ATM Kinase Dead: From Ataxia Telangiectasia Syndrome to Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. ATM Kinase Dead in A-T Patients
3. Lessons from Mouse Models: ATM Knockout and ATM Kinase Dead
4. ATM Kinase Dead in Cancer Patients
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Abraham, R.T. Mammalian target of rapamycin: Immunosuppressive drugs uncover a novel pathway of cytokine receptor signaling. Curr. Opin. Immunol. 1998, 10, 330–336. [Google Scholar] [CrossRef]
- Gatti, R.A.; Berkel, I.; Boder, E.; Braedt, G.; Charmley, P.; Concannon, P.; Ersoy, F.; Foroud, T.; Jaspers, N.G.J.; Lange, K.; et al. Localization of an ataxia-telangiectasia gene to chromosome 11q22-23. Nature 1988, 336, 577–580. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.R.; Welch, C.L.; Warden, C.H.; Lange, E.; Fukao, T.; Lusis, A.J.; Gatti, R.A. Assignment of the mouse ataxia-telangiectasia gene (Atm) to mouse Chromosome 9. Mamm. Genome 1996, 7, 554–555. [Google Scholar] [CrossRef] [PubMed]
- Shiloh, Y.; Ziv, Y. The ATM protein: The importance of being active. J. Cell Biol. 2012, 198, 273–275. [Google Scholar] [CrossRef]
- Savitsky, K.; Bar-Shira, A.; Gilad, S.; Rotman, G.; Ziv, Y.; Vanagaite, L.; Tagle, D.A.; Smith, S.; Uziel, T.; Sfez, S.; et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 1995, 268, 1749–1753. [Google Scholar] [CrossRef]
- Banin, S.; Moyal, L.; Shieh, S.Y.; Taya, Y.; Anderson, C.W.; Chessa, L.; Smorodinsky, N.I.; Prives, C.; Reiss, Y.; Shiloh, Y.; et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 1998, 281, 1674–1677. [Google Scholar] [CrossRef]
- Canman, C.E.; Lim, D.S.; Cimprich, K.A.; Taya, Y.; Tamai, K.; Sakaguchi, K.; Appella, E.; Kastan, M.B.; Siliciano, J.D. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 1998, 281, 1677–1679. [Google Scholar] [CrossRef]
- Bakkenist, C.J.; Kastan, M.B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 2003, 421, 499–506. [Google Scholar] [CrossRef]
- Dupré, A.; Boyer-Chatenet, L.; Gautier, J. Two-step activation of ATM by DNA and the Mre11-Rad50-Nbs1 complex. Nat. Struct. Mol. Biol. 2006, 13, 451–457. [Google Scholar] [CrossRef]
- Lee, J.H.; Paull, T.T. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 2005, 308, 551–554. [Google Scholar] [CrossRef]
- Pellegrini, M.; Celeste, A.; Difilippantonio, S.; Guo, R.; Wang, W.; Feigenbaum, L.; Nussenzweig, A. Autophosphorylation at serine 1987 is dispensable for murine Atm activation in vivo. Nature 2006, 443, 222–225. [Google Scholar] [CrossRef]
- Daniel, J.A.; Pellegrini, M.; Lee, J.H.; Paull, T.T.; Feigenbaum, L.; Nussenzweig, A. Multiple autophosphorylation sites are dispensable for murine ATM activation in vivo. J. Cell Biol. 2008, 183, 777–783. [Google Scholar] [CrossRef]
- Lee, J.H.; Paull, T.T. Cellular functions of the protein kinase ATM and their relevance to human disease. Nat. Rev. Mol. Cell Biol. 2021, 1. [Google Scholar] [CrossRef]
- Shibata, A.; Jeggo, P.A. ATM’s role in the repair of DNA double-strand breaks. Genes 2021, 12, 1370. [Google Scholar] [CrossRef]
- Alexander, A.; Walker, C.L. Differential localization of ATM is correlated with activation of distinct downstream signaling pathways. Cell Cycle 2010, 9, 3709–3710. [Google Scholar] [CrossRef]
