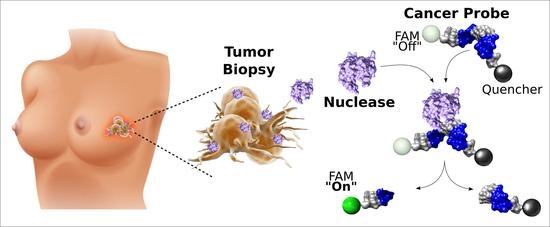
Discovery and Proof-of-Concept Study of Nuclease Activity as a Novel Biomarker for Breast Cancer Tumors
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
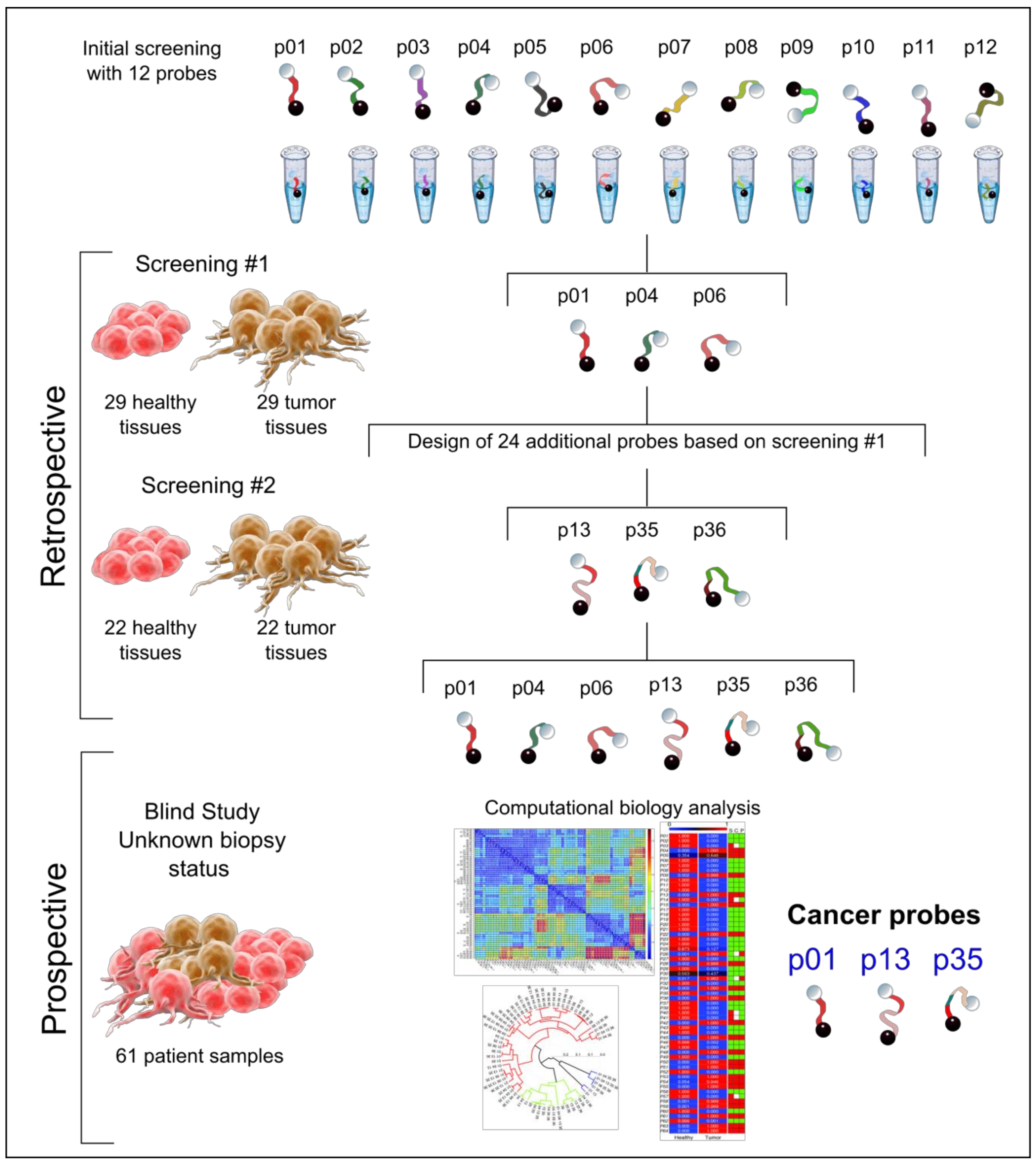
2.1. Retrospective Study
2.1.1. First-Step Retrospective Screening for Detecting the Blueprint of Nuclease Activity in Tumors
2.1.2. Second-Step Retrospective Screening, Using Tailored Probes for Targeting Tumor Nucleases
2.2. Prospective Study
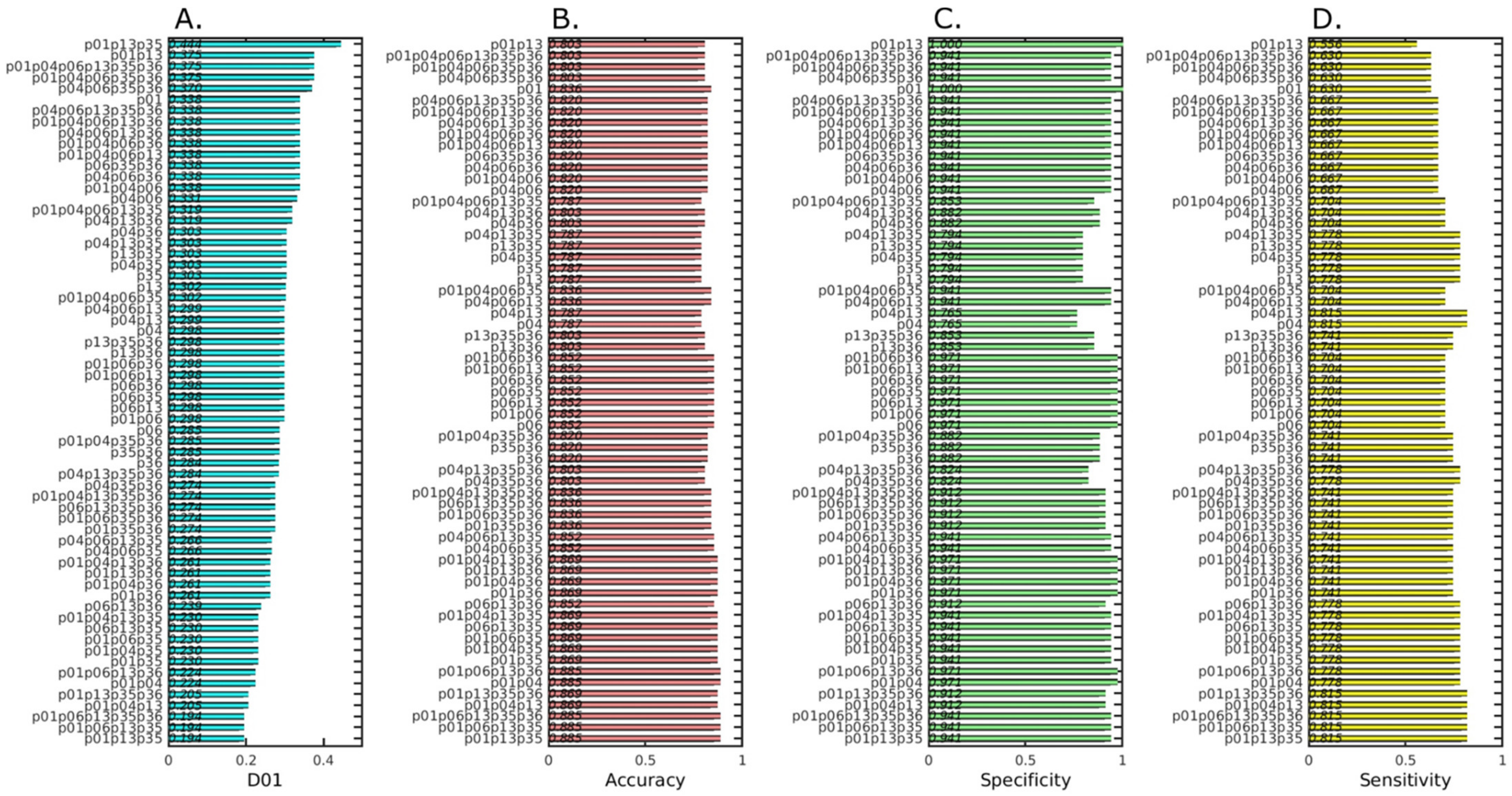
2.3. Pre-Analysis to Identify the Best Performing Probes for Tumor Diagnosis
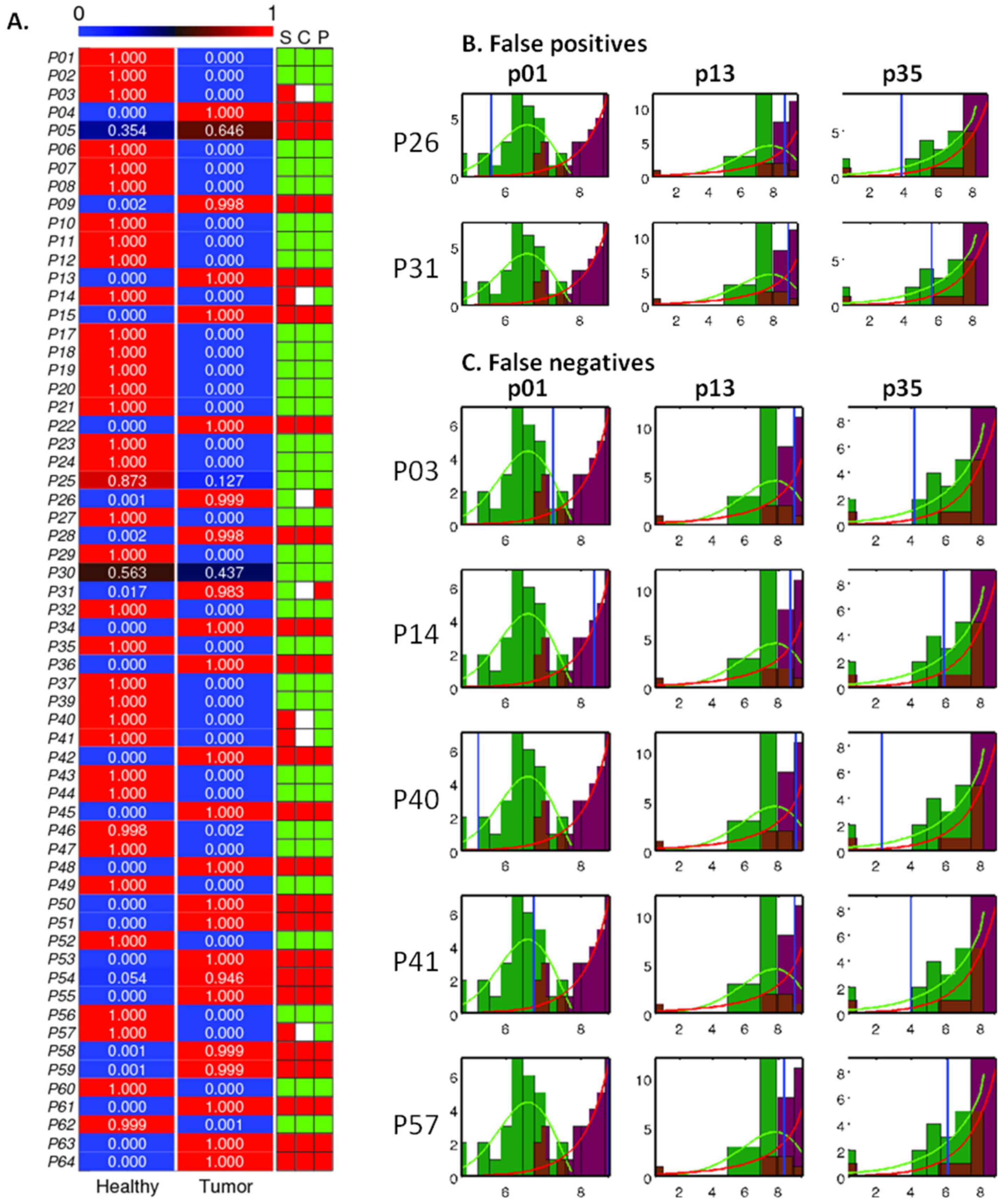
2.4. Analysis and Prediction of the Clinical Status of Each Prospective Sample Using the Three Cancer Probes
2.5. Serum Stability of the Three Cancer Probes
3. Discussion
4. Materials and Methods
4.1. Study Design
4.1.1. Retrospective Studies
Tissue Preparation for the Retrospective Study
4.1.2. Prospective Study
Tissue Preparation for the Prospective Study
4.2. Probe Library Design
4.3. Probes Synthesis
4.4. Nuclease Activity Assay
4.5. Computational Biology Analysis
4.6. Histopathological Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Colomer, R.; Aranda-López, I.; Albanell, J.; García-Caballero, T.; Ciruelos, E.; López-García, M.Á.; Cortés, J.; Rojo, F.; Martín, M.; Palacios-Calvo, J. Biomarkers in breast cancer: A consensus statement by the Spanish society of medical oncology and the Spanish society of pathology. Clin. Transl. Oncol. 2018, 20, 815–826. [Google Scholar] [CrossRef]
- Bertozzi, S.; Londero, A.P.; Seriau, L.; di Vora, R.; Cedolini, C.; Mariuzzi, L. Biomarkers in breast cancer. In Biomarker-Indicator of Abnormal Physiological Process; Begum, G., Ed.; IntechOpen: London, UK, 2018. [Google Scholar] [CrossRef]
- Eliyatkın, N.; Yalçın, E.; Zengel, B.; Aktaş, S.; Vardar, E. Molecular classification of breast carcinoma: From traditional, old-fashioned way to a new age, and a new way. J. Breast Health 2015, 11, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Turashvili, G.; Brogi, E. Tumor heterogeneity in breast cancer. Front. Med. 2017, 4, 227. [Google Scholar] [CrossRef]
