Evaluation of Angiogenesis-Related Genes as Prognostic Biomarkers of Bevacizumab Treated Ovarian Cancer Patients: Results from the Phase IV MITO16A/ManGO OV-2 Translational Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Micro Array (TMA) Building for IHC Analysis
2.2. RNA Extraction, Quality Controls and microRNA Amplification
2.3. Immunohistochemistry Analyses
2.4. Statistical Analyses
3. Results
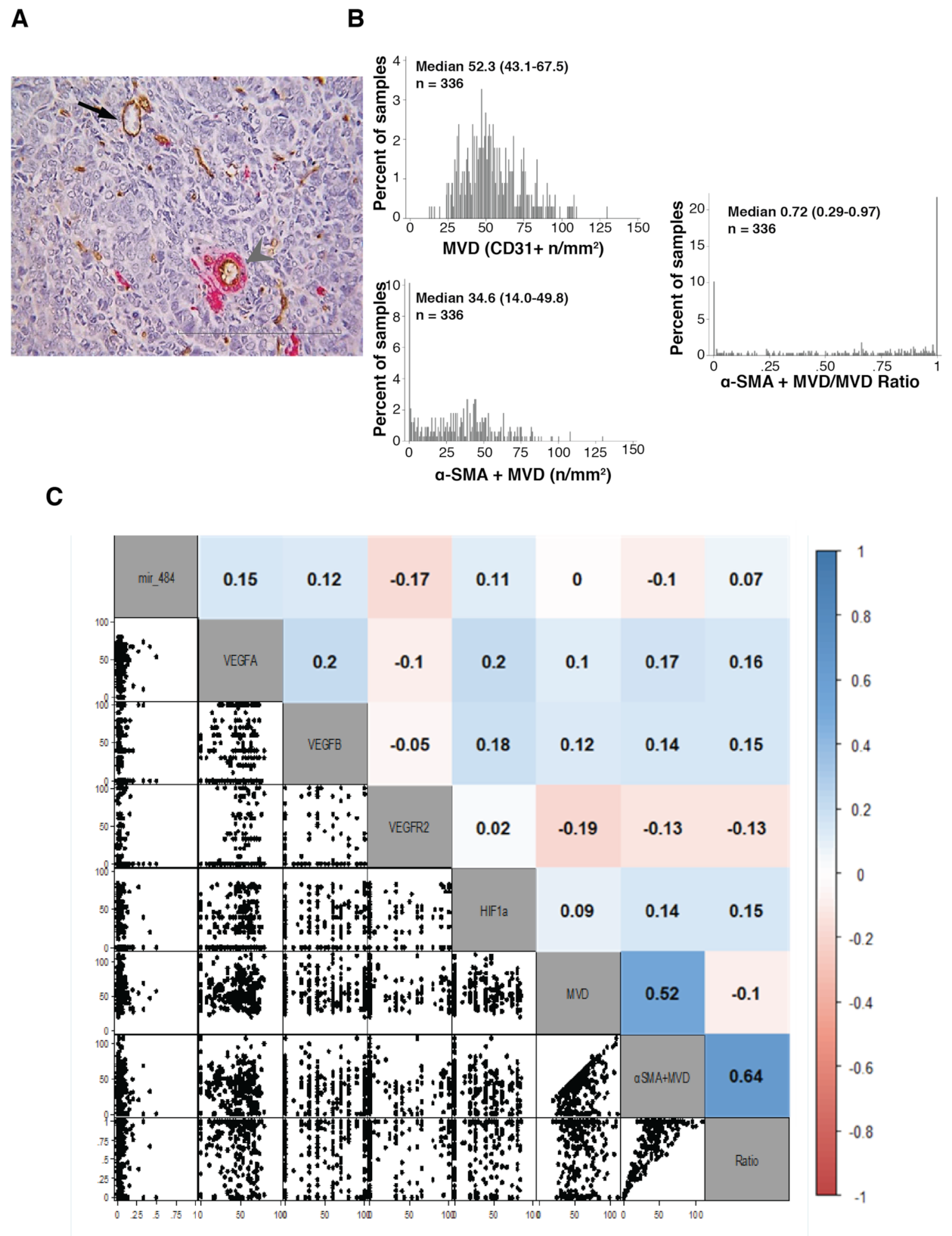
3.1. Immunohistochemistry Evaluation of Tumor Vasculature and VEGF Family Members
3.2. Evaluation of VEGF Family Member Regulators Expression
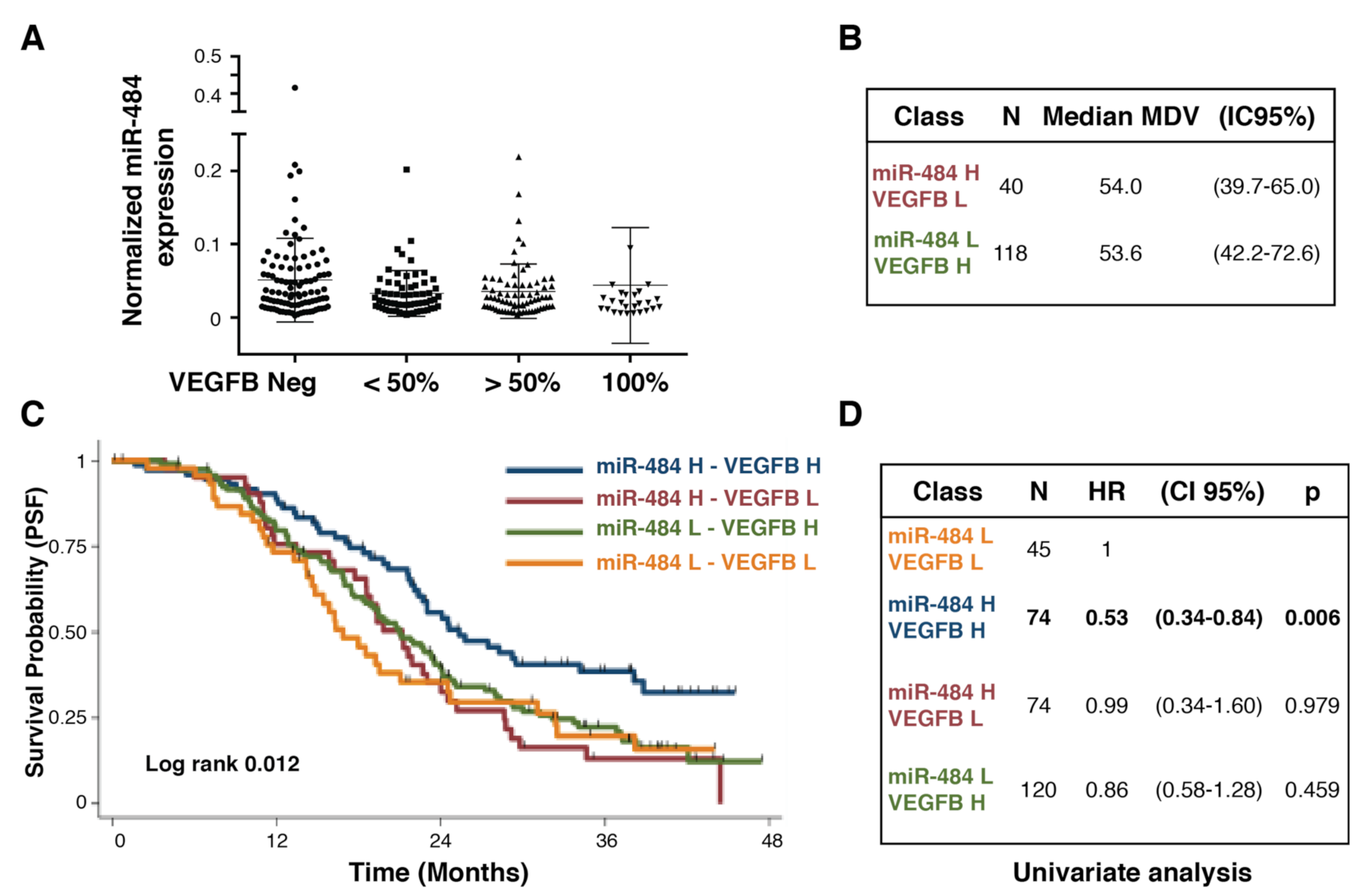
3.3. Coupling Biomarkers Expression with Clinical Variables and Patients’ Outcome
3.4. Testing the Potential of Combining Biomarkers to Predict Patients’ Outcome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jayson, G.C.; Kohn, E.C.; Kitchener, H.C.; Ledermann, J.A. Ovarian Cancer. Lancet 2014, 384, 1376–1388. [Google Scholar] [CrossRef]
- Lheureux, S.; Gourley, C.; Vergote, I.; Oza, A.M. Epithelial Ovarian Cancer. Lancet 2019, 393, 1240–1253. [Google Scholar] [CrossRef]
- Burger, R.A.; Brady, M.F.; Bookman, M.A.; Fleming, G.F.; Monk, B.J.; Huang, H.; Mannel, R.S.; Homesley, H.D.; Fowler, J.; Greer, B.E.; et al. Incorporation of Bevacizumab in the Primary Treatment of Ovarian Cancer. N. Engl. J. Med. 2011, 365, 2473–2483. [Google Scholar] [CrossRef] [PubMed]
- Perren, T.J.; Swart, A.M.; Pfisterer, J.; Ledermann, J.A.; Pujade-Lauraine, E.; Kristensen, G.; Carey, M.S.; Beale, P.; Cervantes, A.; Kurzeder, C.; et al. A Phase 3 Trial of Bevacizumab in Ovarian Cancer. N. Engl. J. Med. 2011, 365, 2484–2496. [Google Scholar] [CrossRef]