- Alexander, A.; Cai, S.L.; Kim, J.; Nanez, A.; Sahin, M.; MacLean, K.H.; Inoki, K.; Guan, K.L.; Shen, J.; Person, M.D.; et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc. Natl. Acad. Sci. USA 2010, 107, 4153–4158. [Google Scholar] [CrossRef]
- Yang, D.Q.; Halaby, M.J.; Li, Y.; Hibma, J.C.; Burn, P. Cytoplasmic ATM protein kinase: An emerging therapeutic target for diabetes, cancer and neuronal degeneration. Drug Discov. Today 2011, 16, 332–338. [Google Scholar] [CrossRef]
- Menolfi, D.; Zha, S. ATM, ATR and DNA-PKcs kinases-the lessons from the mouse models: Inhibition = deletion. Cell Biosci. 2020, 10, 1–15. [Google Scholar] [CrossRef]
- Matsuoka, S.; Ballif, B.A.; Smogorzewska, A.; McDonald, E.R.; Hurov, K.E.; Luo, J.; Bakalarski, C.E.; Zhao, Z.; Solimini, N.; Lerenthal, Y.; et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007, 316, 1160–1166. [Google Scholar] [CrossRef]
- Syllaba, L.; Henner, K. Contribution a l’independance de l’athetose double idiopathique et congenitale. Rev. Neurol. 1926, 1, 541–562. [Google Scholar]
- Rothblum-Oviatt, C.; Wright, J.; Lefton-Greif, M.A.; McGrath-Morrow, S.A.; Crawford, T.O.; Lederman, H.M. Ataxia telangiectasia: A review. Orphanet J. Rare Dis. 2016, 11. [Google Scholar] [CrossRef]
- Boder, E.; Sedgwick, R.P. ATAXIA-TELANGIECTASIA. Pediatrics 1958, 21, 526–554. [Google Scholar]
- Gilad, S.; Chessa, L.; Khosravi, R.; Russell, P.; Galanty, Y.; Piane, M.; Gatti, R.A.; Jorgensen, T.J.; Shiloh, Y.; Bar-Shira, A. Genotype-phenotype relationships in ataxia-telangiectasia and variants. Am. J. Hum. Genet. 1998, 62, 551–561. [Google Scholar] [CrossRef]
- McKinnon, P.J. ATM and the molecular pathogenesis of ataxia telangiectasia. Annu. Rev. Pathol. Mech. Dis. 2012, 7, 303–321. [Google Scholar] [CrossRef]
- Lavin, M.F.; Scott, S.; Gueven, N.; Kozlov, S.; Peng, C.; Chen, P. Functional consequences of sequence alterations in the ATM gene. DNA Repair 2004, 3, 1197–1205. [Google Scholar] [CrossRef]
- Sandoval, N.; Platzer, M.; Rosenthal, A.; Dörk, T.; Bendix, R.; Skawran, B.; Stuhrmann, M.; Wegner, R.D.; Sperling, K.; Banin, S.; et al. Characterization of ATM gene mutations in 66 ataxia telangiectasia families. Hum. Mol. Genet. 1999, 8, 69–79. [Google Scholar] [CrossRef]
- Chun, H.H.; Gatti, R.A. Ataxia-telangiectasia, an evolving phenotype. DNA Repair 2004, 3, 1187–1196. [Google Scholar] [CrossRef]
- Verhagen, M.M.M.; Last, J.I.; Hogervorst, F.B.L.; Smeets, D.F.C.M.; Roeleveld, N.; Verheijen, F.; Catsman-Berrevoets, C.E.; Wulffraat, N.M.; Cobben, J.M.; Hiel, J.; et al. Presence of ATM protein and residual kinase activity correlates with the phenotype in ataxia-telangiectasia: A genotype-phenotype study. Hum. Mutat. 2012, 33, 561–571. [Google Scholar] [CrossRef]
- Reiman, A.; Srinivasan, V.; Barone, G.; Last, J.I.; Wootton, L.L.; Davies, E.G.; Verhagen, M.M.; Willemsen, M.A.; Weemaes, C.M.; Byrd, P.J.; et al. Lymphoid tumours and breast cancer in ataxia telangiectasia; Substantial protective effect of residual ATM kinase activity against childhood tumours. Br. J. Cancer 2011, 105, 586–591. [Google Scholar] [CrossRef]