- McDonald, E.S.; Clark, A.S.; Tchou, J.; Zhang, P.; Freedman, G.M. Clinical diagnosis and management of breast cancer. J. Nucl. Med. 2016, 57, 9S–16S. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Wei, B.; Zheng, Y.; Yin, Y.; Li, K.; Li, S. Breast cancer multi-classification from histopathological images with structured deep learning model. Sci. Rep. 2017, 7, 4172. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Dai, D.; Chen, B.; Tang, H.; Xie, X.; Wei, W. Clinicopathological and prognostic significance of cancer antigen 15-3 and carcinoembryonic antigen in breast cancer: A meta-analysis including 12,993 patients. Dis. Markers 2018, 2018, e9863092. [Google Scholar] [CrossRef] [PubMed]
- Fuzery, A.K.; Levin, J.; Chan, M.M.; Chan, D.W. Translation of proteomic biomarkers into FDA approved cancer diagnostics: Issues and challenges. Clin. Proteom. 2013, 10, e13. [Google Scholar] [CrossRef] [PubMed]
- Inagaki-Kawata, Y.; Yoshida, K.; Kawaguchi-Sakita, N.; Kawashima, M.; Nishimura, T.; Senda, N.; Shiozawa, Y.; Takeuchi, Y.; Inoue, Y.; Sato-Otsubo, A.; et al. Genetic and clinical landscape of breast cancers with germline BRCA1/2 variants. Commun. Biol. 2020, 3, e578. [Google Scholar] [CrossRef]
- Boral, D.; Vishnoi, M.; Liu, H.N.; Yin, W.; Sprouse, M.L.; Scamardo, A.; Hong, D.S.; Tan, T.Z.; Thiery, J.P.; Chang, J.C.; et al. Molecular characterization of breast cancer CTCs associated with brain metastasis. Nat. Commun. 2017, 8, e196. [Google Scholar] [CrossRef]
- Cardoso, F.; Costa, A.; Senkus, E.; Aapro, M.; Andre, F.; Barrios, C.H.; Bergh, J.; Bhattacharyya, G.; Biganzoli, L.; Cardoso, M.J.; et al. ESO-ESMO international consensus guidelines for Advanced Breast Cancer (ABC 3). Breast 2017, 31, 244–259. [Google Scholar] [CrossRef]
