Machine Learning Incorporating Host Factors for Predicting Survival in Head and Neck Squamous Cell Carcinoma Patients
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Cohort
2.2. Machine Learning Models
2.3. Performance Metrics
2.4. Modeling Strategy
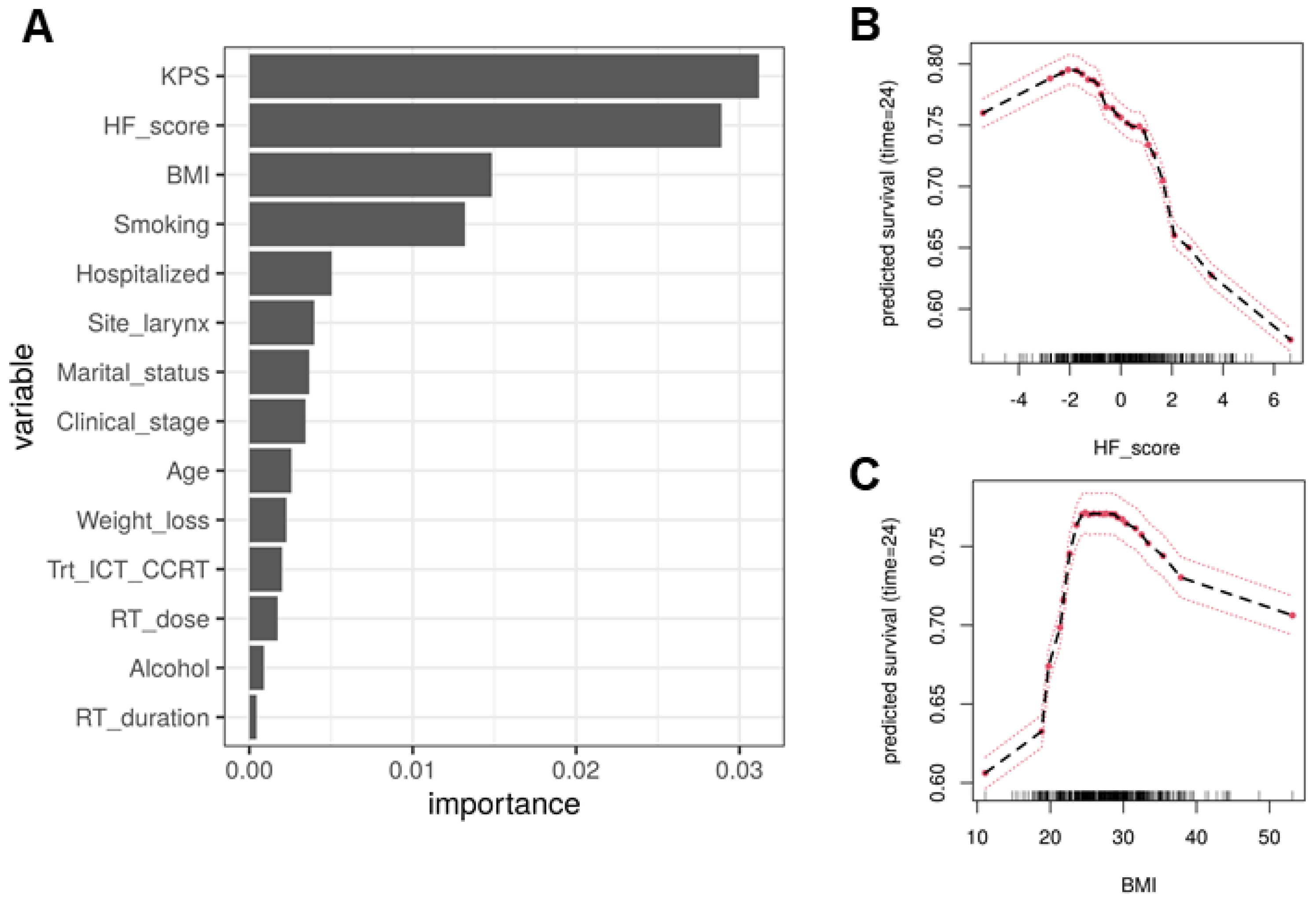
2.5. Model Interpretation
2.6. Statistical Analysis
3. Results
3.1. Patient Characteristics
3.2. Host Factors Organized as Clusters
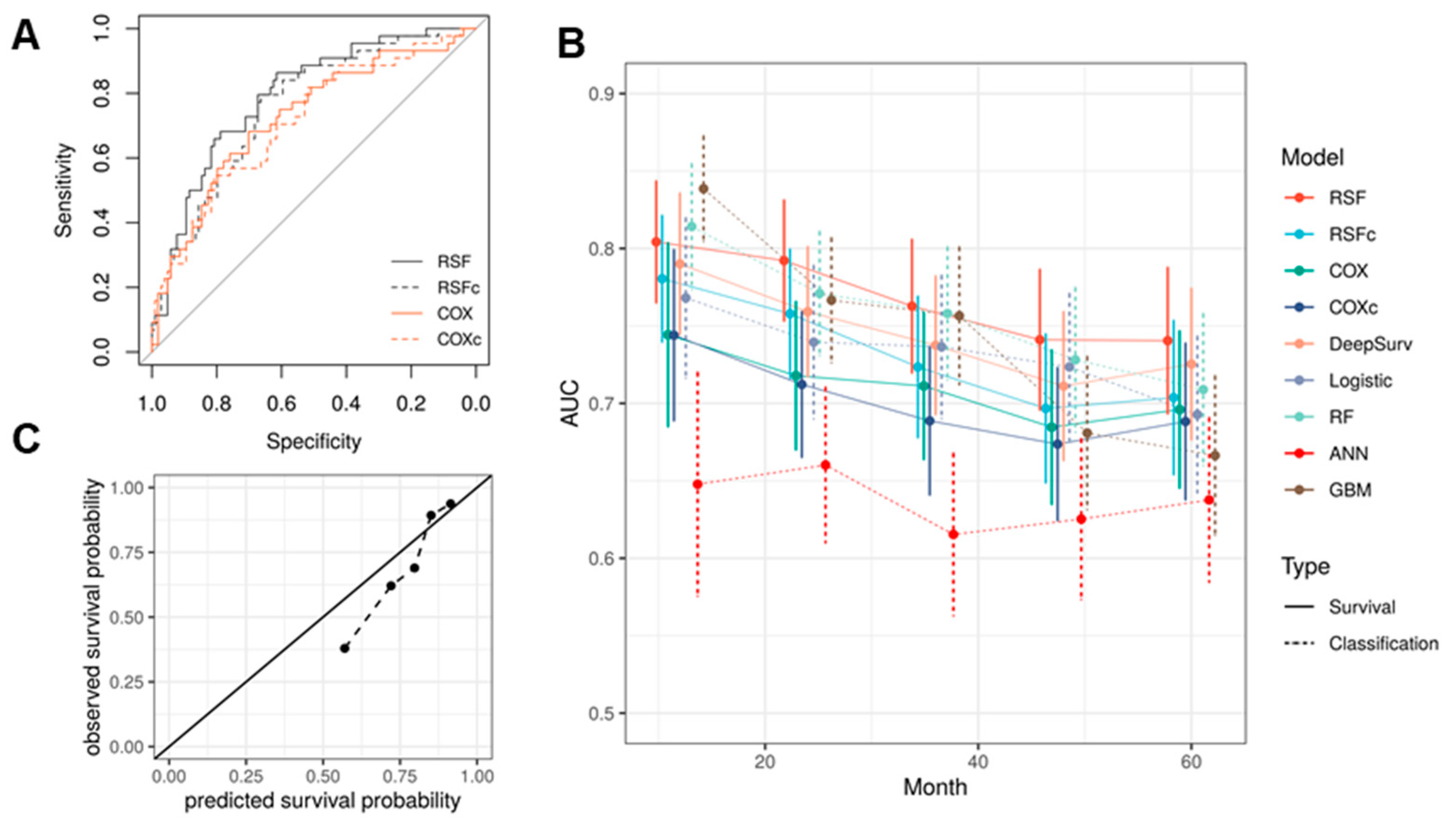
3.3. Model Selection and Evaluation
3.4. Patient Stratification by HF Score and RSF Predicted Risk
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chow, L.Q. Head and neck cancer. N. Engl. J. Med. 2020, 382, 60–72. [Google Scholar] [CrossRef]
- Bonner, J.A.; Harari, P.M.; Giralt, J.; Azarnia, N.; Shin, D.M.; Cohen, R.B.; Jones, C.U.; Sur, R.; Raben, D.; Jassem, J. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N. Engl. J.Med. 2006, 354, 567–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iocca, O.; Farcomeni, A.; Di Rocco, A.; Di Maio, P.; Golusinski, P.; López, S.P.; Savo, A.; Pellini, R.; Spriano, G. Locally advanced squamous cell carcinoma of the head and neck: A systematic review and Bayesian network meta-analysis of the currently available treatment options. Oral Oncol. 2018, 80, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Li, H.; Kaczmar, J.M.; Sharma, A.K.; Day, T.A.; Neskey, D.M.; Pipkorn, P.; Zenga, J.; Graboyes, E.M. Evaluating Adjuvant Therapy With Chemoradiation vs Radiation Alone for Patients With HPV-Negative N2a Head and Neck Cancer. JAMA Otolaryngol. Head Neck Surg. 2020, 146, 1109–1119. [Google Scholar] [CrossRef] [PubMed]