- Becker-Catania, S.G.; Chen, G.; Hwang, M.J.; Wang, Z.; Sun, X.; Sanal, O.; Bernatowska-Matuszkiewicz, E.; Chessa, L.; Lee, E.Y.H.P.; Gatti, R.A. Ataxia-telangiectasia: Phenotype/genotype studies of ATM protein expression, mutations, and radiosensitivity. Mol. Genet. Metab. 2000, 70, 122–133. [Google Scholar] [CrossRef]
- Choi, M.; Kipps, T.; Kurzrock, R. ATM mutations in cancer: Therapeutic implications. Mol. Cancer Ther. 2016, 15, 1781–1791. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Wang, J.; Sprinzen, L.; Xu, J.; Haddock, C.J.; Li, C.; Lee, B.J.; Loredan, D.G.; Jiang, W.; Vindigni, A.; et al. Kinase-dead ATM protein is highly oncogenic and can be preferentially targeted by topo-isomerase I inhibitors. Elife 2016, 5, e14709. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, S.V.; Graham, M.E.; Peng, C.; Chen, P.; Robinson, P.J.; Lavin, M.F. Involvement of novel autophosphorylation sites in ATM activation. EMBO J. 2006, 25, 3504–3514. [Google Scholar] [CrossRef] [PubMed]
- Hiel, J.A.P.; Van Engelen, B.G.M.; Weemaes, C.M.R.; Broeks, A.; Verrips, A.; Ter Laak, H.; Vingerhoets, H.M.; Van Den Heuvel, L.P.W.; Lammens, M.; Gabreëls, F.J.M.; et al. Distal spinal muscular atrophy as a major feature in adult-onset ataxia telangiectasia. Neurology 2006, 67, 346–349. [Google Scholar] [CrossRef]
- Saviozzi, S.; Saluto, A.; Taylor, A.M.R.; Last, J.I.L.; Trebini, F.; Paradiso, M.C.; Grosso, E.; Funaro, A.; Ponzio, G.; Migone, N.; et al. A late onset variant of ataxia-telangiectasia with a compound heterozygous genotype, A8030G/7481 insA. J. Med. Genet. 2002, 39, 57–61. [Google Scholar] [CrossRef]
- Silvestri, G.; Masciullo, M.; Piane, M.; Savio, C.; Modoni, A.; Santoro, M.; Chessa, L. Homozygosity for c 6325T>G transition in the ATM gene causes an atypical, late-onset variant form of ataxia-telangiectasia. J. Neurol. 2010, 257, 1738–1740. [Google Scholar] [CrossRef]
- Simonin, C.; Devos, D.; Vuillaume, I.; De Martinville, B.; Sablonnière, B.; Destée, A.; Stoppa-Lyonnet, D.; Defebvre, L. Attenuated presentation of ataxia-telangiectasia with familial cancer history. J. Neurol. 2008, 255, 1261–1263. [Google Scholar] [CrossRef]
- Sutton, I.J.; Last, J.I.K.; Ritchie, S.J.; Harrington, H.J.; Byrd, P.J.; Taylor, A.M.R. Adult-onset ataxia telangiectasia due to ATM 5762ins137 mutation homozygosity. Ann. Neurol. 2004, 55, 891–895. [Google Scholar] [CrossRef]
- Verhagen, M.M.M.; Abdo, W.F.; Willemsen, M.A.A.P.; Hogervorst, F.B.L.; Smeets, D.F.C.M.; Hiel, J.A.P.; Brunt, E.R.; Van Rijn, M.A.; Majoor Krakauer, D.; Oldenburg, R.A.; et al. Clinical spectrum of ataxia-telangiectasia in adulthood. Neurology 2009, 73, 430–437. [Google Scholar] [CrossRef]
- Lavin, M.F.; Shiloh, Y. The genetic defect in ataxia-telangiectasia. Annu. Rev. Immunol. 1997, 15, 177–202. [Google Scholar] [CrossRef]
- Li, A.; Swift, M. Mutations at the ataxia-telangiectasia locus and clinical phenotypes of A-T patients. Am. J. Med. Genet. 2000, 92, 170–177. [Google Scholar] [CrossRef]
- Taylor, A.M.R.; Lam, Z.; Last, J.I.; Byrd, P.J. Ataxia telangiectasia: More variation at clinical and cellular levels. Clin. Genet. 2015, 87, 199–208. [Google Scholar] [CrossRef]
- Telatar, M.; Wang, Z.; Udar, N.; Liang, T.; Bernatowska-Matuszkiewicz, E.; Lavin, M.; Shiloh, Y.; Concannon, P.; Good, R.A.; Gatti, R.A. Ataxia-telangiectasia: Mutations in ATM cDNA detected by protein- truncation screening. Am. J. Hum. Genet. 1996, 59, 40–44. [Google Scholar]