- Kazarian, A.; Blyuss, O.; Metodieva, G.; Gentry-Maharaj, A.; Ryan, A.; Kiseleva, E.M.; Prytomanova, O.M.; Jacobs, I.J.; Widschwendter, M.; Menon, U.; et al. Testing breast cancer serum biomarkers for early detection and prognosis in pre-diagnosis samples. Br. J. Cancer 2017, 116, 501–508. [Google Scholar] [CrossRef] [PubMed]
- Chau, C.H.; Rixe, O.; McLeod, H.; Figg, W.D. Validation of analytic methods for biomarkers used in drug development. Clin. Cancer Res. 2008, 14, 5967–5976. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; McGookey, M.; Wang, Y.; Cataland, S.R.; Wu, H.M. Effect of blood sampling, processing, and storage on the measurement of complement activation biomarkers. Am. J. Clin. Pathol. 2015, 143, 558–565. [Google Scholar] [CrossRef] [PubMed]
- Byrnes, S.A.; Weigl, B.H. Selecting analytical biomarkers for diagnostic applications: A first principles approach. Expert Rev. Mol. Diagn. 2018, 18, 19–26. [Google Scholar] [CrossRef]
- Sato, S.; Takenaka, S. Highly sensitive nuclease assays based on chemically modified DNA or RNA. Sensors 2014, 14, 12437–12450. [Google Scholar] [CrossRef]
- Li, L.Y.; Luo, X.; Wang, X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature 2001, 412, 95–99. [Google Scholar] [CrossRef]
- Wang, Y.-T.; Chiang, H.-H.; Huang, Y.-S.; Hsu, C.-L.; Yang, P.-J.; Juan, H.-F.; Yang, W.-S. A link between adipogenesis and innate immunity: RNase-L promotes 3T3-L1 adipogenesis by destabilizing Pref-1 mRNA. Cell Death Dis. 2016, 7, e2458. [Google Scholar] [CrossRef]
- Lamarche, B.J.; Orazio, N.I.; Weitzman, M.D. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 2010, 584, 3682–3695. [Google Scholar] [CrossRef]
- Wang, K.; Xie, C.; Chen, D. Flap endonuclease 1 is a promising candidate biomarker in gastric cancer and is involved in cell proliferation and apoptosis. Int. J. Mol. Med. 2014, 33, 1268–1274. [Google Scholar] [CrossRef]
- Yoo, D.G.; Song, Y.J.; Cho, E.J.; Lee, S.K.; Park, J.B.; Yu, J.H.; Lim, S.P.; Kim, J.M.; Jeon, B.H. Alteration of APE1/ref-1 expression in non-small cell lung cancer: The implications of impaired extracellular superoxide dismutase and catalase antioxidant systems. Lung Cancer 2008, 60, 277–284. [Google Scholar] [CrossRef]
