causse, Carmen Garrido, and Jessica Gobbo. 2021. "Tumor-Derived Exosomes: Hidden Players in PD-1/PD-L1 Resistance" Cancers 13, no. 18: 4537. https://doi.org/10.3390/cancers13184537