- Valero, C.; Zanoni, D.K.; Pillai, A.; Ganly, I.; Morris, L.G.; Shah, J.P.; Wong, R.J.; Patel, S.G. Host Factors Independently Associated With Prognosis in Patients With Oral Cavity Cancer. JAMA Otolaryngol. Head Neck Surg. 2020, 146, 699–707. [Google Scholar] [CrossRef]
- Schafer, U.; Micke, O.; Muller, S.B.; Schuller, P.; Willich, N. Hemoglobin as an independent prognostic factor in the radiotherapy of head and neck tumors. Strahlenther. Onkol. 2003, 179, 527–534. [Google Scholar] [CrossRef]
- McCloskey, S.A.; Jaggernauth, W.; Rigual, N.R.; Hicks, W.L., Jr.; Popat, S.R.; Sullivan, M.; Mashtare, T.L., Jr.; Khan, M.K.; Loree, T.R.; Singh, A.K. Radiation treatment interruptions greater than one week and low hemoglobin levels (12 g/dL) are predictors of local regional failure after definitive concurrent chemotherapy and intensity-modulated radiation therapy for squamous cell carcinoma of the head and neck. Am. J. Clin. Oncol. 2009, 32, 587–591. [Google Scholar] [CrossRef]
- Terris, D.J.; Dunphy, E.P. Oxygen tension measurements of head and neck cancers. Arch. Otolaryngol. Head Neck Surg. 1994, 120, 283–287. [Google Scholar] [CrossRef]
- Hoff, C.M. Importance of hemoglobin concentration and its modification for the outcome of head and neck cancer patients treated with radiotherapy. Acta. Oncol. 2012, 51, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wang, H.; Yan, A.; Wang, H.; Li, X.; Liu, J.; Li, W. Pretreatment neutrophil to lymphocyte ratio in determining the prognosis of head and neck cancer: A meta-analysis. BMC Cancer 2018, 18, 383. [Google Scholar] [CrossRef] [PubMed]
