Tumor Heterogeneity and Consequences for Bladder Cancer Treatment
Abstract
Simple Summary
Abstract
1. Introduction
2. Implications of Patient-Patient Tumor Heterogeneity for Therapeutic Intervention
3. Intratumoral Heterogeneity Is Prevalent in Pretreated Primary Bladder Tumors
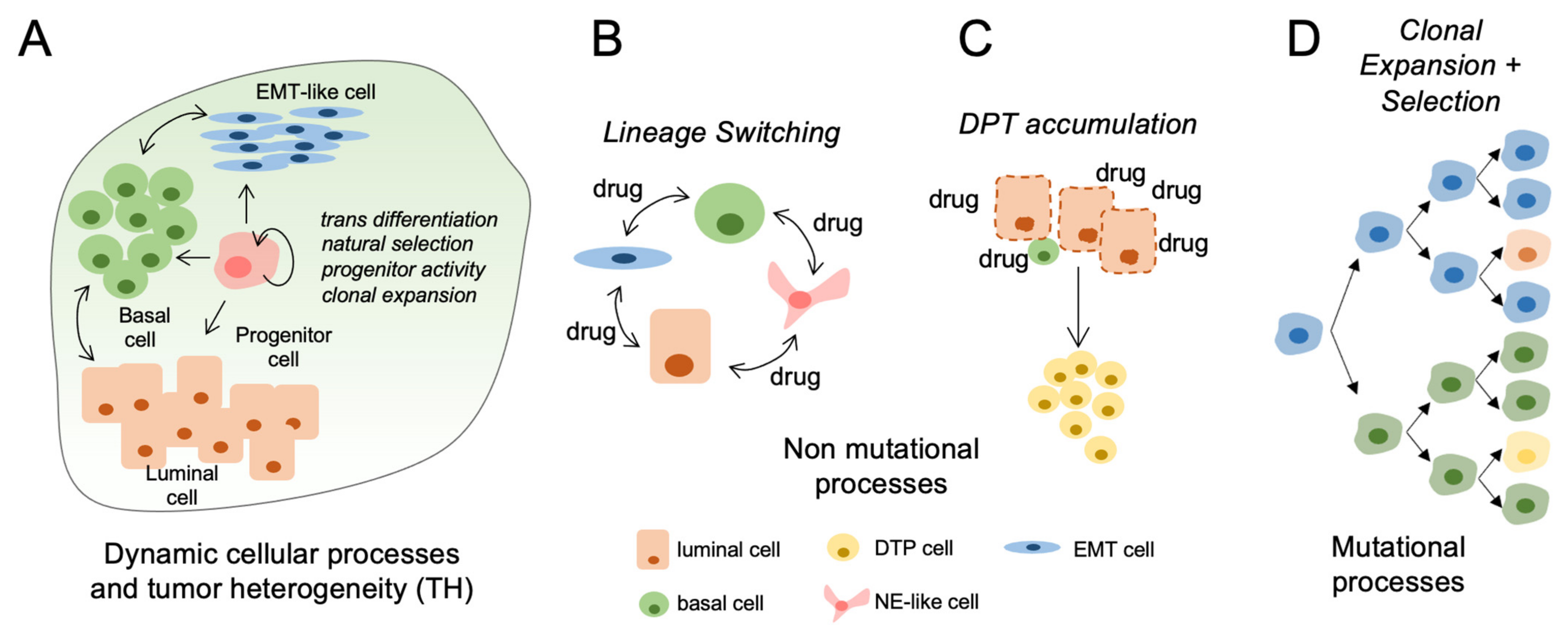
4. Mechanisms Regulating Tumor Heterogeneity
5. Genetic Heterogeneity in Bladder Tumor Epithelia
6. Mutational Heterogeneity and Treatment Response
7. Nonmutational Mechanisms of Tumor Heterogeneity
8. Evidence for Cellular Plasticity and Implications for Treatment Response in Bladder Tumors
9. Epithelial-Mesenchymal Transition (EMT)
10. Neuroendocrine-Like Phenotypes
11. Reversal of Cellular Plasticity as a Therapeutic Strategy
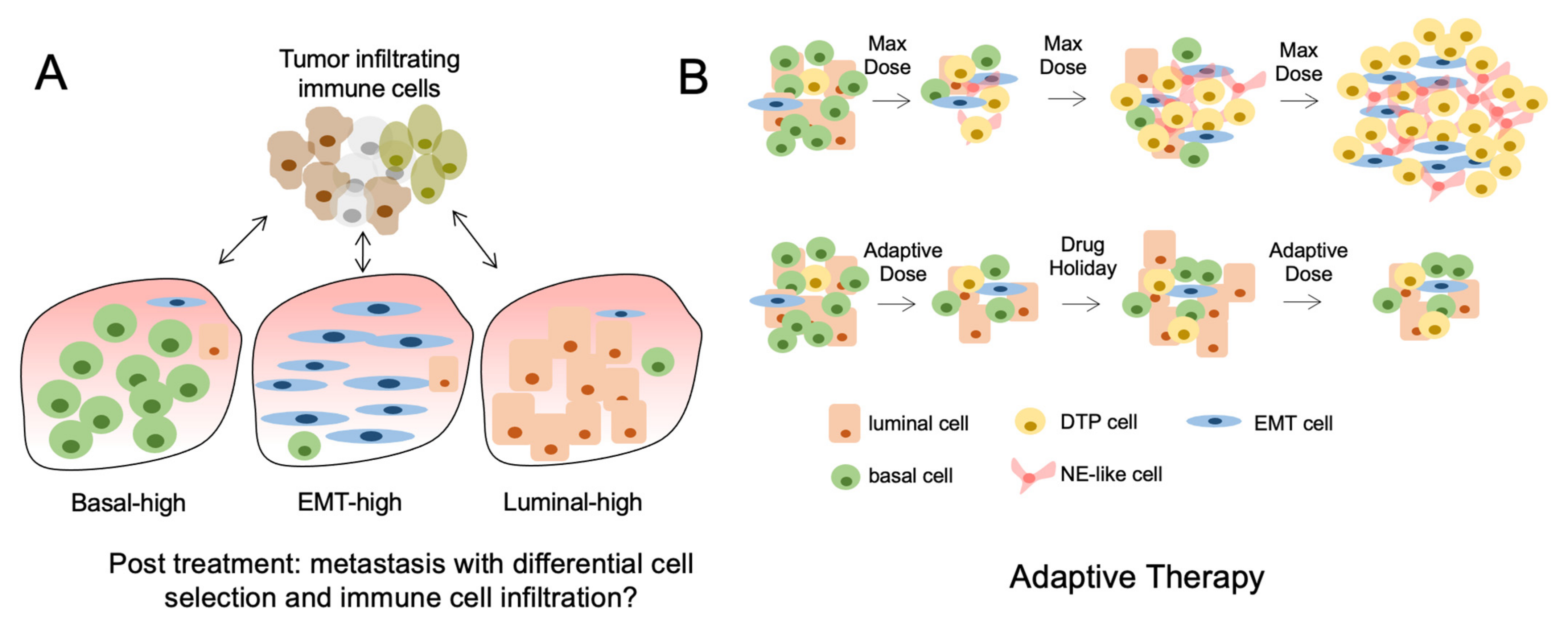
12. Drug-Tolerant Persisters and Post-Treatment Lineage Heterogeneity
13. Follow Up
14. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Marusyk, A.; Janiszewska, M.; Polyak, K. Intratumor Heterogeneity: The Rosetta Stone of Therapy Resistance. Cancer Cell 2020, 37, 471–484. [Google Scholar] [CrossRef]
- Marusyk, A.; Polyak, K. Tumor heterogeneity: Causes and consequences. Biochim. Biophys. Acta 2010, 1805, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Romero, I.; Garrido, C.; Algarra, I.; Chamorro, V.; Collado, A.; Garrido, F.; Garcia-Lora, A.M. MHC Intratumoral Heterogeneity May Predict Cancer Progression and Response to Immunotherapy. Front. Immunol. 2018, 9, 102. [Google Scholar] [CrossRef] [PubMed]
- Shue, Y.T.; Lim, J.S.; Sage, J. Tumor heterogeneity in small cell lung cancer defined and investigated in pre-clinical mouse models. Transl. Lung Cancer Res. 2018, 7, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Tammela, T.; Sage, J. Investigating Tumor Heterogeneity in Mouse Models. Annu. Rev. Cancer Biol. 2020, 4, 99–119. [Google Scholar] [CrossRef] [PubMed]
- Mroz, E.A.; Tward, A.D.; Pickering, C.R.; Myers, J.N.; Ferris, R.L.; Rocco, J.W. High intratumor genetic heterogeneity is related to worse outcome in patients with head and neck squamous cell carcinoma. Cancer 2013, 119, 3034–3042. [Google Scholar] [CrossRef]
- Morris, L.G.; Riaz, N.; Desrichard, A.; Senbabaoglu, Y.; Hakimi, A.A.; Makarov, V.; Reis-Filho, J.S.; Chan, T.A. Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival. Oncotarget 2016, 7, 10051–10063. [Google Scholar] [CrossRef]
- Choi, W.; Porten, S.; Kim, S.; Willis, D.; Plimack, E.R.; Hoffman-Censits, J.; Roth, B.; Cheng, T.; Tran, M.; Lee, I.L.; et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell 2014, 25, 152–165. [Google Scholar] [CrossRef]
- Hinohara, K.; Polyak, K. Intratumoral Heterogeneity: More Than Just Mutations. Trends Cell Biol. 2019, 29, 569–579. [Google Scholar] [CrossRef]
- Sharma, A.; Merritt, E.; Hu, X.; Cruz, A.; Jiang, C.; Sarkodie, H.; Zhou, Z.; Malhotra, J.; Riedlinger, G.M.; De, S. Non-Genetic Intra-Tumor Heterogeneity Is a Major Predictor of Phenotypic Heterogeneity and Ongoing Evolutionary Dynamics in Lung Tumors. Cell Rep. 2019, 29, 2164–2174 e2165. [Google Scholar] [CrossRef]
- Marusyk, A.; Almendro, V.; Polyak, K. Intra-tumour heterogeneity: A looking glass for cancer? Nat. Rev. Cancer 2012, 12, 323–334. [Google Scholar] [CrossRef]
- Merrell, A.J.; Stanger, B.Z. Adult cell plasticity in vivo: De-differentiation and transdifferentiation are back in style. Nat. Rev. Mol. Cell Biol. 2016, 17, 413–425. [Google Scholar] [CrossRef]