- Abbotts, R.; Jewell, R.; Nsengimana, J.; Maloney, D.J.; Simeonov, A.; Seedhouse, C.; Elliott, F.; Laye, J.; Walker, C.; Jadhav, A.; et al. Targeting human apurinic/apyrimidinic endonuclease 1 (APE1) in phosphatase and tensin homolog (PTEN) deficient melanoma cells for personalized therapy. Oncotarget 2014, 5, 3273–3286. [Google Scholar] [CrossRef] [PubMed]
- Yoo, B.K.; Santhekadur, P.K.; Gredler, R.; Chen, D.; Emdad, L.; Bhutia, S.; Pannell, L.; Fisher, P.B.; Sarkar, D. Increased RNA-induced silencing complex (RISC) activity contributes to hepatocellular carcinoma. Hepatology 2011, 53, 1538–1548. [Google Scholar] [CrossRef] [PubMed]
- Kuruma, H.; Kamata, Y.; Takahashi, H.; Igarashi, K.; Kimura, T.; Miki, K.; Miki, J.; Sasaki, H.; Hayashi, N.; Egawa, S. Staphylococcal nuclease domain-containing protein 1 as a potential tissue marker for prostate cancer. Am. J. Pathol. 2009, 174, 2044–2050. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, N.; Ochiai, M.; Nakashima, K.; Ubagai, T.; Sugimura, T.; Nakagama, H. SND1, a component of RNA-induced silencing complex, is up-regulated in human colon cancers and implicated in early stage colon carcinogenesis. Cancer Res. 2007, 67, 9568–9576. [Google Scholar] [CrossRef]
- He, L.; Zhang, Y.; Sun, H.; Jiang, F.; Yang, H.; Wu, H.; Zhou, T.; Hu, S.; Kathera, C.S.; Wang, X.; et al. Targeting DNA flap endonuclease 1 to impede breast cancer progression. EBioMedicine 2016, 14, 32–43. [Google Scholar] [CrossRef]
- Doherty, R.; Madhusudan, S. DNA repair endonucleases: Physiological roles and potential as drug targets. J. Biomol. Screen. 2015, 20, 829–841. [Google Scholar] [CrossRef]
- Singh, P.; Yang, M.; Dai, H.; Yu, D.; Huang, Q.; Tan, W.; Kernstine, K.H.; Lin, D.; Shen, B. Overexpression and hypomethylation of flap endonuclease 1 gene in breast and other cancers. Mol. Cancer Res. 2008, 6, 1710–1717. [Google Scholar] [CrossRef]