- Gilad, S.; Khosravi, R.; Shkedy, D.; Uziel, T.; Ziv, Y.; Savitsky, K.; Rotman, G.; Smith, S.; Chessa, L.; Jorgensen, T.J.; et al. Predominance of null mutations in ataxia-telangiectasia. Hum. Mol. Genet. 1996, 5, 433–439. [Google Scholar] [CrossRef]
- Brown, K.D.; Ziv, Y.; Sadanandan, S.N.; Chessa, L.; Collins, F.S.; Shiloh, Y.; Tagle, D.A. The ataxia-telangiectasia gene product, a constitutively expressed nuclear protein that is not up-regulated following genome damage. Proc. Natl. Acad. Sci. USA 1997, 94, 1840–1845. [Google Scholar] [CrossRef]
- Angèle, S.; Laugé, A.; Fernet, M.; Moullan, N.; Beauvais, P.; Couturier, J.; Stoppa-Lyonnet, D.; Hall, J. Phenotypic cellular characterization of an ataxia telangiectasia patient carrying a causal homozygous missense mutation. Hum. Mutat. 2003, 21, 169–170. [Google Scholar] [CrossRef]
- Van Os, N.J.H.; Chessa, L.; Weemaes, C.M.R.; Van Deuren, M.; Fiévet, A.; Van Gaalen, J.; Mahlaoui, N.; Roeleveld, N.; Schrader, C.; Schindler, D.; et al. Genotype-phenotype correlations in ataxia telangiectasia patients with ATM c.3576G>A and c.8147T>C mutations. J. Med. Genet. 2019, 56, 308–316. [Google Scholar] [CrossRef]
- Barone, G.; Groom, A.; Reiman, A.; Srinivasan, V.; Byrd, P.J.; Taylor, A.M.R. Modeling ATM mutant proteins from missense changes confirms retained kinase activity. Hum. Mutat. 2009, 30, 1222–1230. [Google Scholar] [CrossRef]
- Van Belzen, M.J.; Hiel, J.A.P.; Weemaes, C.M.R.; Gabreëls, F.J.M.; Van Engelen, B.G.M.; Smeets, D.F.C.M.; Van den Heuvel, L.P.W.J. A double missense mutation in the ATM gene of a Dutch family with ataxia telangiectasia. Hum. Genet. 1998, 102, 187–191. [Google Scholar] [CrossRef]
- Stewart, G.S.; Last, J.I.K.; Stankovic, T.; Haites, N.; Kidd, A.M.J.; Byrd, P.J.; Taylor, A.M.R. Residual Ataxia Telangiectasia Mutated Protein Function in Cells from Ataxia Telangiectasia Patients, with 5762ins137 and 7271T→G Mutations, Showing a Less Severe Phenotype. J. Biol. Chem. 2001, 276, 30133–30141. [Google Scholar] [CrossRef]
- Taylor, A.M.R.; Byrd, P.J. Molecular pathology of ataxia telangiectasia. J. Clin. Pathol. 2005, 58, 1009–1015. [Google Scholar] [CrossRef]
- Austen, B.; Barone, G.; Reiman, A.; Byrd, P.J.; Baker, C.; Starczynski, J.; Nobbs, M.C.; Murphy, R.P.; Enright, H.; Chaila, E.; et al. Pathogenic ATM mutations occur rarely in a subset of multiple myeloma patients. Br. J. Haematol. 2008, 142, 925–933. [Google Scholar] [CrossRef]
- Scott, S.P.; Bendix, R.; Chen, P.; Clark, R.; Dörk, T.; Lavin, M.F. Missense mutations but not allelic variants alter the function of ATM by dominant interference in patients with breast cancer. Proc. Natl. Acad. Sci. USA 2002, 99, 925–930. [Google Scholar] [CrossRef]
- Barlow, C.; Hirotsune, S.; Paylor, R.; Liyanage, M.; Eckhaus, M.; Collins, F.; Shiloh, Y.; Crawley, J.N.; Ried, T.; Tagle, D.; et al. ATM deficient mice. Cell 1996, 86. [Google Scholar] [CrossRef]
- Elson, A.; Wang, Y.; Daugherty, C.J.; Morton, C.C.; Zhou, F.; Campos-Torres, J.; Leder, P. Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc. Natl. Acad. Sci. USA 1996, 93, 13084–13089. [Google Scholar] [CrossRef]