- Oza, A.M.; Cibula, D.; Benzaquen, A.O.; Poole, C.; Mathijssen, R.H.J.; Sonke, G.S.; Colombo, N.; Špaček, J.; Vuylsteke, P.; Hirte, H.; et al. Olaparib Combined with Chemotherapy for Recurrent Platinum-Sensitive Ovarian Cancer: A Randomised Phase 2 Trial. Lancet Oncol. 2015, 16, 87–97. [Google Scholar] [CrossRef]
- Mirza, M.R.; Monk, B.J.; Herrstedt, J.; Oza, A.M.; Mahner, S.; Redondo, A.; Fabbro, M.; Ledermann, J.A.; Lorusso, D.; Vergote, I.; et al. Niraparib Maintenance Therapy in Platinum-Sensitive, Recurrent Ovarian Cancer. N. Engl. J. Med. 2016, 375, 2154–2164. [Google Scholar] [CrossRef]
- Pujade-Lauraine, E.; Hilpert, F.; Weber, B.; Reuss, A.; Poveda, A.; Kristensen, G.; Sorio, R.; Vergote, I.; Witteveen, P.; Bamias, A.; et al. Bevacizumab Combined with Chemotherapy for Platinum-Resistant Recurrent Ovarian Cancer: The AURELIA Open-Label Randomized Phase III Trial. J. Clin. Oncol. 2014, 32, 1302–1308. [Google Scholar] [CrossRef]
- Pujade-Lauraine, E.; Ledermann, J.A.; Selle, F.; Gebski, V.; Penson, R.T.; Oza, A.M.; Korach, J.; Huzarski, T.; Poveda, A.; Pignata, S.; et al. Olaparib Tablets as Maintenance Therapy in Patients with Platinum-Sensitive, Relapsed Ovarian Cancer and a BRCA1/2 Mutation (SOLO2/ENGOT-Ov21): A Double-Blind, Randomised, Placebo-Controlled, Phase 3 Trial. Lancet Oncol. 2017, 18, 1274–1284. [Google Scholar] [CrossRef]
- Moore, K.; Colombo, N.; Scambia, G.; Kim, B.-G.; Oaknin, A.; Friedlander, M.; Lisyanskaya, A.; Floquet, A.; Leary, A.; Sonke, G.S.; et al. Maintenance Olaparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2018, 379, 2495–2505. [Google Scholar] [CrossRef]
- González-Martín, A.; Pothuri, B.; Vergote, I.; DePont Christensen, R.; Graybill, W.; Mirza, M.R.; McCormick, C.; Lorusso, D.; Hoskins, P.; Freyer, G.; et al. Niraparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2391–2402. [Google Scholar] [CrossRef]