- Mascarella, M.A.; Mannard, E.; Silva, S.D.; Zeitouni, A. Neutrophil-to-lymphocyte ratio in head and neck cancer prognosis: A systematic review and meta-analysis. Head Neck 2018, 40, 1091–1100. [Google Scholar] [CrossRef]
- Cho, Y.; Kim, J.W.; Yoon, H.I.; Lee, C.G.; Keum, K.C.; Lee, I.J. The Prognostic Significance of Neutrophil-to-Lymphocyte Ratio in Head and Neck Cancer Patients Treated with Radiotherapy. J. Clin. Med. 2018, 7, 512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanahan, D.; Coussens, L.M. Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell 2012, 21, 309–322. [Google Scholar] [CrossRef] [Green Version]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Ishwaran, H.; Kogalur, U.B.; Blackstone, E.H.; Lauer, M.S. Random survival forests. Ann. Appl. Stat. 2008, 2, 841–860. [Google Scholar] [CrossRef]
- Katzman, J.L.; Shaham, U.; Cloninger, A.; Bates, J.; Jiang, T.; Kluger, Y. DeepSurv: Personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC Med. Res. Methodol. 2018, 18, 24. [Google Scholar] [CrossRef] [PubMed]
- Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B Stat. Methodol. 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Chen, T.; He, T.; Benesty, M.; Khotilovich, V.; Tang, Y.; Cho, H.; Chen, K.; Mithcell, R.; Cano, I.; Zhou, T.; et al. xgboost: Extreme Gradient Boosting. R package version 1.4.1.1. Available online: https://cran.r-project.org/package=xgboost (accessed on 1 May 2021).