- Xu, B.; Kim, S.; Kastan, M.B. Involvement of Brca1 in S-Phase and G 2 -Phase Checkpoints after Ionizing Irradiation. Mol. Cell. Biol. 2001, 21, 3445–3450. [Google Scholar] [CrossRef]
- Chiesa, N.; Barlow, C.; Wynshaw-Boris, A.; Strata, P.; Tempia, F. Atm-deficient mice Purkinje cells show age-dependent defects in calcium spike bursts and calcium currents. Neuroscience 2000, 96, 575–583. [Google Scholar] [CrossRef]
- Kuljis, R.O.; Xu, Y.; Aguila, M.C.; Baltimore, D. Degeneration of neurons, synapses, and neuropil and glial activation in a murine Atm knockout model of ataxia-telangiectasia. Proc. Natl. Acad. Sci. USA 1997, 94, 12688–12693. [Google Scholar] [CrossRef]
- Kamsler, A.; Daily, D.; Hochman, A.; Stern, N.; Shiloh, Y.; Rotman, G.; Barzilai, A. Increased oxidative stress in ataxia telangiectasia evidenced by alterations in redox state of brains from Atm-deficient mice. Cancer Res. 2001, 61, 1849–1854. [Google Scholar]
- Chen, P.; Peng, C.; Luff, J.; Spring, K.; Watters, D.; Bottle, S.; Furuya, S.; Lavin, M.F. Oxidative Stress is Responsible for Deficient Survival and Dendritogenesis in Purkinje Neurons from Ataxia-Telangiectasia Mutated Mutant Mice. J. Neurosci. 2003, 23, 11453–11460. [Google Scholar] [CrossRef]
- Quick, K.L.; Dugan, L.L. Superoxide stress identifies neurons at risk in a model of ataxia-telangiectasia. Ann. Neurol. 2001, 49, 627–635. [Google Scholar] [CrossRef] [PubMed]
- Gueven, N.; Luff, J.; Peng, C.; Hosokawa, K.; Bottle, S.E.; Lavin, M.F. Dramatic extension of tumor latency and correction of neurobehavioral phenotype in Atm-mutant mice with a nitroxide antioxidant. Free Radic. Biol. Med. 2006, 41, 992–1000. [Google Scholar] [CrossRef] [PubMed]
- Stern, N.; Hochman, A.; Zemach, N.; Weizman, N.; Hammel, I.; Shiloh, Y.; Rotman, G.; Barzilai, A. Accumulation of DNA damage and reduced levels of nicotine adenine dinucleotide in the brains of Atm-deficient mice. J. Biol. Chem. 2002, 277, 602–608. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Wang, Y.; Jiang, W.; Liu, X.; Dubois, R.L.; Lin, C.S.; Ludwig, T.; Bakkenist, C.J.; Zha, S. Kinase-dead ATM protein causes genomic instability and early embryonic lethality in mice. J. Cell Biol. 2012, 198, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Daniel, J.A.; Pellegrini, M.; Lee, B.S.; Guo, Z.; Filsuf, D.; Belkina, N.V.; You, Z.; Paull, T.T.; Sleckman, B.P.; Feigenbaum, L.; et al. Loss of ATM kinase activity leads to embryonic lethality in mice. J. Cell Biol. 2012, 198, 295–304. [Google Scholar] [CrossRef] [PubMed]
- White, J.S.; Choi, S.; Bakkenist, C.J. Transient ATM kinase inhibition disrupts DNA damage-induced sister chromatid exchange. Sci. Signal. 2010, 3, ra44. [Google Scholar] [CrossRef] [PubMed]
- Tal, E.; Alfo, M.; Zha, S.; Barzilai, A.; De Zeeuw, C.I.; Ziv, Y.; Shiloh, Y. Inactive Atm abrogates DSB repair in mouse cerebellum more than does Atm loss, without causing a neurological phenotype. DNA Repair 2018, 72, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Milanovic, M.; Shao, Z.; Estes, V.M.; Wang, X.S.; Menolfi, D.; Lin, X.; Lee, B.J.; Xu, J.; Cupo, O.M.; Wang, D.; et al. FATC Domain Deletion Compromises ATM Protein Stability, Blocks Lymphocyte Development, and Promotes Lymphomagenesis. J. Immunol. 2021, 206, 1228–1239. [Google Scholar] [CrossRef] [PubMed]