- Meacham, C.E.; Morrison, S.J. Tumour heterogeneity and cancer cell plasticity. Nature 2013, 501, 328–337. [Google Scholar] [CrossRef]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J.; et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017, 9, 34. [Google Scholar] [CrossRef]
- Robertson, A.G.; Kim, J.; Al-Ahmadie, H.; Bellmunt, J.; Guo, G.; Cherniack, A.D.; Hinoue, T.; Laird, P.W.; Hoadley, K.A.; Akbani, R.; et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2017, 171, 540–556.e25. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research, N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014, 507, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Marzouka, N.A.; Eriksson, P.; Rovira, C.; Liedberg, F.; Sjodahl, G.; Hoglund, M. A validation and extended description of the Lund taxonomy for urothelial carcinoma using the TCGA cohort. Sci. Rep. 2018, 8, 3737. [Google Scholar] [CrossRef] [PubMed]
- Damrauer, J.S.; Hoadley, K.A.; Chism, D.D.; Fan, C.; Tiganelli, C.J.; Wobker, S.E.; Yeh, J.J.; Milowsky, M.I.; Iyer, G.; Parker, J.S.; et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc. Natl. Acad. Sci. USA 2014, 111, 3110–3115. [Google Scholar] [CrossRef] [PubMed]
- Rebouissou, S.; Bernard-Pierrot, I.; de Reynies, A.; Lepage, M.L.; Krucker, C.; Chapeaublanc, E.; Herault, A.; Kamoun, A.; Caillault, A.; Letouze, E.; et al. EGFR as a potential therapeutic target for a subset of muscle-invasive bladder cancers presenting a basal-like phenotype. Sci. Transl. Med. 2014, 6, 244ra291. [Google Scholar] [CrossRef] [PubMed]
- Hurst, C.D.; Alder, O.; Platt, F.M.; Droop, A.; Stead, L.F.; Burns, J.E.; Burghel, G.J.; Jain, S.; Klimczak, L.J.; Lindsay, H.; et al. Genomic Subtypes of Non-invasive Bladder Cancer with Distinct Metabolic Profile and Female Gender Bias in KDM6A Mutation Frequency. Cancer Cell 2017, 32, 701–715.e7. [Google Scholar] [CrossRef]
- Kamoun, A.; de Reynies, A.; Allory, Y.; Sjodahl, G.; Robertson, A.G.; Seiler, R.; Hoadley, K.A.; Groeneveld, C.S.; Al-Ahmadie, H.; Choi, W.; et al. A Consensus Molecular Classification of Muscle-invasive Bladder Cancer. Eur. Urol. 2020, 77, 420–433. [Google Scholar] [CrossRef] [PubMed]
- Seiler, R.; Ashab, H.A.D.; Erho, N.; van Rhijn, B.W.G.; Winters, B.; Douglas, J.; Van Kessel, K.E.; Fransen van de Putte, E.E.; Sommerlad, M.; Wang, N.Q.; et al. Impact of Molecular Subtypes in Muscle-invasive Bladder Cancer on Predicting Response and Survival after Neoadjuvant Chemotherapy. Eur. Urol. 2017, 72, 544–554. [Google Scholar] [CrossRef]
- Necchi, A.; de Jong, J.J.; Raggi, D.; Briganti, A.; Marandino, L.; Gallina, A.; Bandini, M.; Dabbas, B.; Davicioni, E.; Capitanio, U.; et al. Molecular Characterization of Residual Bladder Cancer after Neoadjuvant Pembrolizumab. Eur. Urol. 2021, 80, 149–159. [Google Scholar] [CrossRef]
- Seiler, R.; Gibb, E.A.; Wang, N.Q.; Oo, H.Z.; Lam, H.M.; van Kessel, K.E.; Voskuilen, C.S.; Winters, B.; Erho, N.; Takhar, M.M.; et al. Divergent Biological Response to Neoadjuvant Chemotherapy in Muscle-invasive Bladder Cancer. Clin. Cancer Res. 2019, 25, 5082–5093. [Google Scholar] [CrossRef] [PubMed]
- Faltas, B.M.; Prandi, D.; Tagawa, S.T.; Molina, A.M.; Nanus, D.M.; Sternberg, C.; Rosenberg, J.; Mosquera, J.M.; Robinson, B.; Elemento, O.; et al. Clonal evolution of chemotherapy-resistant urothelial carcinoma. Nat. Genet. 2016, 48, 1490–1499. [Google Scholar] [CrossRef] [PubMed]
- Minoli, M.; Kiener, M.; Thalmann, G.N.; Kruithof-de Julio, M.; Seiler, R. Evolution of Urothelial Bladder Cancer in the Context of Molecular Classifications. Int. J. Mol. Sci. 2020, 21, 5670. [Google Scholar] [CrossRef]