- Günther, F.; Fritsch, S. Neuralnet: Training of neural networks. R J. 2010, 2, 30–38. [Google Scholar] [CrossRef] [Green Version]
- Ishwaran, H.; Kogalur, U.B. randomForestSRC: Fast Unified Random Forests for Survival, Regression and Classification (RF-SRC). R package version 2.12.0. Available online: https://cran.r-project.org/package=randomForestSRC (accessed on 9 July 2021).
- Hastie, T.; Qian, J. glmnet: Lasso and Elastic-Net Regularized Generalized Linear Models. R package version 4.1-2. Available online: https://cran.r-project.org/package=glmnet (accessed on 27 June 2021).
- Fotso, S. PySurvival: Open source package for survival analysis modeling. 2019. Available online: https://www.pysurvival.io/ (accessed on 1 June 2021).
- Howard, F.M.; Kochanny, S.; Koshy, M.; Spiotto, M.; Pearson, A.T. Machine Learning–Guided Adjuvant Treatment of Head and Neck Cancer. JAMA Netw. Open 2020, 3, e2025881. [Google Scholar] [CrossRef]
- Bice, N.; Kirby, N.; Bahr, T.; Rasmussen, K.; Saenz, D.; Wagner, T.; Papanikolaou, N.; Fakhreddine, M. Deep learning-based survival analysis for brain metastasis patients with the national cancer database. J. Appl. Clin. Med. Phys. 2020, 21, 187–192. [Google Scholar] [CrossRef]
- Chen, J.-B.; Yang, H.-S.; Moi, S.-H.; Chuang, L.-Y.; Yang, C.-H. Identification of mortality-risk-related missense variant for renal clear cell carcinoma using deep learning. Ther. Adv. Chronic Dis. 2021, 12, 2040622321992624. [Google Scholar] [CrossRef]
- Diamant, A.; Chatterjee, A.; Vallières, M.; Shenouda, G.; Seuntjens, J. Deep learning in head & neck cancer outcome prediction. Sci. Rep. 2019, 9, 1–10. [Google Scholar]
- Cozzi, L.; Franzese, C.; Fogliata, A.; Franceschini, D.; Navarria, P.; Tomatis, S.; Scorsetti, M. Predicting survival and local control after radiochemotherapy in locally advanced head and neck cancer by means of computed tomography based radiomics. Strahlenther. Onkol. 2019, 195, 805–818. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Lee, S.; Kwon, S.; Nam, W.; Cha, I.-H.; Kim, H.J. Deep learning-based survival prediction of oral cancer patients. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hung, M.; Park, J.; Hon, E.S.; Bounsanga, J.; Moazzami, S.; Ruiz-Negrón, B.; Wang, D. Artificial intelligence in dentistry: Harnessing big data to predict oral cancer survival. World J. Clin. Oncol. 2020, 11, 918–934. [Google Scholar] [CrossRef] [PubMed]
- Reddy, J.; Lindsay, W.; Berlind, C.; Ahern, C.; Holmes, A.; Smith, B.; Phan, J.; Frank, S.; Gunn, G.; Rosenthal, D. Applying a machine learning approach to predict acute radiation toxicities for head and neck cancer patients. Int. J. Radiat. Oncol. Biol. Phys. 2019, 105, S69. [Google Scholar] [CrossRef]