- Milanovic, M.; Houghton, L.M.; Menolfi, D.; Lee, J.H.; Yamamoto, K.; Li, Y.; Lee, B.J.; Xu, J.; Estes, V.M.; Wang, D.; et al. The Cancer-Associated ATM R3008H Mutation Reveals the Link between ATM Activation and Its Exchange. Cancer Res. 2021, 81, 426–437. [Google Scholar] [CrossRef]
- Spring, K.; Ahangari, F.; Scott, S.P.; Waring, P.; Purdie, D.M.; Chen, P.C.; Hourigan, K.; Ramsay, J.; McKinnon, P.J.; Swift, M.; et al. Mice heterozygous for mutation in Atm, the gene involved in ataxia-telangiectasia, have heightened susceptibility to cancer. Nat. Genet. 2002, 32, 185–190. [Google Scholar] [CrossRef]
- Lavin, M.F. Ataxia-telangiectasia: From a rare disorder to a paradigm for cell signalling and cancer. Nat. Rev. Mol. Cell Biol. 2008, 9, 759–769. [Google Scholar] [CrossRef]
- Huang, K.L.; Mashl, R.J.; Wu, Y.; Ritter, D.I.; Wang, J.; Oh, C.; Paczkowska, M.; Reynolds, S.; Wyczalkowski, M.A.; Oak, N.; et al. Pathogenic Germline Variants in 10,389 Adult Cancers. Cell 2018, 173, 355–370.e14. [Google Scholar] [CrossRef]
- Jette, N.R.; Kumar, M.; Radhamani, S.; Arthur, G.; Goutam, S.; Yip, S.; Kolinsky, M.; Williams, G.J.; Bose, P.; Lees-Miller, S.P. ATM-deficient cancers provide new opportunities for precision oncology. Cancers 2020, 12, 687. [Google Scholar] [CrossRef]
- Stankovic, T.; Stewart, G.S.; Byrd, P.; Fegan, C.; Moss, P.A.H.; Taylor, A.M.R. ATM mutations in sporadic lymphoid tumours. Leuk. Lymphoma 2002, 43, 1563–1571. [Google Scholar] [CrossRef]
- Gatti, R.A.; Tward, A.; Concannon, P. Cancer risk in ATM heterozygotes: A model of phenotypic and mechanistic differences between missense and truncating mutations. Mol. Genet. Metab. 1999, 68, 419–423. [Google Scholar] [CrossRef]
- Lim, D.S.; Kim, S.T.; Xu, B.; Maser, R.S.; Lin, J.; Petrini, J.H.J.; Kastan, M.B. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature 2000, 404, 613–617. [Google Scholar] [CrossRef]
- Chenevix-Trench, G.; Spurdle, A.B.; Gatei, M.; Kelly, H.; Marsh, A.; Chen, X.; Donn, K.; Cummings, M.; Nyholt, D.; Jenkins, M.A.; et al. Dominant negative ATM mutations in breast cancer families. J. Natl. Cancer Inst. 2002, 94, 205–215. [Google Scholar] [CrossRef]
- Camacho, E.; Hernández, L.; Hernández, S.; Tort, F.; Bellosillo, B.; Beà, S.; Bosch, F.; Montserrat, E.; Cardesa, A.; Fernández, P.L.; et al. ATM gene inactivation in mantle cell lymphoma mainly occurs by truncating mutations and missense mutations involving the phosphatidylinositol-3 kinase domain and is associated with increasing numbers of chromosomal imbalances. Blood 2002, 99, 238–244. [Google Scholar] [CrossRef]
- Randon, G.; Fucà, G.; Rossini, D.; Raimondi, A.; Pagani, F.; Perrone, F.; Tamborini, E.; Busico, A.; Peverelli, G.; Morano, F.; et al. Prognostic impact of ATM mutations in patients with metastatic colorectal cancer. Sci. Rep. 2019, 9, 2858. [Google Scholar] [CrossRef]
- Stankovic, T.; Kidd, A.M.J.; Sutcliffe, A.; McGuire, G.M.; Robinson, P.; Weber, P.; Bedenham, T.; Bradwell, A.R.; Easton, D.F.; Lennox, G.G.; et al. ATM mutations and phenotypes in ataxia-telangiectasia families in the British Isles: Expression of mutant ATM and the risk of leukemia, lymphoma, and breast cancer. Am. J. Hum. Genet. 1998, 62, 334–345. [Google Scholar] [CrossRef]