- Ray-Coquard, I.; Pautier, P.; Pignata, S.; Pérol, D.; González-Martín, A.; Berger, R.; Fujiwara, K.; Vergote, I.; Colombo, N.; Mäenpää, J.; et al. Olaparib plus Bevacizumab as First-Line Maintenance in Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2416–2428. [Google Scholar] [CrossRef] [PubMed]
- Pignata, S.; Lorusso, D.; Joly, F.; Gallo, C.; Colombo, N.; Sessa, C.; Bamias, A.; Salutari, V.; Selle, F.; Frezzini, S.; et al. Carboplatin-Based Doublet plus Bevacizumab beyond Progression versus Carboplatin-Based Doublet Alone in Patients with Platinum-Sensitive Ovarian Cancer: A Randomised, Phase 3 Trial. Lancet Oncol. 2021, 22, 267–276. [Google Scholar] [CrossRef]
- Daniele, G.; Raspagliesi, F.; Scambia, G.; Pisano, C.; Colombo, N.; Frezzini, S.; Tognon, G.; Artioli, G.; Gadducci, A.; Lauria, R.; et al. Bevacizumab, Carboplatin, and Paclitaxel in the First Line Treatment of Advanced Ovarian Cancer Patients: The Phase IV MITO-16A/MaNGO-OV2A Study. Int. J. Gynecol. Cancer 2021, 31, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, N. Microvascular Density as a Predictive Biomarker for Bevacizumab Survival Benefit in Ovarian Cancer: Back to First Principles? JNCI J. Natl. Cancer Inst. 2017, 109. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Baluk, P.; Hashizume, H.; McDonald, D.M. Cellular Abnormalities of Blood Vessels as Targets in Cancer. Curr. Opin. Genet. Dev. 2005, 15, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Armulik, A.; Genové, G.; Betsholtz, C. Pericytes: Developmental, Physiological, and Pathological Perspectives, Problems, and Promises. Dev. Cell 2011, 21, 193–215. [Google Scholar] [CrossRef]