- Warrick, J.I.; Sjodahl, G.; Kaag, M.; Raman, J.D.; Merrill, S.; Shuman, L.; Chen, G.; Walter, V.; DeGraff, D.J. Intratumoral Heterogeneity of Bladder Cancer by Molecular Subtypes and Histologic Variants. Eur. Urol. 2019, 75, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.B.; Fu, L.W. Application of single-cell technology in cancer research. Biotechnol. Adv. 2017, 35, 443–449. [Google Scholar] [CrossRef]
- Lei, Y.; Tang, R.; Xu, J.; Wang, W.; Zhang, B.; Liu, J.; Yu, X.; Shi, S. Applications of single-cell sequencing in cancer research: Progress and perspectives. J. Hematol. Oncol. 2021, 14, 91. [Google Scholar] [CrossRef]
- Landau, D.A.; Carter, S.L.; Stojanov, P.; McKenna, A.; Stevenson, K.; Lawrence, M.S.; Sougnez, C.; Stewart, C.; Sivachenko, A.; Wang, L.; et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 2013, 152, 714–726. [Google Scholar] [CrossRef]
- Zhang, J.; Fujimoto, J.; Zhang, J.; Wedge, D.C.; Song, X.; Zhang, J.; Seth, S.; Chow, C.W.; Cao, Y.; Gumbs, C.; et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 2014, 346, 256–259. [Google Scholar] [CrossRef]
- Ramon, Y.C.S.; Sese, M.; Capdevila, C.; Aasen, T.; De Mattos-Arruda, L.; Diaz-Cano, S.J.; Hernandez-Losa, J.; Castellvi, J. Clinical implications of intratumor heterogeneity: Challenges and opportunities. J. Mol. Med. 2020, 98, 161–177. [Google Scholar] [CrossRef]
- Sfakianos, J.P.; Daza, J.; Hu, Y.; Anastos, H.; Bryant, G.; Bareja, R.; Badani, K.K.; Galsky, M.D.; Elemento, O.; Faltas, B.M.; et al. Epithelial plasticity can generate multi-lineage phenotypes in human and murine bladder cancers. Nat. Commun. 2020, 11, 2540. [Google Scholar] [CrossRef]
- Sirab, N.; Drubay, D.; Maille, P.; Popova, T.; Ngo, C.; Gentien, D.; Moktefi, A.; Soyeux-Porte, P.; Pelletier, R.; Reyes, C.; et al. Multilayer spectrum of intratumoral heterogeneity in basal bladder cancer. J. Pathol. 2021. [Google Scholar] [CrossRef]
- Bell, C.C.; Gilan, O. Principles and mechanisms of non-genetic resistance in cancer. Br. J. Cancer 2020, 122, 465–472. [Google Scholar] [CrossRef]
- Cha, E.K.; Sfakianos, J.P.; Al-Ahmadie, H.; Scott, S.N.; Kim, P.H.; Iyer, G.; Bajorin, D.F.; Rosenberg, J.E.; Berger, M.F.; Bochner, B.H.; et al. Branched evolution and intratumor heterogeneity of urothelial carcinoma of the bladder. J. Clin. Oncol. 2014, 32, 293. [Google Scholar] [CrossRef]
- Meeks, J.J.; Al-Ahmadie, H.; Faltas, B.M.; Taylor, J.A., 3rd; Flaig, T.W.; DeGraff, D.J.; Christensen, E.; Woolbright, B.L.; McConkey, D.J.; Dyrskjot, L. Genomic heterogeneity in bladder cancer: Challenges and possible solutions to improve outcomes. Nat. Rev. Urol. 2020, 17, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Fujita, K.; Hayashi, Y.; Hatano, K.; Kawashima, A.; McConkey, D.J.; Nonomura, N. Mutational Landscape and Environmental Effects in Bladder Cancer. Int. J. Mol. Sci. 2020, 21, 6072. [Google Scholar] [CrossRef] [PubMed]
- da Costa, J.B.; Gibb, E.A.; Nykopp, T.K.; Mannas, M.; Wyatt, A.W.; Black, P.C. Molecular tumor heterogeneity in muscle invasive bladder cancer: Biomarkers, subtypes, and implications for therapy. Urol. Oncol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Pouessel, D.; Neuzillet, Y.; Mertens, L.S.; van der Heijden, M.S.; de Jong, J.; Sanders, J.; Peters, D.; Leroy, K.; Manceau, A.; Maille, P.; et al. Tumor heterogeneity of fibroblast growth factor receptor 3 (FGFR3) mutations in invasive bladder cancer: Implications for perioperative anti-FGFR3 treatment. Ann. Oncol. 2016, 27, 1311–1316. [Google Scholar] [CrossRef]