- Xue, Y.; Marvin, M.E.; Ivanova, I.G.; Lydall, D.; Louis, E.J.; Maringele, L. Rif1 and Exo1 regulate the genomic instability following telomere losses. Aging Cell 2016, 15, 553–562. [Google Scholar] [CrossRef]
- Blanco, M.A.; Aleckovic, M.; Hua, Y.; Li, T.; Wei, Y.; Xu, Z.; Cristea, I.M.; Kang, Y. Identification of staphylococcal nuclease domain-containing 1 (SND1) as a Metadherin-interacting protein with metastasis-promoting functions. J. Biol. Chem. 2011, 286, 19982–19992. [Google Scholar] [CrossRef]
- Jariwala, N.; Rajasekaran, D.; Srivastava, J.; Gredler, R.; Akiel, M.A.; Robertson, C.L.; Emdad, L.; Fisher, P.B.; Sarkar, D. Role of the staphylococcal nuclease and tudor domain containing 1 in oncogenesis (review). Int. J. Oncol. 2015, 46, 465–473. [Google Scholar] [CrossRef]
- Yang, W. Nucleases: Diversity of structure, function and mechanism. Q. Rev. Biophys. 2011, 44, 1–93. [Google Scholar] [CrossRef] [PubMed]
- Kelley, M.R.; Cheng, L.; Foster, R.; Tritt, R.; Jiang, J.; Broshears, J.; Koch, M. Elevated and altered expression of the multifunctional DNA base excision repair and redox enzyme Ape1/ref-1 in prostate cancer. Clin. Cancer Res. 2001, 7, 824–830. [Google Scholar] [PubMed]
- Kumar, S.; Peng, X.; Daley, J.; Yang, L.; Shen, J.; Nguyen, N.; Bae, G.; Niu, H.; Peng, Y.; Hsieh, H.J.; et al. Inhibition of DNA2 nuclease as a therapeutic strategy targeting replication stress in cancer cells. Oncogenesis 2017, 6, e319. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Salas, E.; Peracaula, R.; Frazier, M.L.; de Llorens, R. Ribonucleases expressed by human pancreatic adenocarcinoma cell lines. Eur. J. Biochem. 2000, 267, 1484–1494. [Google Scholar] [CrossRef]
- Kottel, R.H.; Hoch, S.O.; Parsons, R.G.; Hoch, J.A. Serum ribonuclease activity in cancer patients. Br. J. Cancer 1978, 38, 280–286. [Google Scholar] [CrossRef]
- Basso, D.; Fabris, C.; Meani, A.; Del Favero, G.; Panucci, A.; Vianello, D.; Piccoli, A.; Naccarato, R. Serum deoxyribonuclease and ribonuclease in pancreatic cancer and chronic pancreatitis. Tumori J. 1985, 71, 529–532. [Google Scholar] [CrossRef]
