Epithelial to Mesenchymal Transition: Key Regulator of Pancreatic Ductal Adenocarcinoma Progression and Chemoresistance
Abstract
:Simple Summary
Abstract
1. Introduction
2. Epithelial to Mesenchymal Transition and Cancer
3. Epithelial to Mesenchymal Transition in PDAC
4. Pathways Regulating Epithelial to Mesenchymal Transition in PDAC
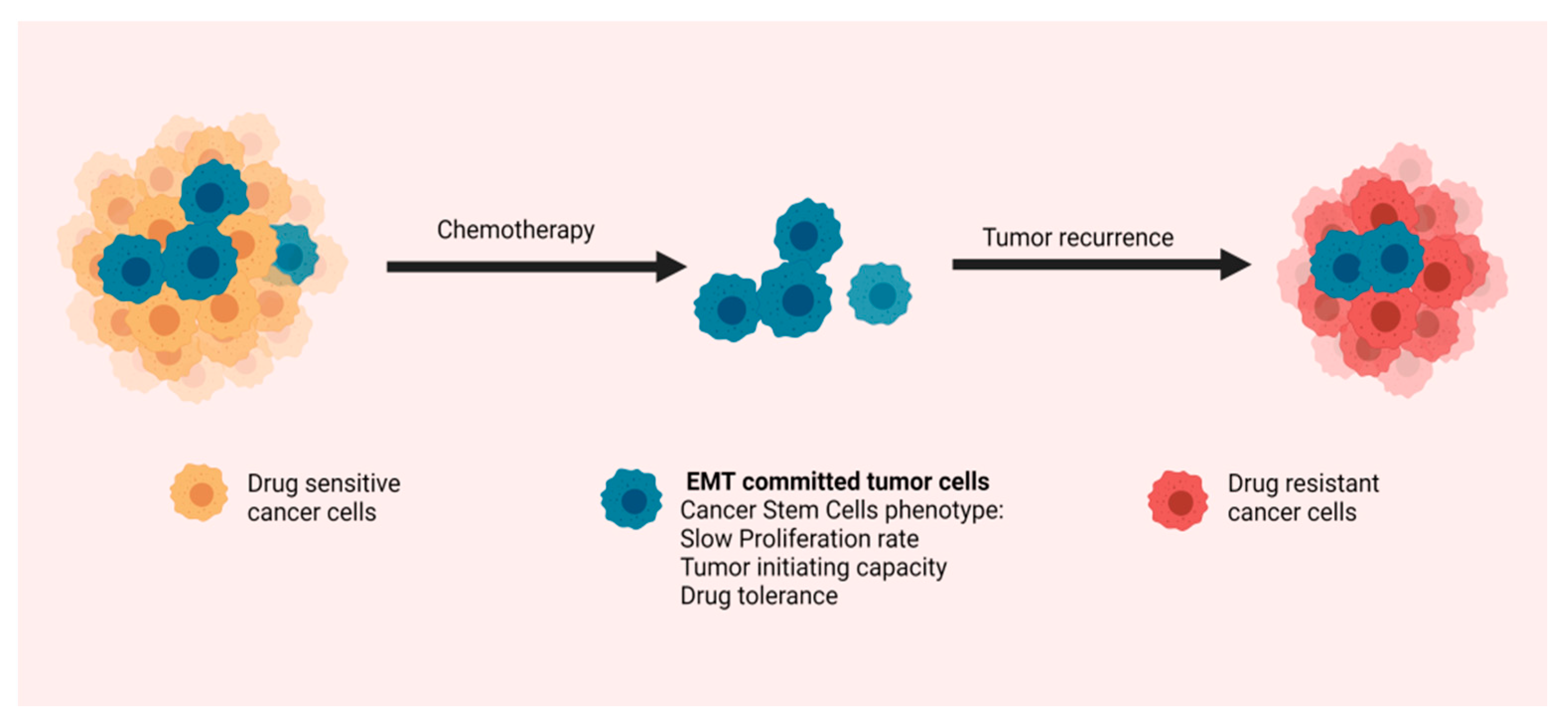
5. Epithelial-to-Mesenchymal Transition and Chemoresistance in Pancreatic Ductal Adenocarcinoma
6. Pharmacological Targeting of Epithelial to Mesenchymal Transition
7. Conclusions—Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.D.; Fidler-Benaoudia, M.; Keegan, T.H.; Hipp, H.S.; Jemal, A.; Siegel, R.L. Cancer statistics for adolescents and young adults, 2020. CA Cancer J. Clin. 2020, 70, 443–459. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P.; Sunkara, T.; Gaduputi, V. Epidemiology of Pancreatic Cancer: Global Trends, Etiology and Risk Factors. World J. Oncol. 2019, 10, 10–27. [Google Scholar] [CrossRef] [PubMed]
- Kleeff, J.; Korc, M.; Apte, M.; La Vecchia, C.; Johnson, C.D.; Biankin, A.V.; Neale, R.E.; Tempero, M.; Tuveson, D.A.; Hruban, R.H.; et al. Pancreatic cancer. Nat. Rev. Dis. Primers 2016, 2, 16022. [Google Scholar] [CrossRef]
- Kuehn, B.M. Looking to Long-term Survivors for Improved Pancreatic Cancer Treatment. JAMA 2020, 324, 2242–2244. [Google Scholar] [CrossRef]
- Picozzi, V.J.; Oh, S.Y.; Edwards, A.; Mandelson, M.T.; Dorer, R.; Rocha, F.G.; Alseidi, A.; Biehl, T.; Traverso, L.W.; Helton, W.S.; et al. Five-Year Actual Overall Survival in Resected Pancreatic Cancer: A Contemporary Single-Institution Experience from a Multidisciplinary Perspective. Ann. Surg. Oncol. 2017, 24, 1722–1730. [Google Scholar] [CrossRef]
- Conroy, T.; Hammel, P.; Hebbar, M.; Ben Abdelghani, M.; Wei, A.C.; Raoul, J.L.; Choné, L.; Francois, E.; Artru, P.; Biagi, J.J.; et al. Canadian Cancer Trials Group and the Unicancer-GI–PRODIGE Group. FOLFIRINOX or Gemcitabine as Adjuvant Therapy for Pancreatic Cancer. N. Engl. J. Med. 2018, 379, 2395–2406. [Google Scholar] [CrossRef]
- Amrutkar, M.; Gladhaug, I.P. Pancreatic Cancer Chemoresistance to Gemcitabine. Cancers 2017, 9, 157. [Google Scholar] [CrossRef] [Green Version]
- Neoptolemos, J.P.; Kleeff, J.; Michl, P.; Costello, E.; Greenhalf, W.; Palmer, D.H. Therapeutic developments in pancreatic cancer: Current and future perspectives. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 333–348. [Google Scholar] [CrossRef]
- Zeng, S.; Pöttler, M.; Lan, B.; Grützmann, R.; Pilarsky, C.; Yang, H. Chemoresistance in Pancreatic Cancer. Int. J. Mol. Sci. 2019, 20, 4504. [Google Scholar] [CrossRef] [Green Version]
- Hosein, A.N.; Brekken, R.A.; Maitra, A. Pancreatic cancer stroma: An update on therapeutic targeting strategies. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 487–505. [Google Scholar] [CrossRef]
- Nagathihalli, N.S.; Castellanos, J.A.; Shi, C.; Beesetty, Y.; Reyzer, M.L.; Caprioli, R.; Chen, X.; Walsh, A.; Skala, M.C.; Moses, H.L.; et al. Signal Transducer and Activator of Transcription 3, Mediated Remodeling of the Tumor Microenvironment Results in Enhanced Tumor Drug Delivery in a Mouse Model of Pancreatic Cancer. Gastroenterology 2015, 149, 1932–1943.e9. [Google Scholar] [CrossRef] [Green Version]
- Olive, K.P.; Jacobetz, M.A.; Davidson, C.J.; Gopinathan, A.; McIntyre, D.; Honess, D.; Madhu, B.; Goldgraben, M.A.; Caldwell, M.E.; Allard, D.; et al. Inhibition of Hedgehog Signaling Enhances Delivery of Chemotherapy in a Mouse Model of Pancreatic Cancer. Science 2009, 324, 1457–1461. [Google Scholar] [CrossRef] [Green Version]
- Provenzano, P.P.; Cuevas, C.; Chang, A.E.; Goel, V.K.; Von Hoff, D.D.; Hingorani, S.R. Enzymatic Targeting of the Stroma Ablates Physical Barriers to Treatment of Pancreatic Ductal Adenocarcinoma. Cancer Cell 2012, 21, 418–429. [Google Scholar] [CrossRef] [Green Version]
- Hezel, A.F.; Kimmelman, A.C.; Stanger, B.Z.; Bardeesy, N.; Depinho, R.A. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006, 20, 1218–1249. [Google Scholar] [CrossRef] [Green Version]
- Makohon-Moore, A.; Iacobuzio-Donahue, C.A. Pancreatic cancer biology and genetics from an evolutionary perspective. Nat. Rev. Cancer 2016, 16, 553–565. [Google Scholar] [CrossRef] [Green Version]
- Rooman, I.; Real, F.X. Pancreatic ductal adenocarcinoma and acinar cells: A matter of differentiation and development? Gut 2011, 61, 449–458. [Google Scholar] [CrossRef]
- Stanger, B.Z.; Stiles, B.; Lauwers, G.Y.; Bardeesy, N.; Mendoza, M.; Wang, Y.; Greenwood, A.; Cheng, K.-H.; McLaughlin, M.; Brown, D.; et al. Pten constrains centroacinar cell expansion and malignant transformation in the pancreas. Cancer Cell 2005, 8, 185–195. [Google Scholar] [CrossRef] [Green Version]