- Sowter, H.M.; Corps, A.N.; Evans, A.L.; Clark, D.E.; Charnock-Jones, D.S.; Smith, S.K. Expression and Localization of the Vascular Endothelial Growth Factor Family in Ovarian Epithelial Tumors. Lab. Investig. 1997, 77, 607–614. [Google Scholar] [PubMed]
- van der Bilt, A.R.M.; van der Zee, A.G.J.; de Vries, E.G.E.; de Jong, S.; Timmer-Bosscha, H.; ten Hoor, K.A.; den Dunnen, W.F.A.; Hollema, H.; Reyners, A.K.L. Multiple VEGF Family Members Are Simultaneously Expressed in Ovarian Cancer: A Proposed Model for Bevacizumab Resistance. Curr. Pharm. Des. 2012, 18, 3784–3792. [Google Scholar] [CrossRef]
- Karaman, S.; Leppänen, V.-M.; Alitalo, K. Vascular Endothelial Growth Factor Signaling in Development and Disease. Development 2018, 145, dev151019. [Google Scholar] [CrossRef]
- Vecchione, A.; Belletti, B.; Lovat, F.; Volinia, S.; Chiappetta, G.; Giglio, S.; Sonego, M.; Cirombella, R.; Onesti, E.C.; Pellegrini, P.; et al. A MicroRNA Signature Defines Chemoresistance in Ovarian Cancer through Modulation of Angiogenesis. Proc. Natl. Acad. Sci. USA. 2013, 110, 9845–9850. [Google Scholar] [CrossRef] [PubMed]
- Califano, D.; Russo, D.; Scognamiglio, G.; Losito, N.S.; Spina, A.; Bello, A.M.; Capiluongo, A.; Galdiero, F.; De Cecio, R.; Bevilacqua, S.; et al. Ovarian Cancer Translational Activity of the Multicenter Italian Trial in Ovarian Cancer (MITO) Group: Lessons Learned in 10 Years of Experience. Cells 2020, 9, 903. [Google Scholar] [CrossRef]
- Pimentel, R.S.; Niewiadomska-Bugaj, M.; Wang, J.-C. Association of Zero-Inflated Continuous Variables. Stat. Probab. Lett. 2015, 96, 61–67. [Google Scholar] [CrossRef]