- Spring, K.; Cross, S.; Li, C.; Watters, D.; Ben-Senior, L.; Waring, P.; Ahangari, F.; Lu, S.I.; Chen, P.; Misko, I.; et al. Atm knock-in mice harboring an in-frame deletion corresponding to the human ATM 7636de19 common mutation exhibit a variant phenotype. Cancer Res. 2001, 61, 4561–4568. [Google Scholar]
- Navrkalova, V.; Sebejova, L.; Zemanova, J.; Kminkova, J.; Kubesova, B.; Malcikova, J.; Mraz, M.; Smardova, J.; Pavlova, S.; Doubek, M.; et al. Atm mutations uniformly lead to ATM dysfunction in chronic lymphocytic leukemia: Application of functional test using doxorubicin. Haematologica 2013, 98, 1124–1131. [Google Scholar] [CrossRef][Green Version]
- Tamaichi, H.; Sato, M.; Porter, A.C.G.; Shimizu, T.; Mizutani, S.; Takagi, M. Ataxia telangiectasia mutated-dependent regulation of topoisomerase II alpha expression and sensitivity to topoisomerase II inhibitor. Cancer Sci. 2013, 104, 178–184. [Google Scholar] [CrossRef]
- Mei, L.; Zhang, J.; He, K.; Zhang, J. Ataxia telangiectasia and Rad3-related inhibitors and cancer therapy: Where we stand. J. Hematol. Oncol. 2019, 12. [Google Scholar] [CrossRef]
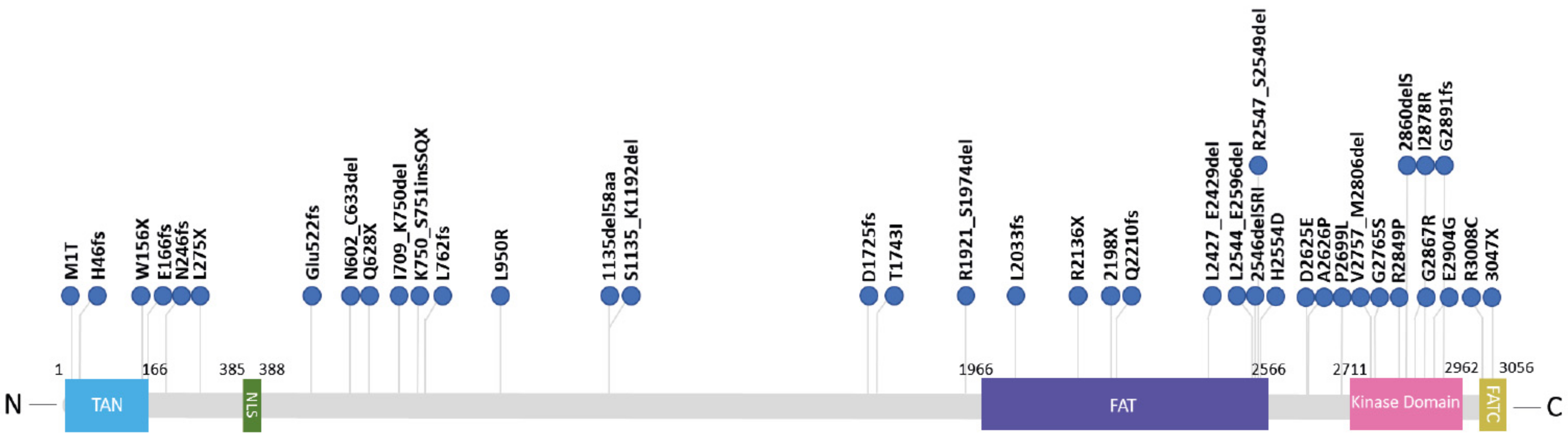
| Patients | Mutations | Amino Acids | %Protein Expression Relative to Normal Level | Associated Tumors | References |
|---|---|---|---|---|---|
| A, B (hom) | 7875T→G; 7876G→C | D2625E-A2626P | ND | [28,49] (15I,15II) | |
| Aa034 (het) | 8546G→C | R2849P | 0% | [26] | |
| Aa027 (het) | 8599G→C | G2867R | 2% | ||
| AT41RM (hom) | 8711A→G | E2904G | ND | [44] * | |
| AT9RM (hom) | 9139C→T | 3047X | 16.8% | [23] * | |
| AT31RM, A-T2RM, AT35RM (het) | 3403del174; 3576G→A | 1135del58aa | 4.8%, 5.4%, 1.0% | [23] * | |
| AT53RM | 6572ins7 | 2198X | 11.7% | [23] * | |
| 39-4 LCL | 7636del9 | 2546delSRI | ND ** | T-ALL, TCL | [33,50] |
| AT173 (het) | 9022C→T | R3008C | 50% | [46] | |
| (het) | 8189A→C | Q2730P | ≈100% | [51] | |
| (het) | 9022C→T; IVS10-6T→G | R3008C | ND | Myeloma | [52] |
| 9 (het) | 7875T→G; 7876G→C | D2625E-A2626P | ND | [28] | |
| 10, 11. I, 11.II (hom) | 3576G→A | S1135_K1192del | Dermatofibrosarcoma | ||
| 12I, 12II ND | 5762-2 A→T | R1921_S1974del | |||
| 13 (het) | 6629delA; 8578_8580delTCT | Q2210fs 2860delS | |||
| 14.I, 14.II, 15.I, 15.II (hom) | 7875T→G; 7876G→C | D2625E-A2626P | T-ALL, lymphoma | ||
| 16 (het) | 7875T→G; 7876G→C; 8578_8580delTCT | D2625E-A2626P; S2860del | Breast cancer | ||
| 17.I, 17.II ND | 8633T→G | I2878R | Hodgkin | ||
| AT73 | 1563_1564delAG; ND | Glu522fs | >50% | B cell lymphoma C859 | [29] |
| AT76 | 7660C→G; 824delT | H2554D; L275X | B-cell lymphoma | ||
| AT82 | 5172dupA; ND | D1725fs | Hodgkin’s lymphoma T-ALL C813, C910, C845 | ||
| AT95-1 | 2250G→A; 8786+1G→A | I709_K750del; G2891fs | Lymphoma C835, C857 | ||
| AT130 | 468G→A; ND | W156X | Ganglioglioma | ||
| AT134 | 1898+2T→G; Large Genomic Deletion | N602_C633del | Leukaemia/lymphoma C910 | ||
| AT37 | 6198+1G→A; ND | L2033fs | Burkitt lymphoma C837 | ||
| AT123 | 1882C→T; 8418+5_8delGTGA | Q628X; V2757_M2806del | Burkitt like lymphoma C833 | ||
| AT72-2 | 2284_2285delCT; 7280_7288del9 | L762fs; L2427_E2429del | T-PLL C910 | ||
| AT21 | 497-ND 662+ND del; 7638_7646del9 | E166fs; R2547_S2549del | Thyroid tumour C739 | ||