- Weickmann, J.L.; Olson, E.M.; Glitz, D.G. Immunological assay of pancreatic ribonuclease in serum as an indicator of pancreatic cancer. Cancer Res. 1984, 44, 1682–1687. [Google Scholar]
- Huang, W.; Zhao, M.; Wei, N.; Wang, X.; Cao, H.; Du, Q.; Liang, Z. Site-specific RNase A activity was dramatically reduced in serum from multiple types of cancer patients. PLoS ONE 2014, 9, e96490. [Google Scholar] [CrossRef]
- Gallo, M.; Montserrat, J.M.; Iribarren, A.M. Design and applications of modified oligonucleotides. Braz. J. Med Biol. Res. 2003, 36, 143–151. [Google Scholar] [CrossRef]
- Balian, A.; Gonzalez, J.G.; Bastida, N.; Akhtar, K.K.; Borsa, B.A.; Hernandez, F.J. Kinetic screening of nuclease activity using nucleic acid probes. J. Vis. Exp. 2019. [Google Scholar] [CrossRef]
- Hernandez, L.I.; Ozalp, V.C.; Hernandez, F.J. Nuclease activity as a specific biomarker for breast cancer. Chem. Commun. 2016, 52, 12346–12349. [Google Scholar] [CrossRef] [PubMed]
- Kruspe, S.; Dickey, D.D.; Urak, K.T.; Blanco, G.N.; Miller, M.J.; Clark, K.C.; Burghardt, E.; Gutierrez, W.R.; Phadke, S.D.; Kamboj, S.; et al. Rapid and sensitive detection of breast cancer cells in patient blood with nuclease-activated probe technology. Mol. Ther.-Nucleic Acids 2017, 8, 542–557. [Google Scholar] [CrossRef]
- Garcia-Gutierrez, S.; Orive, M.; Sarasqueta, C.; Legarreta, M.J.; Gonzalez, N.; Redondo, M.; Rivero, A.; Serrano-Aguilar, P.; Castells, X.; Quintana, J.M.; et al. Health services research in patients with breast cancer (CAMISS-prospective): Study protocol for an observational prospective study. BMC Cancer 2018, 18, e54. [Google Scholar] [CrossRef] [PubMed]
- Perry, N.; Broeders, M.; de Wolf, C.; Tornberg, S.; Holland, R.; von Karsa, L. European guidelines for quality assurance in breast cancer screening and diagnosis. Fourth edition--summary document. Ann. Oncol. 2008, 19, 614–622. [Google Scholar] [CrossRef] [PubMed]
- Hulley, S.B.; Cummings, S.R.; Browner, W.S. Designing Clinical Research: An Epidemiologic Approach; Lippincott, Williams & Wilkins: Baltimore, MD, USA, 1988. [Google Scholar]