- Holländer, N.; Sauerbrei, W.; Schumacher, M. Confidence Intervals for the Effect of a Prognostic Factor after Selection of an “optimal” Cutpoint. Stat. Med. 2004, 23, 1701–1713. [Google Scholar] [CrossRef]
- Pezzuto, A.; Carico, E. Role of HIF-1 in Cancer Progression: Novel Insights. A Review. Curr. Mol. Med. 2018, 18, 343–351. [Google Scholar] [CrossRef]
- Nakayama, K.; Kanzaki, A.; Hata, K.; Katabuchi, H.; Okamura, H.; Miyazaki, K.; Fukumoto, M.; Takebayashi, Y. Hypoxia-Inducible Factor 1 Alpha (HIF-1 Alpha) Gene Expression in Human Ovarian Carcinoma. Cancer Lett. 2002, 176, 215–223. [Google Scholar] [CrossRef]
- Tewari, K.S.; Burger, R.A.; Enserro, D.; Norquist, B.M.; Swisher, E.M.; Brady, M.F.; Bookman, M.A.; Fleming, G.F.; Huang, H.; Homesley, H.D.; et al. Final Overall Survival of a Randomized Trial of Bevacizumab for Primary Treatment of Ovarian Cancer. JCO J. Clin. Oncol. 2019, 37, 2317–2328. [Google Scholar] [CrossRef]
- Bais, C.; Mueller, B.; Brady, M.F.; Mannel, R.S.; Burger, R.A.; Wei, W.; Marien, K.M.; Kockx, M.M.; Husain, A.; Birrer, M.J.; et al. Tumor Microvessel Density as a Potential Predictive Marker for Bevacizumab Benefit: GOG-0218 Biomarker Analyses. JNCI J. Natl. Cancer Inst. 2017, 109. [Google Scholar] [CrossRef]
- Guyot, M.; Pagès, G. VEGF Splicing and the Role of VEGF Splice Variants: From Physiological-Pathological Conditions to Specific Pre-MRNA Splicing. Methods Mol. Biol. 2015, 1332, 3–23. [Google Scholar] [CrossRef]
- Pentheroudakis, G.; Mavroeidis, L.; Papadopoulou, K.; Koliou, G.-A.; Bamia, C.; Chatzopoulos, K.; Samantas, E.; Mauri, D.; Efstratiou, I.; Pectasides, D.; et al. Angiogenic and Antiangiogenic VEGFA Splice Variants in Colorectal Cancer: Prospective Retrospective Cohort Study in Patients Treated With Irinotecan-Based Chemotherapy and Bevacizumab. Clin. Colorectal Cancer 2019, 18, e370–e384. [Google Scholar] [CrossRef]