- Zhang, L.; Vijg, J. Somatic Mutagenesis in Mammals and Its Implications for Human Disease and Aging. Annu. Rev. Genet. 2018, 52, 397–419. [Google Scholar] [CrossRef]
- Roberts, S.A.; Lawrence, M.S.; Klimczak, L.J.; Grimm, S.A.; Fargo, D.; Stojanov, P.; Kiezun, A.; Kryukov, G.V.; Carter, S.L.; Saksena, G.; et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 2013, 45, 970–976. [Google Scholar] [CrossRef]
- Hedegaard, J.; Lamy, P.; Nordentoft, I.; Algaba, F.; Hoyer, S.; Ulhoi, B.P.; Vang, S.; Reinert, T.; Hermann, G.G.; Mogensen, K.; et al. Comprehensive Transcriptional Analysis of Early-Stage Urothelial Carcinoma. Cancer Cell 2016, 30, 27–42. [Google Scholar] [CrossRef]
- Nordentoft, I.; Lamy, P.; Birkenkamp-Demtroder, K.; Shumansky, K.; Vang, S.; Hornshoj, H.; Juul, M.; Villesen, P.; Hedegaard, J.; Roth, A.; et al. Mutational context and diverse clonal development in early and late bladder cancer. Cell Rep. 2014, 7, 1649–1663. [Google Scholar] [CrossRef] [PubMed]
- Glaser, A.P.; Fantini, D.; Wang, Y.; Yu, Y.; Rimar, K.J.; Podojil, J.R.; Miller, S.D.; Meeks, J.J. APOBEC-mediated mutagenesis in urothelial carcinoma is associated with improved survival, mutations in DNA damage response genes, and immune response. Oncotarget 2018, 9, 4537–4548. [Google Scholar] [CrossRef] [PubMed]
- Mullane, S.A.; Werner, L.; Rosenberg, J.; Signoretti, S.; Callea, M.; Choueiri, T.K.; Freeman, G.J.; Bellmunt, J. Correlation of Apobec Mrna Expression with overall Survival and pd-l1 Expression in Urothelial Carcinoma. Sci. Rep. 2016, 6, 27702. [Google Scholar] [CrossRef]
- Vlachostergios, P.J.; Faltas, B.M. Treatment resistance in urothelial carcinoma: An evolutionary perspective. Nat. Rev. Clin. Oncol. 2018, 15, 495–509. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Shirakawa, K.; Yokoyama, M.; Fukuda, H.; Sarca, A.D.; Koyabu, S.; Yamazaki, H.; Kazuma, Y.; Matsui, H.; Maruyama, W.; et al. Protein kinase A inhibits tumor mutator APOBEC3B through phosphorylation. Sci. Rep. 2019, 9, 8307. [Google Scholar] [CrossRef]
- Kvach, M.V.; Barzak, F.M.; Harjes, S.; Schares, H.A.M.; Jameson, G.B.; Ayoub, A.M.; Moorthy, R.; Aihara, H.; Harris, R.S.; Filichev, V.V.; et al. Inhibiting APOBEC3 Activity with Single-Stranded DNA Containing 2’-Deoxyzebularine Analogues. Biochemistry 2019, 58, 391–400. [Google Scholar] [CrossRef]
- Feng, W.; Wang, Y.; Chen, S.; Zhu, X. Intra-tumoral heterogeneity and immune responses predicts prognosis of gastric cancer. Aging 2020, 12, 24333–24344. [Google Scholar] [CrossRef]
- Lamy, P.; Nordentoft, I.; Birkenkamp-Demtroder, K.; Thomsen, M.B.; Villesen, P.; Vang, S.; Hedegaard, J.; Borre, M.; Jensen, J.B.; Hoyer, S.; et al. Paired Exome Analysis Reveals Clonal Evolution and Potential Therapeutic Targets in Urothelial Carcinoma. Cancer Res. 2016, 76, 5894–5906. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Gao, R.; Sei, E.; Brandt, R.; Hartman, J.; Hatschek, T.; Crosetto, N.; Foukakis, T.; Navin, N.E. Chemoresistance Evolution in Triple-Negative Breast Cancer Delineated by Single-Cell Sequencing. Cell 2018, 173, 879–893.e13. [Google Scholar] [CrossRef]
- Brady, S.W.; McQuerry, J.A.; Qiao, Y.; Piccolo, S.R.; Shrestha, G.; Jenkins, D.F.; Layer, R.M.; Pedersen, B.S.; Miller, R.H.; Esch, A.; et al. Combating subclonal evolution of resistant cancer phenotypes. Nat. Commun. 2017, 8, 1231. [Google Scholar] [CrossRef] [PubMed]