- Storz, P. Acinar cell plasticity and development of pancreatic ductal adenocarcinoma. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 296–304. [Google Scholar] [CrossRef]
- Hruban, R.H.; Adsay, N.V.; Albores–Saavedra, J.; Compton, C.; Garrett, E.S.; Goodman, S.N.; Kern, S.E.; Klimstra, D.S.; Klöppel, G.; Longnecker, D.S.; et al. Pancreatic intraepithelial neoplasia: A new nomenclature and classification system for pancreatic duct lesions. Am. J. Surg. Pathol. 2001, 25, 579–586. [Google Scholar] [CrossRef]
- Hruban, R.H.; Goggins, M.; Parsons, J.; Kern, E.S. Progression model for pancreatic cancer. Clin. Cancer Res. 2000, 6, 2969–2972. [Google Scholar]
- Aguirre, A.J.; Bardeesy, N.; Sinha, M.; Lopez, L.; Tuveson, D.A.; Horner, J.; Redston, M.S.; Depinho, R.A. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003, 17, 3112–3126. [Google Scholar] [CrossRef] [Green Version]
- Clark, C.E.; Hingorani, S.R.; Mick, R.; Combs, C.; Tuveson, D.A.; Vonderheide, R.H.; Hitchins, M.P.; Ap Lin, V.; Buckle, A.; Cheong, K.; et al. Dynamics of the Immune Reaction to Pancreatic Cancer from Inception to Invasion. Cancer Res. 2007, 67, 9518–9527. [Google Scholar] [CrossRef] [Green Version]
- Neesse, A.; Bauer, C.A.; Öhlund, D.; Lauth, M.; Buchholz, M.; Michl, P.; Tuveson, D.A.; Gress, T. Stromal biology and therapy in pancreatic cancer: Ready for clinical translation? Gut 2019, 68, 159–171. [Google Scholar] [CrossRef]
- Hay, E.D. The mesenchymal cell, its role in the embryo, and the remarkable signaling mechanisms that create it. Dev. Dyn. 2005, 233, 706–720. [Google Scholar] [CrossRef]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428, Erratum in: J. Clin. Investig. 2010, 120, 1786. [Google Scholar] [CrossRef] [Green Version]
- Nieto, M.A.; Huang, R.Y.-J.; Jackson, R.A.; Thiery, J.P. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef] [Green Version]
- Shook, D.; Keller, R. Mechanisms, mechanics and function of epithelial–mesenchymal transitions in early development. Mech. Dev. 2003, 120, 1351–1383. [Google Scholar] [CrossRef]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [Green Version]
- De Craene, B.; Berx, G. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer 2013, 13, 97–110. [Google Scholar] [CrossRef]
- Lambert, A.W.; Pattabiraman, D.R.; Weinberg, R.A. Emerging Biological Principles of Metastasis. Cell 2017, 168, 670–691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pastushenko, I.; Blanpain, C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol. 2019, 29, 212–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pastushenko, I.; Brisebarre, A.; Sifrim, A.; Fioramonti, M.; Revenco, T.; Boumahdi, S.; Van Keymeulen, A.; Brown, D.; Moers, V.; Lemaire, S.; et al. Identification of the tumour transition states occurring during EMT. Nature 2018, 556, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Oskarsson, T.; Batlle, E.; Massagué, J. Metastatic Stem Cells: Sources, Niches, and Vital Pathways. Cell Stem Cell 2014, 14, 306–321. [Google Scholar] [CrossRef] [Green Version]
- Shibue, T.; Weinberg, T.S.R.A. EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 2017, 14, 611–629. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.; Vanner, R.; Dirks, P.B.; Eaves, C.J. Cancer stem cells: An evolving concept. Nat. Rev. Cancer 2012, 12, 133–143. [Google Scholar] [CrossRef]
- Mani, S.A.; Guo, W.; Liao, M.-J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The Epithelial-Mesenchymal Transition Generates Cells with Properties of Stem Cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef] [Green Version]
- Morel, A.-P.; Lièvre, M.; Thomas, C.; Hinkal, G.; Ansieau, S.; Puisieux, A. Generation of Breast Cancer Stem Cells through Epithelial-Mesenchymal Transition. PLoS ONE 2008, 3, e2888. [Google Scholar] [CrossRef]
- Friedl, P.; Locker, J.; Sahai, E.; Segall, J.E. Classifying collective cancer cell invasion. Nat. Cell Biol. 2012, 14, 777–783. [Google Scholar] [CrossRef]
- Chung, Y.-C.; Wei, W.-C.; Kuo, J.-F.; Hsu, C.-P.; Chang, K.-J.; Chao, W.-T.; Hung, C.-N. Rab11 collaborates E-cadherin to promote collective cell migration and indicates a poor prognosis in colorectal carcinoma. Eur. J. Clin. Investig. 2016, 46, 1002–1011. [Google Scholar] [CrossRef]
- Veracini, L.; Grall, D.; Schaub, S.; Divonne, S.B.-D.L.F.; Etienne-Grimaldi, M.-C.; Milano, G.; Bozec, A.; Babin, E.; Sudaka, A.; Thariat, J.; et al. Elevated Src family kinase activity stabilizes E-cadherin-based junctions and collective movement of head and neck squamous cell carcinomas. Oncotarget 2015, 6, 7570–7583. [Google Scholar] [CrossRef] [Green Version]