- Chow, S.C.; Wang, H.; Shao, J. Sample Size Calculations in Clinical Research; CRC Press: Boca Raton, FL, USA, 2003. [Google Scholar]
- Hernandez, F.J.; Huang, L.; Olson, M.E.; Powers, K.M.; Hernandez, L.I.; Meyerholz, D.K.; Thedens, D.R.; Behlke, M.A.; Horswill, A.R.; McNamara, J.O. Noninvasive imaging of Staphylococcus aureus infections with a nuclease-activated probe. Nat. Med. 2014, 20, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, F.J.; Stockdale, K.R.; Huang, L.; Horswill, A.R.; Behlke, M.A.; McNamara, J.O. Degradation of nuclease-stabilized RNA oligonucleotides in Mycoplasma-contaminated cell culture media. Nucleic Acid Ther. 2012, 22, 58–68. [Google Scholar] [CrossRef]
- Choi, J.S.; Kim, S.; Yoo, D.; Shin, T.H.; Kim, H.; Gomes, M.D.; Kim, S.H.; Pines, A.; Cheon, J. Distance-dependent magnetic resonance tuning as a versatile MRI sensing platform for biological targets. Nat. Mater. 2017, 16, 537–542. [Google Scholar] [CrossRef]
- Carril, M. Activatable probes for diagnosis and biomarker detection by MRI. J. Mater. Chem. B 2017, 5, 4332–4347. [Google Scholar] [CrossRef]
- Shin, T.H.; Kang, S.; Park, S.; Choi, J.S.; Kim, P.K.; Cheon, J. A magnetic resonance tuning sensor for the MRI detection of biological targets. Nat. Protoc. 2018, 13, 2664–2684. [Google Scholar] [CrossRef]
- Hincapié, F.J.H.; Hernandez, L.I. Agents for Use in the Detection of Nuclease Activity. U.S. Patent Application No. 15/520,828, 4 January 2018. [Google Scholar]

| Retrospective | Prospective | |
|---|---|---|
| n = 51 | n = 61 | |
| Age (years) | ||
| Mean | 62 | 56 |
| Median (range) | 63 (36–85) | 52 (40–77) |
| Clinical tumor size | ||
| <20 mm | 34 (66.7%) | |
| >20 mm and ≤50 mm | 12 (23.5%) | |
| > 50 mm | 5 (9.8%) | |
| Unknown | 0 | |
| Grading | ||
| G1 | 0 | 8(29.6%) |
| G2 | 12 (23.5%) | 14 (51.9%) |
| G3 | 32 (62.7%) | 2 (7.4%) |
| Unknonwn | 7 (13.7%) | 3 (11.1%) |
| ER/PR status | ||
| Both negative | 24 (47.1%) | 4 (14.8%) |