- Bhang, H.E.; Ruddy, D.A.; Krishnamurthy Radhakrishna, V.; Caushi, J.X.; Zhao, R.; Hims, M.M.; Singh, A.P.; Kao, I.; Rakiec, D.; Shaw, P.; et al. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat. Med. 2015, 21, 440–448. [Google Scholar] [CrossRef] [PubMed]
- Vasan, N.; Baselga, J.; Hyman, D.M. A view on drug resistance in cancer. Nature 2019, 575, 299–309. [Google Scholar] [CrossRef]
- Wolf, Y.; Bartok, O.; Patkar, S.; Eli, G.B.; Cohen, S.; Litchfield, K.; Levy, R.; Jimenez-Sanchez, A.; Trabish, S.; Lee, J.S.; et al. UVB-Induced Tumor Heterogeneity Diminishes Immune Response in Melanoma. Cell 2019, 179, 219–235.e21. [Google Scholar] [CrossRef]
- Gejman, R.S.; Chang, A.Y.; Jones, H.F.; DiKun, K.; Hakimi, A.A.; Schietinger, A.; Scheinberg, D.A. Rejection of immunogenic tumor clones is limited by clonal fraction. Elife 2018, 7, e41090. [Google Scholar] [CrossRef]
- Marine, J.C.; Dawson, S.J.; Dawson, M.A. Non-genetic mechanisms of therapeutic resistance in cancer. Nat. Rev. Cancer 2020, 20, 743–756. [Google Scholar] [CrossRef]
- Qin, S.; Jiang, J.; Lu, Y.; Nice, E.C.; Huang, C.; Zhang, J.; He, W. Emerging role of tumor cell plasticity in modifying therapeutic response. Signal Transduct. Target. Ther. 2020, 5, 228. [Google Scholar] [CrossRef]
- Shaffer, S.M.; Dunagin, M.C.; Torborg, S.R.; Torre, E.A.; Emert, B.; Krepler, C.; Beqiri, M.; Sproesser, K.; Brafford, P.A.; Xiao, M.; et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 2017, 546, 431–435. [Google Scholar] [CrossRef]
- Kurata, T.; Tamura, K.; Kaneda, H.; Nogami, T.; Uejima, H.; Asai Go, G.; Nakagawa, K.; Fukuoka, M. Effect of re-treatment with gefitinib (′Iressa′, ZD1839) after acquisition of resistance. Ann. Oncol. 2004, 15, 173–174. [Google Scholar] [CrossRef]
- Nooka, A.K.; Joseph, N.S.; Kaufman, J.L.; Heffner, L.T.; Gupta, V.A.; Gleason, C.; Boise, L.H.; Lonial, S. Clinical efficacy of daratumumab, pomalidomide, and dexamethasone in patients with relapsed or refractory myeloma: Utility of re-treatment with daratumumab among refractory patients. Cancer 2019, 125, 2991–3000. [Google Scholar] [CrossRef]
- Xu, B.; Hu, X.; Zheng, H.; Wang, X.; Zhang, Q.; Cui, S.; Liu, D.; Liao, N.; Luo, R.; Sun, Q.; et al. Outcomes of re-treatment with first-line trastuzumab plus a taxane in HER2 positive metastatic breast cancer patients after (neo)adjuvant trastuzumab: A prospective multicenter study. Oncotarget 2016, 7, 50643–50655. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bruchovsky, N.; Rennie, P.S.; Coldman, A.J.; Goldenberg, S.L.; To, M.; Lawson, D. Effects of androgen withdrawal on the stem cell composition of the Shionogi carcinoma. Cancer Res. 1990, 50, 2275–2282. [Google Scholar] [PubMed]
- Crook, J.M.; O′Callaghan, C.J.; Duncan, G.; Dearnaley, D.P.; Higano, C.S.; Horwitz, E.M.; Frymire, E.; Malone, S.; Chin, J.; Nabid, A.; et al. Intermittent androgen suppression for rising PSA level after radiotherapy. N. Engl. J. Med. 2012, 367, 895–903. [Google Scholar] [CrossRef]
- Yuan, S.; Norgard, R.J.; Stanger, B.Z. Cellular Plasticity in Cancer. Cancer Discov. 2019, 9, 837–851. [Google Scholar] [CrossRef] [PubMed]
- Fanelli, G.N.; Naccarato, A.G.; Scatena, C. Recent Advances in Cancer Plasticity: Cellular Mechanisms, Surveillance Strategies, and Therapeutic Optimization. Front. Oncol. 2020, 10, 569. [Google Scholar] [CrossRef] [PubMed]
- Batlle, E.; Clevers, H. Cancer stem cells revisited. Nat. Med. 2017, 23, 1124–1134. [Google Scholar] [CrossRef]
- Krivtsov, A.V.; Twomey, D.; Feng, Z.; Stubbs, M.C.; Wang, Y.; Faber, J.; Levine, J.E.; Wang, J.; Hahn, W.C.; Gilliland, D.G.; et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 2006, 442, 818–822. [Google Scholar] [CrossRef]