- Friedl, P.; Gilmour, D. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 2009, 10, 445–457. [Google Scholar] [CrossRef]
- Westcott, J.M.; Prechtl, A.M.; Maine, E.A.; Dang, T.; Esparza, M.; Sun, H.; Zhou, Y.; Xie, Y.; Pearson, G.W. An epigenetically distinct breast cancer cell subpopulation promotes collective invasion. J. Clin. Investig. 2015, 125, 1927–1943. [Google Scholar] [CrossRef]
- Ye, X.; Tam, W.L.; Shibue, T.; Kaygusuz, Y.; Reinhardt, F.; Eaton, E.N.; Weinberg, R.A. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nat. Cell Biol. 2015, 525, 256–260. [Google Scholar] [CrossRef] [Green Version]
- Grigore, A.D.; Jolly, M.K.; Jia, D.; Farach-Carson, M.C.; Levine, H. Tumor Budding: The Name is EMT. Partial EMT. J. Clin. Med. 2016, 5, 51. [Google Scholar] [CrossRef]
- Lugli, A.; Zlobec, I.; Berger, M.D.; Kirsch, R.; Nagtegaal, I.D. Tumour budding in solid cancers. Nat. Rev. Clin. Oncol. 2021, 18, 101–115. [Google Scholar] [CrossRef]
- Attramadal, C.G.; Kumar, S.; Boysen, M.E.; Dhakal, H.P.; Nesland, J.M.; Bryne, M. Tumor Budding, EMT and Cancer Stem Cells in T1-2/N0 Oral Squamous Cell Carcinomas. Anticancer. Res. 2015, 35, 6111–6120. [Google Scholar]
- Jensen, D.; Dabelsteen, E.; Specht, L.; Fiehn, A.; Therkildsen, M.; Jønson, L.; Vikesaa, J.; Nielsen, F.; von Buchwald, C. Molecular profiling of tumour budding implicates TGFβ-mediated epithelial-mesenchymal transition as a therapeutic target in oral squamous cell carcinoma. J. Pathol. 2015, 236, 505–516. [Google Scholar] [CrossRef]
- Nakagawa, Y.; Ohira, M.; Kubo, N.; Yamashita, Y.; Sakurai, K.; Toyokawa, T.; Tanaka, H.; Muguruma, K.; Shibutani, M.; Yamazoe, S.; et al. Tumor budding and E-cadherin expression are useful predictors of nodal involvement in T1 esophageal squamous cell carcinoma. Anticancer. Res. 2013, 33, 5023–5029. [Google Scholar]
- Koyuncuoglu, M.; Okyay, E.; Saatli, B.; Olgan, S.; Akin, M.; Saygili, U. Tumor budding and E-Cadherin expression in endometrial carcinoma: Are they prognostic factors in endometrial cancer? Gynecol. Oncol. 2012, 125, 208–213. [Google Scholar] [CrossRef]
- Lee, S.-J.; Choi, S.Y.; Kim, W.-J.; Ji, M.; Lee, T.-G.; Son, B.-R.; Yoon, S.M.; Sung, R.; Lee, E.J.; Youn, S.J.; et al. Combined aberrant expression of E-cadherin and S100A4, but not β-catenin is associated with disease-free survival and overall survival in colorectal cancer patients. Diagn. Pathol. 2013, 8, 99. [Google Scholar] [CrossRef] [Green Version]
- Zlobec, I.; Lugli, A.; Baker, K.; Roth, S.; Minoo, P.; Hayashi, S.; Terracciano, L.; Jass, J.R. Role of APAF-1, E-cadherin and peritumoural lymphocytic infiltration in tumour budding in colorectal cancer. J. Pathol. 2007, 212, 260–268. [Google Scholar] [CrossRef]
- Kohler, I.; Bronsert, P.; Timme, S.; Werner, M.; Brabletz, T.; Hopt, U.T.; Schilling, O.; Bausch, D.; Keck, T.; Wellner, U.F. Detailed analysis of epithelial-mesenchymal transition and tumor budding identifies predictors of long-term survival in pancreatic ductal adenocarcinoma. J. Gastroenterol. Hepatol. 2015, 30 (Suppl. 1), 78–84. [Google Scholar] [CrossRef]
- Galván, J.A.; Zlobec, I.; Wartenberg, M.; Lugli, A.; Gloor, B.; Perren, A.; Karamitopoulou, E. Expression of E-cadherin repressors SNAIL, ZEB1 and ZEB2 by tumour and stromal cells influences tumour-budding phenotype and suggests heterogeneity of stromal cells in pancreatic cancer. Br. J. Cancer 2015, 112, 1944–1950. [Google Scholar] [CrossRef] [Green Version]
- Hotz, B.; Arndt, M.; Dullat, S.; Bhargava, S.; Buhr, H.-J.; Hotz, H.G. Epithelial to Mesenchymal Transition: Expression of the Regulators Snail, Slug, and Twist in Pancreatic Cancer. Clin. Cancer Res. 2007, 13, 4769–4776. [Google Scholar] [CrossRef] [Green Version]
- Stemmler, M.P.; Eccles, R.L.; Brabletz, S.; Brabletz, T. Non-redundant functions of EMT transcription factors. Nat. Cell Biol. 2019, 21, 102–112. [Google Scholar] [CrossRef]
- Yamada, S.; Fuchs, B.C.; Fujii, T.; Shimoyama, Y.; Sugimoto, H.; Nomoto, S.; Takeda, S.; Tanabe, K.K.; Kodera, Y.; Nakao, A. Epithelial-to-mesenchymal transition predicts prognosis of pancreatic cancer. Surgery 2013, 154, 946–954. [Google Scholar] [CrossRef]
- Lawlor, R.T.; Veronese, N.; Nottegar, A.; Malleo, G.; Smith, L.; Demurtas, J.; Cheng, L.; Wood, L.D.; Silvestris, N.; Salvia, R.; et al. Prognostic Role of High-Grade Tumor Budding in Pancreatic Ductal Adenocarcinoma: A Systematic Review and Meta-Analysis with a Focus on Epithelial to Mesenchymal Transition. Cancers 2019, 11, 113. [Google Scholar] [CrossRef] [Green Version]