| Variable | Value | Frequency/Median | Percentage/IQR | Total |
|---|---|---|---|---|
| Age | 60.49 | (54.22, 66.86) | 591 | |
| BMI | 27.09 | (23.82, 30.24) | 573 | |
| Weight loss | 6 | (2.5, 9.8) | 565 | |
| Karnofsky Performance Status | 9 | (8,10) | 591 | |
| Dose of primary radiotherapy | 70 | (70, 70) | 587 | |
| Radiotherapy duration | 46 | (45, 46) | 591 | |
| Gender | Male | 483 | 81.7% | 591 |
| Female | 108 | 18.3% | ||
| Marital status | Married | 298 | 50.4% | 591 |
| Other | 293 | 49.6% | ||
| Anti-coagulants | No | 546 | 92.4% | 591 |
| Yes | 45 | 7.6% | ||
| NSAIDs | No | 322 | 54.5% | 591 |
| Yes | 269 | 45.5% | ||
| Alcohol consumption | Never | 105 | 18.6% | 566 |
| Former | 124 | 21.9% | ||
| Current | 337 | 59.5% | ||
| Smoking status | Never | 131 | 22.2% | 591 |
| Former | 301 | 50.9% | ||
| Current | 159 | 26.9% | ||
| Site | Oral cavity/lip | 67 | 11.3% | 591 |
| Oropharynx | 257 | 43.5% | ||
| Hypopharynx | 43 | 7.3% | ||
| Nasopharynx | 15 | 2.5% | ||
| Larynx | 142 | 24.0% | ||
| Salivary gland | 9 | 1.5% | ||
| Not specified | 52 | 8.8% | ||
| Other | 6 | 1% | ||
| Clinical stage | I | 18 | 3.1% | 575 |
| II | 46 | 8% | ||
| III | 454 | 79% | ||
| IV | 57 | 9.9% | ||
| Pathological grading | Well differentiated | 40 | 8.4% | |
| Moderately differentiated | 227 | 47.4% | ||
| Poorly differentiated | 204 | 42.6% | ||
| Undifferentiated | 8 | 1.7% | ||
| HPV | Negative | 131 | 39.3% | 333 |
| Positive | 202 | 60.7% | ||
| Treatment type | RT only | 33 | 5.6% | 591 |
| CCRT | 364 | 61.6% | ||
| Surgery + CCRT | 106 | 17.9% | ||
| Surgery + RT | 25 | 4.2% | ||
| CCRT + Neck Dissection | 7 | 1.2% | ||
| ICT + CCRT | 56 | 9.5% | ||
| Primary chemotherapy type | Other or no chemotherapy | 136 | 23% | 591 |
| Cisplatin | 455 | 77% | ||
| Radiotherapy delayed | No | 565 | 96.6% | 585 |
| Yes | 20 | 3.4% | ||
| Type of radiation | Definitive | 474 | 80.2% | 591 |
| Post-operative (adjuvant) | 117 | 19.8% | ||
| Laterality of radiation | Unilateral | 99 | 32.6% | 304 |
| Bilateral | 205 | 67.4% | ||
| Feeding tube type | No | 250 | 42.3% | 591 |
| Yes | 341 | 57.7% | ||
| Hospitalized | No | 463 | 78.5% | 590 |
| Yes | 127 | 21.5% |
| Variable | N | Median | IQR |
|---|---|---|---|
| WBC | 591 | 7.25 | (6.05, 9.13) |
| HGB | 591 | 13.5 | (12.1, 14.75) |
| HCT | 591 | 40.1 | (36.4, 43.3) |
| RBC | 591 | 4.44 | (3.96, 4.81) |
| MCV | 591 | 90.7 | (87.1, 93.9) |
| MCH | 590 | 30.7 | (29.5, 31.9) |
| MCHC | 591 | 33.8 | (33, 34.4) |
| Neutrophil (%) | 591 | 65.8 | (59.15, 72.4) |
| Lymphocyte (%) | 591 | 23.9 | (18.6, 30) |