- Schwitalla, S.; Fingerle, A.A.; Cammareri, P.; Nebelsiek, T.; Goktuna, S.I.; Ziegler, P.K.; Canli, O.; Heijmans, J.; Huels, D.J.; Moreaux, G.; et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 2013, 152, 25–38. [Google Scholar] [CrossRef]
- Tata, P.R.; Mou, H.; Pardo-Saganta, A.; Zhao, R.; Prabhu, M.; Law, B.M.; Vinarsky, V.; Cho, J.L.; Breton, S.; Sahay, A.; et al. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature 2013, 503, 218–223. [Google Scholar] [CrossRef]
- Suva, M.L.; Rheinbay, E.; Gillespie, S.M.; Patel, A.P.; Wakimoto, H.; Rabkin, S.D.; Riggi, N.; Chi, A.S.; Cahill, D.P.; Nahed, B.V.; et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell 2014, 157, 580–594. [Google Scholar] [CrossRef] [PubMed]
- Jopling, C.; Boue, S.; Izpisua Belmonte, J.C. Dedifferentiation, transdifferentiation and reprogramming: Three routes to regeneration. Nat. Rev. Mol. Cell Biol. 2011, 12, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Tata, P.R.; Rajagopal, J. Cellular plasticity: 1712 to the present day. Curr. Opin. Cell Biol. 2016, 43, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Li, C.; Fan, Z.; Liu, H.; Zhang, X.; Cai, Z.; Xu, L.; Luo, J.; Huang, Y.; He, L.; et al. Single-cell Sequencing Reveals Variants in ARID1A, GPRC5A and MLL2 Driving Self-renewal of Human Bladder Cancer Stem Cells. Eur. Urol. 2017, 71, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Hu, W.; Matulay, J.T.; Silva, M.V.; Owczarek, T.B.; Kim, K.; Chua, C.W.; Barlow, L.J.; Kandoth, C.; Williams, A.B.; et al. Tumor Evolution and Drug Response in Patient-Derived Organoid Models of Bladder Cancer. Cell 2018, 173, 515–528.e17. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.B.; Fillmore, C.M.; Jiang, G.; Shapira, S.D.; Tao, K.; Kuperwasser, C.; Lander, E.S. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell 2011, 146, 633–644. [Google Scholar] [CrossRef]
- Walter, R.J.; Sonnentag, S.J.; Orian-Rousseau, V.; Munoz-Sagredo, L. Plasticity in Colorectal Cancer: Why Cancer Cells Differentiate. Cancers 2021, 13, 918. [Google Scholar] [CrossRef]
- Smigiel, J.M.; Taylor, S.E.; Bryson, B.L.; Tamagno, I.; Polak, K.; Jackson, M.W. Cellular plasticity and metastasis in breast cancer: A pre- and post-malignant problem. J. Cancer Metastasis Treat. 2019, 5, 47. [Google Scholar] [CrossRef]
- Sousa, B.; Ribeiro, A.S.; Paredes, J. Heterogeneity and Plasticity of Breast Cancer Stem Cells. Adv. Exp. Med. Biol. 2019, 1139, 83–103. [Google Scholar] [CrossRef]
- Liu, S.; Cong, Y.; Wang, D.; Sun, Y.; Deng, L.; Liu, Y.; Martin-Trevino, R.; Shang, L.; McDermott, S.P.; Landis, M.D.; et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Rep. 2014, 2, 78–91. [Google Scholar] [CrossRef]
- Marjanovic, N.D.; Hofree, M.; Chan, J.E.; Canner, D.; Wu, K.; Trakala, M.; Hartmann, G.G.; Smith, O.C.; Kim, J.Y.; Evans, K.V.; et al. Emergence of a High-Plasticity Cell State during Lung Cancer Evolution. Cancer Cell 2020, 38, 229–246.e13. [Google Scholar] [CrossRef]
- Bocci, F.; Gearhart-Serna, L.; Boareto, M.; Ribeiro, M.; Ben-Jacob, E.; Devi, G.R.; Levine, H.; Onuchic, J.N.; Jolly, M.K. Toward understanding cancer stem cell heterogeneity in the tumor microenvironment. Proc. Natl. Acad. Sci. USA 2019, 116, 148–157. [Google Scholar] [CrossRef]
- Gupta, P.B.; Pastushenko, I.; Skibinski, A.; Blanpain, C.; Kuperwasser, C. Phenotypic Plasticity: Driver of Cancer Initiation, Progression, and Therapy Resistance. Cell Stem Cell 2019, 24, 65–78. [Google Scholar] [CrossRef]