- Chouat, E.; Zehani, A.; Chelly, I.; Njima, M.; Maghrebi, H.; Bani, M.A.; Njim, L.; Zakhama, A.; Haouet, S.; Kchir, N. Tumor budding is a prognostic factor linked to epithelial mesenchymal transition in pancreatic ductal adenocarcinoma. Study report and literature review. Pancreatology 2018, 18, 79–84. [Google Scholar] [CrossRef]
- Lapshyn, H.; Bolm, L.; Kohler, I.; Werner, M.; Billmann, F.G.; Bausch, D.; Hopt, U.T.; Makowiec, F.; Wittel, U.A.; Keck, T.; et al. Histopathological tumor invasion of the mesenterico-portal vein is characterized by aggressive biology and stromal fibroblast activation. HPB 2017, 19, 67–74. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.-N.; Lv, A.; Tian, Z.-H.; Tian, X.-Y.; Guan, X.-Y.; Dong, B.; Zhao, M.; Hao, C.-Y. Superior mesenteric artery margin in pancreaticoduodenectomy for pancreatic adenocarcinoma. Oncotarget 2017, 8, 7766–7776. [Google Scholar] [CrossRef] [PubMed]
- Wartenberg, M.; Cibin, S.; Zlobec, I.; Vassella, E.; Eppenberger-Castori, S.; Terracciano, L.; Eichmann, M.; Worni, M.; Gloor, B.; Perren, A.; et al. Integrated Genomic and Immunophenotypic Classification of Pancreatic Cancer Reveals Three Distinct Subtypes with Prognostic/Predictive Significance. Clin. Cancer Res. 2018, 24, 4444–4454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karamitopoulou, E.; Zlobec, I.; Born, D.; Kondi-Pafiti, A.; Lykoudis, P.; Mellou, A.; Gennatas, K.; Gloor, B.; Lugli, A. Tumour budding is a strong and independent prognostic factor in pancreatic cancer. Eur. J. Cancer 2013, 49, 1032–1039. [Google Scholar] [CrossRef] [PubMed]
- Lohneis, P.; Sinn, M.; Klein, F.; Bischoff, S.; Striefler, J.K.; Wislocka, L.; Sinn, B.V.; Pelzer, U.; Oettle, H.; Riess, H.; et al. Tumour buds determine prognosis in resected pancreatic ductal adenocarcinoma. Br. J. Cancer 2018, 118, 1485–1491. [Google Scholar] [CrossRef]
- O’Connor, K.; Li-Chang, H.H.; Kalloger, S.E.; Peixoto, R.D.; Webber, D.L.; Owen, D.A.; Driman, D.K.; Kirsch, R.; Serra, S.; Scudamore, C.H.; et al. Tumor Budding Is an Independent Adverse Prognostic Factor in Pancreatic Ductal Adenocarcinoma. Am. J. Surg. Pathol. 2015, 39, 472–478. [Google Scholar] [CrossRef]
- Zhang, L.; Department of General Surgery; Guo, L.; Tao, M.; Fu, W.; Xiu, D. Parasympathetic neurogenesis is strongly associated with tumor budding and correlates with an adverse prognosis in pancreatic ductal adenocarcinoma. Chin. J. Cancer Res. 2016, 28, 180–186. [Google Scholar] [CrossRef]
- Cairns, J. Mutation selection and the natural history of cancer. Nat. Cell Biol. 1975, 255, 197–200. [Google Scholar] [CrossRef]
- Hellman, S. Karnofsky Memorial Lecture. Natural history of small breast cancers. J. Clin. Oncol. 1994, 12, 2229–2234. [Google Scholar] [CrossRef]
- Klein, C.A. Parallel progression of primary tumours and metastases. Nat. Rev. Cancer 2009, 9, 302–312. [Google Scholar] [CrossRef]
- Sänger, N.; Effenberger, K.E.; Riethdorf, S.; Van Haasteren, V.; Gauwerky, J.; Wiegratz, I.; Strebhardt, K.; Kaufmann, M.; Pantel, K. Disseminated tumor cells in the bone marrow of patients with ductal carcinoma in situ. Int. J. Cancer 2011, 129, 2522–2526. [Google Scholar] [CrossRef]
- Hingorani, S.R.; Wang, L.; Multani, A.S.; Combs, C.; Deramaudt, T.B.; Hruban, R.H.; Rustgi, A.K.; Chang, S.; Tuveson, D.A. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 2005, 7, 469–483. [Google Scholar] [CrossRef] [Green Version]
- Rhim, A.D.; Mirek, E.T.; Aiello, N.M.; Maitra, A.; Bailey, J.M.; McAllister, F.; Reichert, M.; Beatty, G.L.; Rustgi, A.K.; Vonderheide, R.H.; et al. EMT and Dissemination Precede Pancreatic Tumor Formation. Cell 2012, 148, 349–361. [Google Scholar] [CrossRef] [Green Version]
- Yachida, S.; Jones, S.; Bozic, I.; Antal, T.; Leary, R.; Fu, B.; Kamiyama, M.; Hruban, R.H.; Eshleman, J.R.; Nowak, M.A.; et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nat. Cell Biol. 2010, 467, 1114–1117. [Google Scholar] [CrossRef] [Green Version]
- Guerra, C.; Collado, M.; Navas, C.; Schuhmacher, A.J.; Hernández-Porras, I.; Cañamero, M.; Rodriguez-Justo, M.; Serrano, M.; Barbacid, M. Pancreatitis-Induced Inflammation Contributes to Pancreatic Cancer by Inhibiting Oncogene-Induced Senescence. Cancer Cell 2011, 19, 728–739. [Google Scholar] [CrossRef] [Green Version]
- Guerra, C.; Schuhmacher, A.J.; Cañamero, M.; Grippo, P.J.; Verdaguer, L.; Pérez-Gallego, L.; Dubus, P.; Sandgren, E.P.; Barbacid, M. Chronic Pancreatitis Is Essential for Induction of Pancreatic Ductal Adenocarcinoma by K-Ras Oncogenes in Adult Mice. Cancer Cell 2007, 11, 291–302. [Google Scholar] [CrossRef] [Green Version]
- Bulle, A.; Lim, K.-H. Beyond just a tight fortress: Contribution of stroma to epithelial-mesenchymal transition in pancreatic cancer. Signal Transduct. Target. Ther. 2020, 5, 1–12. [Google Scholar] [CrossRef]