| One or both positives | 27 (52.9%) | 15 (55.6%) |
| Unknown | 0 | 8 (29.6%) |
| HER2 status | ||
| Negative | 40 (78.4%) | 13 (48.1%) |
| Positive | 11 (21.6%) | 6 (22.2%) |
| Unknown | 0 | 8 (29.6%) |
| Lymphovascular invasion | ||
| No | 46 (90.2%) | 15 (55.6%) |
| Yes | 5 (9.8%) | 7 (25.9%) |
| Unknown | 0 | 5 (18.5%) |
| Histological tumor type | ||
| Ductal invasive | 38 (74.5%) | 21 (34.4%) |
| Lobular invasive | 3 (5.9%) | 1 (1.6%) |
| Other | 7 (13.7%) | 5 (8.2%) |
| Unknown | 3 (5.9%) | 0 |
| Benign | NA | 34 (55.7%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernandez, L.I.; Araúzo-Bravo, M.J.; Gerovska, D.; Solaun, R.R.; Machado, I.; Balian, A.; Botero, J.; Jiménez, T.; Zuriarrain Bergara, O.; Larburu Gurruchaga, L.; et al. Discovery and Proof-of-Concept Study of Nuclease Activity as a Novel Biomarker for Breast Cancer Tumors. Cancers 2021, 13, 276. https://doi.org/10.3390/cancers13020276
Hernandez LI, Araúzo-Bravo MJ, Gerovska D, Solaun RR, Machado I, Balian A, Botero J, Jiménez T, Zuriarrain Bergara O, Larburu Gurruchaga L, et al. Discovery and Proof-of-Concept Study of Nuclease Activity as a Novel Biomarker for Breast Cancer Tumors. Cancers. 2021; 13(2):276. https://doi.org/10.3390/cancers13020276
Chicago/Turabian StyleHernandez, Luiza I., Marcos J. Araúzo-Bravo, Daniela Gerovska, Ricardo Rezola Solaun, Isabel Machado, Alien Balian, Juliana Botero, Tania Jiménez, Olaia Zuriarrain Bergara, Lide Larburu Gurruchaga, and et al. 2021. "Discovery and Proof-of-Concept Study of Nuclease Activity as a Novel Biomarker for Breast Cancer Tumors" Cancers 13, no. 2: 276. https://doi.org/10.3390/cancers13020276
APA StyleHernandez, L. I., Araúzo-Bravo, M. J., Gerovska, D., Solaun, R. R., Machado, I., Balian, A., Botero, J., Jiménez, T., Zuriarrain Bergara, O., Larburu Gurruchaga, L., Urruticoechea, A., & Hernandez, F. J. (2021). Discovery and Proof-of-Concept Study of Nuclease Activity as a Novel Biomarker for Breast Cancer Tumors. Cancers, 13(2), 276. https://doi.org/10.3390/cancers13020276