- Pellarin, I.; Belletti, B.; Baldassarre, G. RNA Splicing Alteration in the Response to Platinum Chemotherapy in Ovarian Cancer: A Possible Biomarker and Therapeutic Target. Med. Res. Rev. 2021, 41, 586–615. [Google Scholar] [CrossRef]
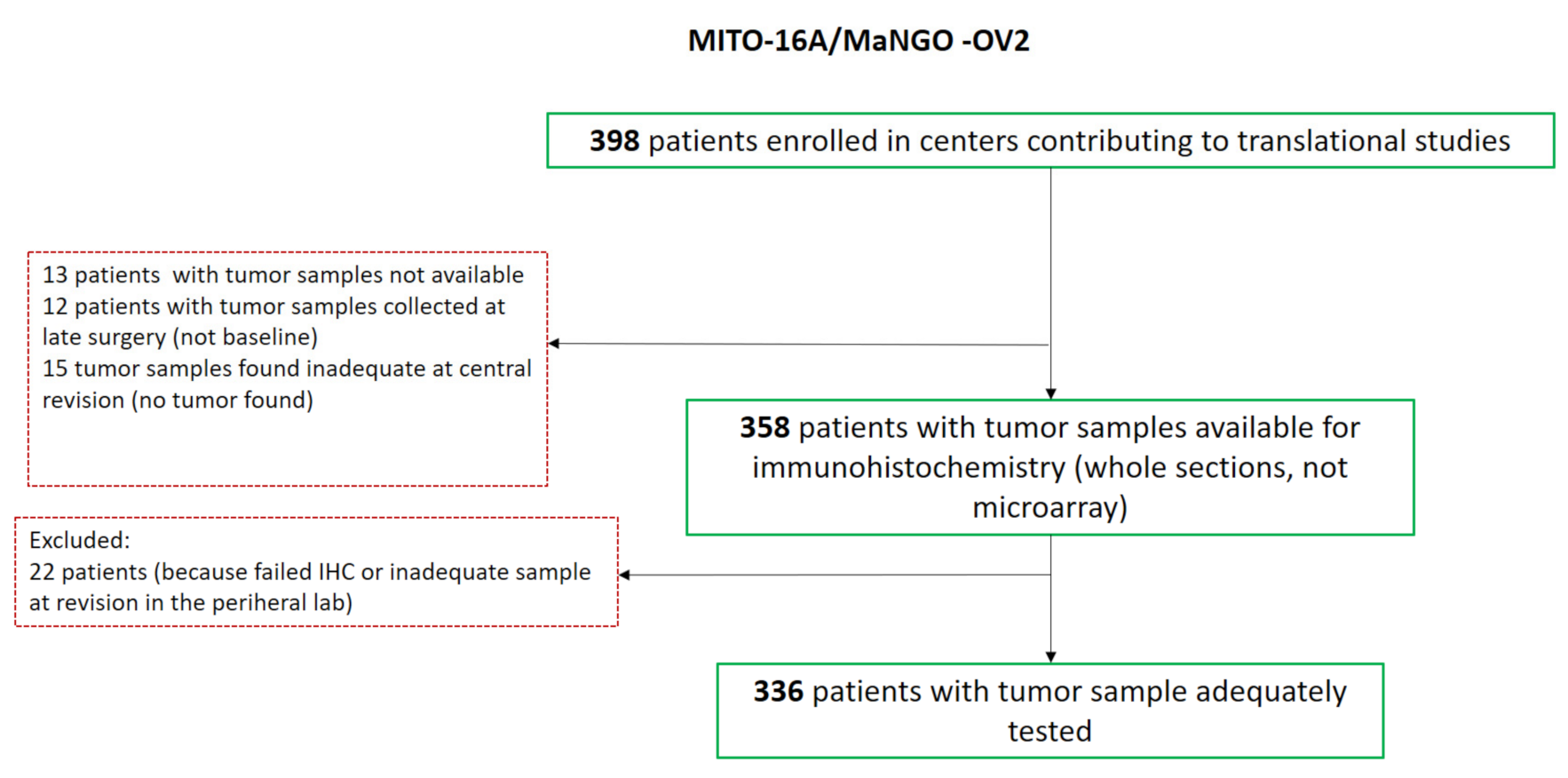
| Variables | Patients in Analysis (n = 336) | MITO16A Population (n = 398) | ||
|---|---|---|---|---|
| Age, median (IQR) | 59.3 | (50.0; 66.5) | 59.2 | (49.9; 66.5) |
| Age category, n(%) | ||||
| <65 | 234 | (69.6) | 278 | (70.0) |
| ≥65 | 102 | (30.4) | 120 | (30) |
| ECOG performance status, n(%) | ||||
| 0 | 265 | (78.9) | 315 | (79.2) |
| 1–2 | 71 | (22.1) | 69 | (17.3) |
| Missing | 14 | (3.5) | ||
| Residual disease, n(%) | ||||
| None | 129 | (38.4) | 153 | (38.4) |
| ≤1 cm | 68 | (20.2) | 72 | (18.1) |
| >1 cm/ | 102 | (30.4) | 120 | (30.2) |
| not operated | 37 | (11.0) | 53 | (13.3) |
| FIGO stage, n(%) | ||||
| IIIB | 31 | (9.2) | 36 | (9.1) |
| IIIC | 233 | (69.3) | 275 | (69.1) |
| IV | 72 | (21.4) | 87 | (21.9) |
| Tumor histology, n(%) | ||||
| High grade serous | 290 | (86.3) | 333 | (83.7) |
| Low grade serous | 12 | (3.6) | 13 | (3.3) |
| Endometrioid | 9 | (2.7) | 9 | (2.3) |
| Clear cell | 10 | (3.0) | 11 | (2.8) |
| Mucinous | 1 | (0.3) | 3 | (0.8) |
| Mixed | 2 | (0.6) | 4 | (1.0) |
| Other | 12 | (3.6) | 25 | (6.3) |
| Baseline hypertension, n(%) | ||||
| No hypertension | 56 | (16.6) | 70 | (17.6) |
| Prehypertension | 176 | (52.4) | 199 | (50.0) |
| On-AHT | 104 | (31.0) | 122 | (30.7) |
| Missing | 0 | - | 7 | (1.8) |
| PFS | ||||||