| Monocyte (%) | 591 | 6.2 | (5.1, 7.5) |
| Eosinophil (%) | 591 | 2.5 | (1.5, 3.6) |
| Basophil (%) | 590 | 0.5 | (0.4, 0.7) |
| Model | Validation C-Index |
|---|---|
| RSFc | 0.707 (0.032) |
| RSF-1 | 0.721 (0.013) |
| RSF-2 | 0.717 (0.013) |
| RSF-3 | 0.717 (0.009) |
| RSF-ALL | 0.705 (0.015) |
| COXc | 0.671 (0.042) |
| COX-1 | 0.690 (0.024) |
| COX-2 | 0.690 (0.024) |
| COX-3 | 0.690 (0.024) |
| COX-ALL | 0.686 (0.021) |
| Model | C-Index | AUC | Specificity | Sensitivity | PPV | NPV |
|---|---|---|---|---|---|---|
| RSF | 0.729 (0.027) | 0.792 (0.039) | 0.615 | 0.864 | 0.487 | 0.914 |
| RSFc | 0.703 (0.029) | 0.758 (0.042) | 0.663 | 0.795 | 0.500 | 0.885 |
| COX | 0.679 (0.035) | 0.718 (0.048) | 0.817 | 0.568 | 0.568 | 0.817 |
| COXc | 0.636 (0.036) | 0.712 (0.047) | 0.808 | 0.545 | 0.545 | 0.808 |
| DeepSurv | 0.712 (0.029) | 0.759 (0.042) | 0.731 | 0.705 | 0.525 | 0.854 |
| RF | 0.719 (0.029) | 0.771 (0.040) | 0.625 | 0.818 | 0.480 | 0.89 |
| Logistic | 0.697 (0.035) | 0.740 (0.050) | 0.760 | 0.682 | 0.545 | 0.849 |
| ANN | 0.640 (0.037) | 0.660 (0.050) | 0.750 | 0.568 | 0.490 | 0.804 |
| GBM | 0.717 (0.030) | 0.767 (0.040) | 0.683 | 0.750 | 0.500 | 0.866 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, H.; Ma, S.J.; Farrugia, M.; Iovoli, A.J.; Wooten, K.E.; Gupta, V.; McSpadden, R.P.; Kuriakose, M.A.; Markiewicz, M.R.; Chan, J.M.; et al. Machine Learning Incorporating Host Factors for Predicting Survival in Head and Neck Squamous Cell Carcinoma Patients. Cancers 2021, 13, 4559. https://doi.org/10.3390/cancers13184559
Yu H, Ma SJ, Farrugia M, Iovoli AJ, Wooten KE, Gupta V, McSpadden RP, Kuriakose MA, Markiewicz MR, Chan JM, et al. Machine Learning Incorporating Host Factors for Predicting Survival in Head and Neck Squamous Cell Carcinoma Patients. Cancers. 2021; 13(18):4559. https://doi.org/10.3390/cancers13184559
Chicago/Turabian StyleYu, Han, Sung Jun Ma, Mark Farrugia, Austin J. Iovoli, Kimberly E. Wooten, Vishal Gupta, Ryan P. McSpadden, Moni A. Kuriakose, Michael R. Markiewicz, Jon M. Chan, and et al. 2021. "Machine Learning Incorporating Host Factors for Predicting Survival in Head and Neck Squamous Cell Carcinoma Patients" Cancers 13, no. 18: 4559. https://doi.org/10.3390/cancers13184559
APA StyleYu, H., Ma, S. J., Farrugia, M., Iovoli, A. J., Wooten, K. E., Gupta, V., McSpadden, R. P., Kuriakose, M. A., Markiewicz, M. R., Chan, J. M., Hicks, W. L., Jr., Platek, M. E., & Singh, A. K. (2021). Machine Learning Incorporating Host Factors for Predicting Survival in Head and Neck Squamous Cell Carcinoma Patients. Cancers, 13(18), 4559. https://doi.org/10.3390/cancers13184559