- Carstens, J.L.; Yang, S.; de Sampaio, P.C.; Zheng, X.; Barua, S.; McAndrews, K.M.; Rao, A.; Burks, J.K.; Rhim, A.D.; Kalluri, R. Stabilized epithelial phenotype of cancer cells in primary tumors leads to increased colonization of liver metastasis in pancreatic cancer. Cell Rep. 2021, 35, 108990. [Google Scholar] [CrossRef]
- Li, C.; Heidt, D.G.; Dalerba, P.; Burant, C.F.; Zhang, L.; Adsay, V.; Wicha, M.; Clarke, M.F.; Simeone, D.M. Identification of Pancreatic Cancer Stem Cells. Cancer Res. 2007, 67, 1030–1037. [Google Scholar] [CrossRef] [Green Version]
- Rao, C.V.; Mohammed, A. New insights into pancreatic cancer stem cells. World J. Stem Cells 2015, 7, 547–555. [Google Scholar] [CrossRef]
- Bailey, J.M.; Alsina, J.; Rasheed, Z.A.; McAllister, F.M.; Fu, Y.-Y.; Plentz, R.; Zhang, H.; Pasricha, P.J.; Bardeesy, N.; Matsui, W.; et al. DCLK1 Marks a Morphologically Distinct Subpopulation of Cells With Stem Cell Properties in Preinvasive Pancreatic Cancer. Gastroenterology 2014, 146, 245–256. [Google Scholar] [CrossRef] [Green Version]
- Hermann, P.C.; Huber, S.L.; Herrler, T.; Aicher, A.; Ellwart, J.W.; Guba, M.; Bruns, C.J.; Heeschen, C. Distinct Populations of Cancer Stem Cells Determine Tumor Growth and Metastatic Activity in Human Pancreatic Cancer. Cell Stem Cell 2007, 1, 313–323. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.P.; Fleming, J.B.; Wang, H.; Abbruzzese, J.L.; Choi, W.; Kopetz, S.; McConkey, D.J.; Evans, D.B.; Gallick, G.E. ALDH Activity Selectively Defines an Enhanced Tumor-Initiating Cell Population Relative to CD133 Expression in Human Pancreatic Adenocarcinoma. PLoS ONE 2011, 6, e20636. [Google Scholar] [CrossRef]
- Ohara, Y.; Oda, T.; Sugano, M.; Hashimoto, S.; Enomoto, T.; Yamada, K.; Akashi, Y.; Miyamoto, R.; Kobayashi, A.; Fukunaga, K.; et al. Histological and prognostic importance of CD44+/CD24+/EpCAM+expression in clinical pancreatic cancer. Cancer Sci. 2013, 104, 1127–1134. [Google Scholar] [CrossRef]
- Li, C.; Wu, J.; Hynes, M.; Dosch, J.; Sarkar, B.; Welling, T.H.; di Magliano, M.P.; Simeone, D.M. c-Met Is a Marker of Pancreatic Cancer Stem Cells and Therapeutic Target. Gastroenterology 2011, 141, 2218–2227.e5. [Google Scholar] [CrossRef]
- Helms, E.; Onate, M.K.; Sherman, M.H. Fibroblast Heterogeneity in the Pancreatic Tumor Microenvironment. Cancer Discov. 2020, 10, 648–656. [Google Scholar] [CrossRef] [Green Version]
- Öhlund, D.; Handly-Santana, A.; Biffi, G.; Elyada, E.; Almeida, A.S.; Ponz-Sarvise, M.; Corbo, V.; Oni, T.E.; Hearn, S.A.; Lee, E.J.; et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 2017, 214, 579–596. [Google Scholar] [CrossRef]
- Biffi, G.; Oni, T.E.; Spielman, B.; Hao, Y.; Elyada, E.; Park, Y.; Preall, J.; Tuveson, D.A. IL1-Induced JAK/STAT Signaling Is Antagonized by TGFβ to Shape CAF Heterogeneity in Pancreatic Ductal Adenocarcinoma. Cancer Discov. 2019, 9, 282–301. [Google Scholar] [CrossRef] [Green Version]
- Elyada, E.; Bolisetty, M.; Laise, P.; Flynn, W.F.; Courtois, E.T.; Burkhart, R.A.; Teinor, J.A.; Belleau, P.; Biffi, G.; Lucito, M.S.; et al. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discov. 2019, 9, 1102–1123. [Google Scholar] [CrossRef] [Green Version]
- Özdemir, B.C.; Pentcheva-Hoang, T.; Carstens, J.L.; Zheng, X.; Wu, C.-C.; Simpson, T.R.; Laklai, H.; Sugimoto, H.; Kahlert, C.; Novitskiy, S.V.; et al. Depletion of Carcinoma-Associated Fibroblasts and Fibrosis Induces Immunosuppression and Accelerates Pancreas Cancer with Reduced Survival. Cancer Cell 2014, 25, 719–734. [Google Scholar] [CrossRef] [Green Version]
- Ligorio, M.; Sil, S.; Malagon-Lopez, J.; Nieman, L.; Misale, S.; Di Pilato, M.; Ebright, R.Y.; Karabacak, N.M.; Kulkarni, A.S.; Liu, A.; et al. Stromal Microenvironment Shapes the Intratumoral Architecture of Pancreatic Cancer. Cell 2019, 178, 160–175.e27. [Google Scholar] [CrossRef]
- Jang, J.-E.; Hajdu, C.H.; Liot, C.; Miller, G.; Dustin, M.L.; Bar-Sagi, D. Crosstalk between Regulatory T Cells and Tumor-Associated Dendritic Cells Negates Anti-tumor Immunity in Pancreatic Cancer. Cell Rep. 2017, 20, 558–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helm, O.; Held-Feindt, J.; Grage-Griebenow, E.; Reiling, N.; Ungefroren, H.; Vogel, I.; Krüger, U.; Becker, T.; Ebsen, M.; Röcken, C.; et al. Tumor-associated macrophages exhibit pro- and anti-inflammatory properties by which they impact on pancreatic tumorigenesis. Int. J. Cancer 2014, 135, 843–861. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Liu, Q.; Liao, Q. Tumor-Associated Macrophages in Pancreatic Ductal Adenocarcinoma: Origin, Polarization, Function, and Reprogramming. Front. Cell Dev. Biol. 2021, 8, 607209. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.Y.; Xu, J.Y.; Shi, X.Y.; Huang, W.; Ruan, T.Y.; Xie, P.; Ding, J.L. M2-polarized tumor-associated macrophages promoted epithelial–mesenchymal transition in pancreatic cancer cells, partially through TLR4/IL-10 signaling pathway. Lab. Investig. 2013, 93, 844–854. [Google Scholar] [CrossRef] [Green Version]