|---|---|---|---|---|---|---|
| Original Coefficients | Shrunken Coefficient | |||||
| HR | (95% CI) | p | HR | (95% CI) | p | |
| MVD: | ||||||
| >31.2 | 0.74 | (0.49–1.13) | 0.165 | 0.87 | (0.4–1.88) | 0.715 |
| SMA_MVD: | ||||||
| >64.1 | 0.80 | (0.53–1.22) | 0.295 | 0.98 | (0.44–2.19) | 0.962 |
| Ratio: | ||||||
| >0.86 | 0.89 | (0.68–1.17) | 0.404 | 1.05 | (0.42–2.61) | 0.915 |
| MIR 484: | ||||||
| >0.017 | 0.71 | (0.53–0.96) | 0.023 | 0.76 | (0.39–1.48) | 0.421 |
| VEGFA: | ||||||
| >56.7 | 1.32 | (0.99–1.75) | 0.056 | 1.22 | (0.63–2.38) | 0.557 |
| VEGFB: | ||||||
| >45.0 | 0.69 | (0.52–0.92) | 0.011 | 0.73 | (0.33–1.65) | 0.453 |
| VGFR2: | ||||||
| >60.0 | 1.09 | (0.75–1.60) | 0.642 | 0.72 | (0.17–2.99) | 0.651 |
| HIF1-α: | ||||||
| >0.0 | 1.06 | (0.79–1.40) | 0.709 | 0.71 | (0.23–2.19) | 0.557 |
| OS | ||||||
| HR | (95% CI) | p | HR | (95% CI) | p | |
| MVD: | ||||||
| >31.2 | 0.86 | (0.48–1.53) | 0.601 | 1.51 | (0.18–12.91) | 0.707 |
| SMA_MVD: | ||||||
| >64.1 | 0.74 | (0.37–1.49) | 0.406 | 1.14 | (0.16–8.37) | 0.897 |
| Ratio: | ||||||
| >0.86 | 0.86 | (0.57–1.30) | 0.484 | 1.16 | (0.25–5.5) | 0.848 |
| MIR 484: | ||||||
| >0.017 | 0.59 | (0.39–0.89) | 0.012 | 0.64 | (0.20–2.08) | 0.455 |
| VEGFA: | ||||||
| >56.7 | 0.85 | (0.55–1.31) | 0.451 | 1.14 | (0.13–10.07) | 0.909 |
| VEGFB: | ||||||
| >45.0 | 0.70 | (0.46–1.06) | 0.089 | 0.79 | (0.18–3.52) | 0.755 |
| VGFR2: | ||||||
| >60.0 | 1.19 | (0.69–2.03) | 0.531 | 0.77 | (0.12–4.88) | 0.779 |
| HIF1-α: | ||||||