| AT103 | 2T→C; 3760ins2 | M1T | lymphoma | ||
| AT5-2 and AT5-1 | 2T→C; 6405_6406insTT | M1T; R2136X | Pancreatic C259; T-PLL | ||
| AT19 | 5228C→T;2251-10T→G | T1743I K750_S751insSQX | T-PLL C913 | ||
| AT22-3 | 7638_7646del9; ND | R2547_S2549del | T-ALL | ||
| AT8 | 8418+5_8delGTGA; 2849T→G | V2757_M2806del; L950R | T-ALL C910 | ||
| AT39-1 and AT39-2 | 138_141delTTCA; 7638_7646del9 | H46fs; R2547_S2549del | T-cell lymphoma C832; C850 | ||
| ATNe45-2 (hom) | 3576G→A | S1135_K1192del | >50% | Dermatofibrosarcoma protuberans | |
| ATNe42-1 And ATNe42-2 | 633T →G; ND | I2878R | M.Hodgkin | ||
| ATNe31 | 738_739delinsA; 7875_7876delinsGC | N246fs; A2626P | B cell lymphoma | ||
| ATNe10-2 And ATNe34-2 (hom) | 7875_7876delinsGC | A2626P | ALL; Lymphoma | ||
| ATNe33 | 7875_7876delinsGC; 8578_8580delTCT | A2626P; S2860del | Breast | ||
| AT47 | 7630-ND 7788+NDdel; 8293G→A | L2544_E2596del; G2765S | ≈80% | Breast C509 | |
| AT111 | 2249A→G; 8293G→A | I709_K750del; G2765S | Myeloid leukaemia | ||
| AT153 | 8096C→T; 2250G→A | P2699L; I709_K750del | ≈50% | Lymphoma C835 |
| Mutations | Amino Acids | References |
|---|---|---|
| 8546G→C | R2849P | [53] |
| 8599G→C | G2867R | |
| 7987del GTT | V2662del | |
| 7636del9 | 2546delSRI | |
| 7987delGTT | V2662del | |
| 6056A→G | Y2019C | [48] |
| 7181C→T | S2394L | |
| 7660C→G | H2554D | |
| 8189A→C | Q2730P | |
| 8565_8566 delTGinsAA | SV2855RI | |
| 7278_7283del6 | 2426delLR | |
| 7638_7646del9 | 2546delSRI | |
| 7013T→C | L2338P | |
| 7355T→C | L2452P | |
| 8096C→T | P2699L | |
| 8293G→A | G2765S | |
| 9022C>T | R3008C | |
| 8264_8268del5 | 2717delGL | |
| 9139C→T | R3047X |
| Murine Amino Acid Substitution (Human Amino Acid Substitution) | References |
|---|---|
| D2880A/N2885K (D2870A/N2875K) | [32,64,67] |
| Q2740P (Q2730P) | [65] |
| D2899A (D2889A) | [65] |
| 2556delSRI (2546delSRI) | [70] |
| R3016 (R3008H) | [69] * |
| R3057X (R3047X) | [68] * |
| Kinase Dead Mutations in Oncologic Patients | Amino Acid | Tumors | References |
|---|---|---|---|
| 7271T→G | V2424G | Breast B-CLL | [74,77] |
| 7775C→G | S2592C | Breast | [53] |
| 7636del9 9022C→T 9023G→A * 9139C→T * 8084G→C * 8266A→T * 8174A→T | 2546delSRI R3008C R3008H R3047X G2695A K2756X D2725V | T-PLL | [74] |
| 7181C→T | S2394L | Myeloma | [52] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Putti, S.; Giovinazzo, A.; Merolle, M.; Falchetti, M.L.; Pellegrini, M. ATM Kinase Dead: From Ataxia Telangiectasia Syndrome to Cancer. Cancers 2021, 13, 5498. https://doi.org/10.3390/cancers13215498
Putti S, Giovinazzo A, Merolle M, Falchetti ML, Pellegrini M. ATM Kinase Dead: From Ataxia Telangiectasia Syndrome to Cancer. Cancers. 2021; 13(21):5498. https://doi.org/10.3390/cancers13215498
Chicago/Turabian StylePutti, Sabrina, Alessandro Giovinazzo, Matilde Merolle, Maria Laura Falchetti, and Manuela Pellegrini. 2021. "ATM Kinase Dead: From Ataxia Telangiectasia Syndrome to Cancer" Cancers 13, no. 21: 5498. https://doi.org/10.3390/cancers13215498
APA StylePutti, S., Giovinazzo, A., Merolle, M., Falchetti, M. L., & Pellegrini, M. (2021). ATM Kinase Dead: From Ataxia Telangiectasia Syndrome to Cancer. Cancers, 13(21), 5498. https://doi.org/10.3390/cancers13215498