- Nieto, M.A.; Huang, R.Y.; Jackson, R.A.; Thiery, J.P. Emt: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef] [PubMed]
- Garg, M. Urothelial cancer stem cells and epithelial plasticity: Current concepts and therapeutic implications in bladder cancer. Cancer Metastasis Rev. 2015, 34, 691–701. [Google Scholar] [CrossRef] [PubMed]
- Chin, V.L.; Lim, C.L. Epithelial-mesenchymal plasticity-engaging stemness in an interplay of phenotypes. Stem Cell Investig. 2019, 6, 25. [Google Scholar] [CrossRef]
- Singh, R.; Ansari, J.A.; Maurya, N.; Mandhani, A.; Agrawal, V.; Garg, M. Epithelial-To-Mesenchymal Transition and Its Correlation With Clinicopathologic Features in Patients With Urothelial Carcinoma of the Bladder. Clin. Genitourin. Cancer 2017, 15, e187–e197. [Google Scholar] [CrossRef] [PubMed]
- Yun, S.J.; Kim, W.J. Role of the epithelial-mesenchymal transition in bladder cancer: From prognosis to therapeutic target. Korean J. Urol. 2013, 54, 645–650. [Google Scholar] [CrossRef]
- Liu, B.; Miyake, H.; Nishikawa, M.; Fujisawa, M. Expression profile of epithelial-mesenchymal transition markers in non-muscle-invasive urothelial carcinoma of the bladder: Correlation with intravesical recurrence following transurethral resection. Urol. Oncol. 2015, 33, 110. e111–e118. [Google Scholar] [CrossRef]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef]
- Chen, Y.; Li, L.; Zeng, J.; Wu, K.; Zhou, J.; Guo, P.; Zhang, D.; Xue, Y.; Liang, L.; Wang, X.; et al. Twist confers chemoresistance to anthracyclines in bladder cancer through upregulating P-glycoprotein. Chemotherapy 2012, 58, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Davies, A.H.; Beltran, H.; Zoubeidi, A. Cellular plasticity and the neuroendocrine phenotype in prostate cancer. Nat. Rev. Urol. 2018, 15, 271–286. [Google Scholar] [CrossRef] [PubMed]
- Rubin, M.A.; Bristow, R.G.; Thienger, P.D.; Dive, C.; Imielinski, M. Impact of Lineage Plasticity to and from a Neuroendocrine Phenotype on Progression and Response in Prostate and Lung Cancers. Mol. Cell 2020, 80, 562–577. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Bondaruk, J.; Cogdell, D.; Wang, Z.; Lee, S.; Lee, J.G.; Zhang, S.; Choi, W.; Wang, Y.; Liang, Y.; et al. Urothelial-to-Neural Plasticity Drives Progression to Small Cell Bladder Cancer. iScience 2020, 23, 101201. [Google Scholar] [CrossRef] [PubMed]
- Malek, R.; Wang, H.; Taparra, K.; Tran, P.T. Therapeutic Targeting of Epithelial Plasticity Programs: Focus on the Epithelial-Mesenchymal Transition. Cells Tissues Organs 2017, 203, 114–127. [Google Scholar] [CrossRef]
- Cho, E.S.; Kang, H.E.; Kim, N.H.; Yook, J.I. Therapeutic implications of cancer epithelial-mesenchymal transition (EMT). Arch. Pharm. Res. 2019, 42, 14–24. [Google Scholar] [CrossRef]
- Ramesh, V.; Brabletz, T.; Ceppi, P. Targeting EMT in Cancer with Repurposed Metabolic Inhibitors. Trends Cancer 2020, 6, 942–950. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Cao, R.; Wang, Y.; Qian, G.; Dan, H.C.; Jiang, W.; Ju, L.; Wu, M.; Xiao, Y.; Wang, X. Simvastatin induces cell cycle arrest and inhibits proliferation of bladder cancer cells via PPARgamma signalling pathway. Sci. Rep. 2016, 6, 35783. [Google Scholar] [CrossRef]
- Kochanowski, K.; Morinishi, L.; Altschuler, S.; Wu, L. Drug persistence—From antibiotics to cancer therapies. Curr. Opin. Syst. Biol. 2018, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Swayden, M.; Chhouri, H.; Anouar, Y.; Grumolato, L. Tolerant/Persister Cancer Cells and the Path to Resistance to Targeted Therapy. Cells 2020, 9, 2601. [Google Scholar] [CrossRef]