- Nomura, A.; Gupta, V.K.; Dauer, P.; Sharma, N.S.; Dudeja, V.; Merchant, N.; Saluja, A.K.; Banerjee, S. NFκB-Mediated Invasiveness in CD133+ Pancreatic TICs Is Regulated by Autocrine and Paracrine Activation of IL1 Signaling. Mol. Cancer Res. 2018, 16, 162–172. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Lu, P.; Shen, W.; He, R.; Yang, M.-W.; Fang, Y.; Sun, Y.-W.; Niu, N.; Xue, J. CD90 highly expressed population harbors a stemness signature and creates an immunosuppressive niche in pancreatic cancer. Cancer Lett. 2019, 453, 158–169. [Google Scholar] [CrossRef]
- Tekin, C.; Aberson, H.L.; Waasdorp, C.; Hooijer, G.K.J.; De Boer, O.J.; Dijk, F.; Bijlsma, M.F.; Spek, C.A. Macrophage-secreted MMP9 induces mesenchymal transition in pancreatic cancer cells via PAR1 activation. Cell. Oncol. 2020, 43, 1161–1174. [Google Scholar] [CrossRef]
- Goebel, L.; Grage-Griebenow, E.; Gorys, A.; Helm, O.; Genrich, G.; Lenk, L.; Wesch, D.; Ungefroren, H.; Freitag-Wolf, S.; Sipos, B.; et al. CD4+T cells potently induce epithelial-mesenchymal-transition in premalignant and malignant pancreatic ductal epithelial cells–novel implications of CD4+T cells in pancreatic cancer development. OncoImmunology 2015, 4, e1000083. [Google Scholar] [CrossRef] [Green Version]
- Wartenberg, M.; Zlobec, I.; Perren, A.; Koelzer, V.; Gloor, B.; Lugli, A.; Karamitopoulou, E. Accumulation of FOXP3+T-cells in the tumor microenvironment is associated with an epithelial-mesenchymal-transition-type tumor budding phenotype and is an independent prognostic factor in surgically resected pancreatic ductal adenocarcinoma. Oncotarget 2015, 6, 4190–4201. [Google Scholar] [CrossRef] [Green Version]
- Lanfranca, M.P.; Zhang, Y.; Girgis, A.; Kasselman, S.; Lazarus, J.; Kryczek, I.; Delrosario, L.; Rhim, A.; Koneva, L.; Sartor, M.; et al. Interleukin 22 Signaling Regulates Acinar Cell Plasticity to Promote Pancreatic Tumor Development in Mice. Gastroenterology 2020, 158, 1417–1432.e11. [Google Scholar] [CrossRef]
- Alvarez, M.A.; Freitas, J.P.; Hussain, S.M.; Glazer, E.S. TGF-β Inhibitors in Metastatic Pancreatic Ductal Adenocarcinoma. J. Gastrointest. Cancer 2019, 50, 207–213. [Google Scholar] [CrossRef]
- Nakao, A.; Imamura, T.; Souchelnytskyi, S.; Kawabata, M.; Ishisaki, A.; Oeda, E.; Tamaki, K.; Hanai, J.; Heldin, C.; Miyazono, K.; et al. TGF-beta receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 1997, 16, 5353–5362. [Google Scholar] [CrossRef] [Green Version]
- Siegel, P.M.; Massague, J. Cytostatic and apoptotic actions of TGF-β in homeostasis and cancer. Nat. Rev. Cancer 2003, 3, 807–820. [Google Scholar] [CrossRef]
- Xu, J.; Lamouille, S.; Derynck, R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009, 19, 156–172. [Google Scholar] [CrossRef]
- Principe, D.R.; Decant, B.; Mascariñas, E.; Wayne, E.A.; Diaz, A.M.; Akagi, N.; Hwang, R.; Pasche, B.; Dawson, W.; Fang, D.; et al. TGFβ Signaling in the Pancreatic Tumor Microenvironment Promotes Fibrosis and Immune Evasion to Facilitate Tumorigenesis. Cancer Res. 2016, 76, 2525–2539. [Google Scholar] [CrossRef] [Green Version]
- Ancrile, B.; Lim, K.-H.; Counter, C.M. Oncogenic Ras-induced secretion of IL6 is required for tumorigenesis. Genes Dev. 2007, 21, 1714–1719. [Google Scholar] [CrossRef] [Green Version]
- Nagathihalli, N.S.; Castellanos, J.A.; VanSaun, M.N.; Dai, X.; Ambrose, M.; Guo, Q.; Xiong, Y.; Merchant, N.B. Pancreatic stellate cell secreted IL-6 stimulates STAT3 dependent invasiveness of pancreatic intraepithelial neoplasia and cancer cells. Oncotarget 2016, 7, 65982–65992. [Google Scholar] [CrossRef] [Green Version]
- Corcoran, R.B.; Contino, G.; Deshpande, V.; Tzatsos, A.; Conrad, C.; Benes, C.H.; Levy, D.E.; Settleman, J.; Engelman, J.A.; Bardeesy, N. STAT3 Plays a Critical Role in KRAS-Induced Pancreatic Tumorigenesis. Cancer Res. 2011, 71, 5020–5029. [Google Scholar] [CrossRef] [Green Version]
- Fukuda, A.; Wang, S.; Morris, J.P.; Folias, A.E.; Liou, A.; Kim, G.E.; Akira, S.; Boucher, K.; Firpo, M.A.; Mulvihill, S.J.; et al. Stat3 and MMP7 Contribute to Pancreatic Ductal Adenocarcinoma Initiation and Progression. Cancer Cell 2011, 19, 441–455. [Google Scholar] [CrossRef] [Green Version]