| >0.0 | 1.24 | (0.81–1.90) | 0.332 | 0.99 | (0.14–7.11) | 0.990 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Califano, D.; Gallo, D.; Rampioni Vinciguerra, G.L.; De Cecio, R.; Arenare, L.; Signoriello, S.; Russo, D.; Ferrandina, G.; Citron, F.; Losito, N.S.; et al. Evaluation of Angiogenesis-Related Genes as Prognostic Biomarkers of Bevacizumab Treated Ovarian Cancer Patients: Results from the Phase IV MITO16A/ManGO OV-2 Translational Study. Cancers 2021, 13, 5152. https://doi.org/10.3390/cancers13205152
Califano D, Gallo D, Rampioni Vinciguerra GL, De Cecio R, Arenare L, Signoriello S, Russo D, Ferrandina G, Citron F, Losito NS, et al. Evaluation of Angiogenesis-Related Genes as Prognostic Biomarkers of Bevacizumab Treated Ovarian Cancer Patients: Results from the Phase IV MITO16A/ManGO OV-2 Translational Study. Cancers. 2021; 13(20):5152. https://doi.org/10.3390/cancers13205152
Chicago/Turabian StyleCalifano, Daniela, Daniela Gallo, Gian Luca Rampioni Vinciguerra, Rossella De Cecio, Laura Arenare, Simona Signoriello, Daniela Russo, Gabriella Ferrandina, Francesca Citron, Nunzia Simona Losito, and et al. 2021. "Evaluation of Angiogenesis-Related Genes as Prognostic Biomarkers of Bevacizumab Treated Ovarian Cancer Patients: Results from the Phase IV MITO16A/ManGO OV-2 Translational Study" Cancers 13, no. 20: 5152. https://doi.org/10.3390/cancers13205152
APA StyleCalifano, D., Gallo, D., Rampioni Vinciguerra, G. L., De Cecio, R., Arenare, L., Signoriello, S., Russo, D., Ferrandina, G., Citron, F., Losito, N. S., Gargiulo, P., Simeon, V., Scambia, G., Cecere, S. C., Montella, M., Colombo, N., Tognon, G., Bignotti, E., Zannoni, G. F., ... Baldassarre, G. (2021). Evaluation of Angiogenesis-Related Genes as Prognostic Biomarkers of Bevacizumab Treated Ovarian Cancer Patients: Results from the Phase IV MITO16A/ManGO OV-2 Translational Study. Cancers, 13(20), 5152. https://doi.org/10.3390/cancers13205152