- Dobson, S.M.; Garcia-Prat, L.; Vanner, R.J.; Wintersinger, J.; Waanders, E.; Gu, Z.; McLeod, J.; Gan, O.I.; Grandal, I.; Payne-Turner, D.; et al. Relapse-Fated Latent Diagnosis Subclones in Acute B Lineage Leukemia Are Drug Tolerant and Possess Distinct Metabolic Programs. Cancer Discov. 2020, 10, 568–587. [Google Scholar] [CrossRef] [PubMed]
- De Conti, G.; Dias, M.H.; Bernards, R. Fighting Drug Resistance through the Targeting of Drug-Tolerant Persister Cells. Cancers 2021, 13, 1118. [Google Scholar] [CrossRef] [PubMed]
- Risom, T.; Langer, E.M.; Chapman, M.P.; Rantala, J.; Fields, A.J.; Boniface, C.; Alvarez, M.J.; Kendsersky, N.D.; Pelz, C.R.; Johnson-Camacho, K.; et al. Differentiation-state plasticity is a targetable resistance mechanism in basal-like breast cancer. Nat. Commun. 2018, 9, 3815. [Google Scholar] [CrossRef]
- Sharma, S.V.; Lee, D.Y.; Li, B.; Quinlan, M.P.; Takahashi, F.; Maheswaran, S.; McDermott, U.; Azizian, N.; Zou, L.; Fischbach, M.A.; et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010, 141, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Vallette, F.M.; Olivier, C.; Lezot, F.; Oliver, L.; Cochonneau, D.; Lalier, L.; Cartron, P.F.; Heymann, D. Dormant, quiescent, tolerant and persister cells: Four synonyms for the same target in cancer. Biochem. Pharmacol. 2019, 162, 169–176. [Google Scholar] [CrossRef]
- Enriquez-Navas, P.M.; Kam, Y.; Das, T.; Hassan, S.; Silva, A.; Foroutan, P.; Ruiz, E.; Martinez, G.; Minton, S.; Gillies, R.J.; et al. Exploiting evolutionary principles to prolong tumor control in preclinical models of breast cancer. Sci. Transl. Med. 2016, 8, 327ra24. [Google Scholar] [CrossRef]
- West, J.; You, L.; Zhang, J.; Gatenby, R.A.; Brown, J.S.; Newton, P.K.; Anderson, A.R.A. Towards Multidrug Adaptive Therapy. Cancer Res. 2020, 80, 1578–1589. [Google Scholar] [CrossRef]
- Ishay-Ronen, D.; Diepenbruck, M.; Kalathur, R.K.R.; Sugiyama, N.; Tiede, S.; Ivanek, R.; Bantug, G.; Morini, M.F.; Wang, J.; Hess, C.; et al. Gain Fat-Lose Metastasis: Converting Invasive Breast Cancer Cells into Adipocytes Inhibits Cancer Metastasis. Cancer Cell 2019, 35, 17–32.e6. [Google Scholar] [CrossRef]
- Ishay-Ronen, D.; Christofori, G. Targeting Cancer Cell Metastasis by Converting Cancer Cells into Fat. Cancer Res. 2019, 79, 5471–5475. [Google Scholar] [CrossRef]
- Rusan, M.; Li, K.; Li, Y.; Christensen, C.L.; Abraham, B.J.; Kwiatkowski, N.; Buczkowski, K.A.; Bockorny, B.; Chen, T.; Li, S.; et al. Suppression of Adaptive Responses to Targeted Cancer Therapy by Transcriptional Repression. Cancer Discov. 2018, 8, 59–73. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lavallee, E.; Sfakianos, J.P.; Mulholland, D.J. Tumor Heterogeneity and Consequences for Bladder Cancer Treatment. Cancers 2021, 13, 5297. https://doi.org/10.3390/cancers13215297
Lavallee E, Sfakianos JP, Mulholland DJ. Tumor Heterogeneity and Consequences for Bladder Cancer Treatment. Cancers. 2021; 13(21):5297. https://doi.org/10.3390/cancers13215297
Chicago/Turabian StyleLavallee, Etienne, John P. Sfakianos, and David J. Mulholland. 2021. "Tumor Heterogeneity and Consequences for Bladder Cancer Treatment" Cancers 13, no. 21: 5297. https://doi.org/10.3390/cancers13215297
APA StyleLavallee, E., Sfakianos, J. P., & Mulholland, D. J. (2021). Tumor Heterogeneity and Consequences for Bladder Cancer Treatment. Cancers, 13(21), 5297. https://doi.org/10.3390/cancers13215297