- D’Amico, S.; Shi, J.; Martin, B.; Crawford, H.C.; Petrenko, O.; Reich, N.C. STAT3 is a master regulator of epithelial identity and KRAS-driven tumorigenesis. Genes Dev. 2018, 32, 1175–1187. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Diab, I.; Zhang, X.; Izmailova, E.S.; Zehner, Z.E. Stat3 enhances vimentin gene expression by binding to the antisilencer element and interacting with the repressor protein, ZBP-89. Oncogene 2004, 23, 168–178. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Li, L.; Jiang, H.; Knolhoff, B.L.; Lockhart, A.C.; Wang-Gillam, A.; DeNardo, D.G.; Ruzinova, M.B.; Lim, K.-H. Constitutive IRAK4 Activation Underlies Poor Prognosis and Chemoresistance in Pancreatic Ductal Adenocarcinoma. Clin. Cancer Res. 2017, 23, 1748–1759. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Li, L.; Jiang, H.; Li, Q.; Wang-Gillam, A.; Yu, J.; Head, R.; Liu, J.; Ruzinova, M.B.; Lim, K.H. Tumor–Stroma IL1beta-IRAK4 Feedforward Circuitry Drives Tumor Fibrosis, Chemoresistance, and Poor Prognosis in Pancreatic Cancer. Cancer Res. 2018, 78, 1700–1712. [Google Scholar] [CrossRef] [Green Version]
- Correction: IL1 Receptor Antagonist Inhibits Pancreatic Cancer Growth by Abrogating NF-κB Activation. Clin. Cancer Res. 2017, 23, 868, Erratum for: Clin. Cancer Res. 2016, 22, 1432–1244. [CrossRef] [Green Version]
- Maier, H.J.; Schmidt-Straßburger, U.; Huber, M.A.; Wiedemann, E.M.; Beug, H.; Wirth, T. NF-κB promotes epithelial–mesenchymal transition, migration and invasion of pancreatic carcinoma cells. Cancer Lett. 2010, 295, 214–228. [Google Scholar] [CrossRef]
- Nusse, R.; Clevers, H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017, 169, 985–999. [Google Scholar] [CrossRef]
- Steinhart, Z.; Angers, S. Wnt signaling in development and tissue homeostasis. Development 2018, 145, dev146589. [Google Scholar] [CrossRef] [Green Version]
- Chiba, S. Concise Review: Notch Signaling in Stem Cell Systems. Stem Cells 2006, 24, 2437–2447. [Google Scholar] [CrossRef]
- Siebel, C.; Lendahl, U. Notch Signaling in Development, Tissue Homeostasis, and Disease. Physiol. Rev. 2017, 97, 1235–1294. [Google Scholar] [CrossRef] [Green Version]
- Ingham, P.W. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 2001, 15, 3059–3087. [Google Scholar] [CrossRef] [Green Version]
- Petrova, R.; Joyner, A.L. Roles for Hedgehog signaling in adult organ homeostasis and repair. Development 2014, 141, 3445–3457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, F.-X.; Zhao, B.; Guan, K.L. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell 2015, 163, 811–828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Y.; Pan, D. The Hippo Signaling Pathway in Development and Disease. Dev. Cell 2019, 50, 264–282. [Google Scholar] [CrossRef] [PubMed]
- Scheibner, K.; Bakhti, M.; Bastidas-Ponce, A.; Lickert, H. Wnt signaling: Implications in endoderm development and pancreas organogenesis. Curr. Opin. Cell Biol. 2019, 61, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-Y.; Zhai, W.-J.; Teng, C.-B. Notch Signaling in Pancreatic Development. Int. J. Mol. Sci. 2015, 17, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, J.; Kawahira, H.; Hebrok, M. Hedgehog signaling in pancreas development and disease. Cell. Mol. Life Sci. 2006, 63, 642–652. [Google Scholar] [CrossRef]
- Gao, T.; Zhou, D.; Yang, C.; Singh, T.; Penzo–Méndez, A.; Maddipati, R.; Tzatsos, A.; Bardeesy, N.; Avruch, J.; Stanger, B.Z. Hippo Signaling Regulates Differentiation and Maintenance in the Exocrine Pancreas. Gastroenterology 2013, 144, 1543–1553.e1. [Google Scholar] [CrossRef] [Green Version]
- Yanling, W.; Wang, Y.; Zhu, Y.; Yuan, C.; Wang, D.; Zhang, W.; Qi, B.; Qiu, J.; Song, X.; Ye, J.; et al. The Hippo transducer TAZ promotes epithelial to mesenchymal transition and cancer stem cell maintenance in oral cancer. Mol. Oncol. 2015, 9, 1091–1105. [Google Scholar] [CrossRef] [Green Version]
- Misiorek, J.O.; Przybyszewska-Podstawka, A.; Kałafut, J.; Paziewska, B.; Rolle, K.; Rivero-Müller, A.; Nees, M. Context Matters: NOTCH Signatures and Pathway in Cancer Progression and Metastasis. Cells 2021, 10, 94. [Google Scholar] [CrossRef]
- Park, J.H.; Shin, J.E.; Park, H.W. The Role of Hippo Pathway in Cancer Stem Cell Biology. Mol. Cells 2018, 41, 83–92. [Google Scholar] [CrossRef]
- Takebe, N.; Miele, L.; Harris, P.J.; Jeong, W.; Bando, H.; Kahn, M.G.; Yang, S.X.; Ivy, S.P. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: Clinical update. Nat. Rev. Clin. Oncol. 2015, 12, 445–464. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, V.; Nataraj, R.; Thangaraj, G.S.; Karthikeyan, M.; Gnanasekaran, A.; Kaginelli, S.B.; Kuppanna, G.; Kallappa, C.G.; Basalingappa, K.M. Targeting Notch signalling pathway of cancer stem cells. Stem Cell Investig. 2018, 5, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vincan, E.; Barker, N. The upstream components of the Wnt signalling pathway in the dynamic EMT and MET associated with colorectal cancer progression. Clin. Exp. Metastasis 2008, 25, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Macdonald, B.T.; Tamai, K.; He, X. Wnt/β-Catenin Signaling: Components, Mechanisms, and Diseases. Dev. Cell 2009, 17, 9–26. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Yang, R.; Dong, Y.; Chen, M.; Wang, Y.; Wang, G. Knockdown of FOXO3a induces epithelial-mesenchymal transition and promotes metastasis of pancreatic ductal adenocarcinoma by activation of the β-catenin/TCF4 pathway through SPRY2. J. Exp. Clin. Cancer Res. 2019, 38, 38, Erratum in: J. Exp. Clin. Cancer Res. 2021, 40, 249. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Yang, H.; Abel, E.V.; Ney, G.M.; Palmbos, P.L.; Bednar, F.; Zhang, Y.; Leflein, J.; Waghray, M.; Owens, S.; et al. ATDC induces an invasive switch in KRAS-induced pancreatic tumorigenesis. Genes Dev. 2015, 29, 171–183. [Google Scholar] [CrossRef] [Green Version]
- Bray, S. Notch signalling in context. Nat. Rev. Mol. Cell Biol. 2016, 17, 722–735. [Google Scholar] [CrossRef]
- Aster, J.C.; Pear, W.S.; Blacklow, S.C. The Varied Roles of Notch in Cancer. Annu. Rev. Pathol. Mech. Dis. 2017, 12, 245–275. [Google Scholar] [CrossRef] [Green Version]
- Lobry, C.; Oh, P.; Aifantis, I. Oncogenic and tumor suppressor functions of Notch in cancer: It’s NOTCH what you think. J. Exp. Med. 2011, 208, 1931–1935. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Li, Y.; Kong, D.; Banerjee, S.; Ahmad, A.; Azmi, A.; Ali, S.; Abbruzzese, J.L.; Gallick, G.E.; Sarkar, F.H. Acquisition of Epithelial-Mesenchymal Transition Phenotype of Gemcitabine-Resistant Pancreatic Cancer Cells Is Linked with Activation of the Notch Signaling Pathway. Cancer Res. 2009, 69, 2400–2407. [Google Scholar] [CrossRef] [Green Version]
- Palagani, V.; El Khatib, M.; Kossatz, U.; Bozko, P.; Müller, M.R.; Manns, M.P.; Krech, T.; Malek, N.P.; Plentz, R.R. Epithelial Mesenchymal Transition and Pancreatic Tumor Initiating CD44+/EpCAM+ Cells Are Inhibited by γ-Secretase Inhibitor IX. PLoS ONE 2012, 7, e46514. [Google Scholar] [CrossRef] [Green Version]
- Geng, Y.; Fan, J.; Chen, L.; Zhang, C.; Qu, C.; Qian, L.; Chen, K.; Meng, Z.; Chen, Z.; Wang, P. A Notch-dependent inflammatory feedback circuit between macrophages and cancer cells regulates pancreatic cancer metastasis. Cancer Res. 2020, 81, 64–76. [Google Scholar] [CrossRef]
- Briscoe, J.; Therond, P.P. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 2013, 14, 416–429. [Google Scholar] [CrossRef]
- Di Magliano, M.P.; Sekine, S.; Ermilov, A.; Ferris, J.; Dlugosz, A.A.; Hebrok, M. Hedgehog/Ras interactions regulate early stages of pancreatic cancer. Genes Dev. 2006, 20, 3161–3173. [Google Scholar] [CrossRef] [Green Version]
- Steele, N.G.; Biffi, G.; Kemp, S.B.; Zhang, Y.; Drouillard, D.; Syu, L.; Hao, Y.; Oni, T.E.; Brosnan, E.; Elyada, E.; et al. Inhibition of Hedgehog Signaling Alters Fibroblast Composition in Pancreatic Cancer. Clin. Cancer Res. 2021, 27, 2023–2037. [Google Scholar] [CrossRef]
- Wang, F.; Ma, L.; Zhang, Z.; Liu, X.; Gao, H.; Zhuang, Y.; Yang, P.; Kornmann, M.; Tian, X.; Yang, Y. Hedgehog Signaling Regulates Epithelial-Mesenchymal Transition in Pancreatic Cancer Stem-Like Cells. J. Cancer 2016, 7, 408–417. [Google Scholar] [CrossRef] [Green Version]
- Ma, S.; Meng, Z.; Chen, R.; Guan, K.-L. The Hippo Pathway: Biology and Pathophysiology. Annu. Rev. Biochem. 2019, 88, 577–604. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.; Li, D.; Li, H.; Wang, L.; Tian, G.; Dong, Y. YAP overexpression promotes the epithelial-mesenchymal transition and chemoresistance in pancreatic cancer cells. Mol. Med. Rep. 2015, 13, 237–242. [Google Scholar] [CrossRef] [Green Version]
- Xie, D.; Cui, J.; Xia, T.; Jia, Z.; Wang, L.; Wei, W.; Zhu, A.; Gao, Y.; Xie, K.; Quan, M. Hippo transducer TAZ promotes epithelial mesenchymal transition and supports pancreatic cancer progression. Oncotarget 2015, 6, 35949–35963. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Friedland, S.C.; Guo, B.; O’Dell, M.R.; Alexander, W.; Whitney-Miller, C.L.; Agostini-Vulaj, D.; Huber, A.R.; Myers, J.R.; Ashton, J.; et al. ARID1A, a SWI/SNF subunit, is critical to acinar cell homeostasis and regeneration and is a barrier to transformation and epithelial-mesenchymal transition in the pancreas. Gut 2019, 68, 1245–1258. [Google Scholar] [CrossRef]
- Kadoch, C.; Hargreaves, D.C.; Hodges, H.C.; Elias, L.; Ho, L.; Ranish, J.; Crabtree, G.R. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 2013, 45, 592–601. [Google Scholar] [CrossRef]
- Wang, S.C.; Nassour, I.; Xiao, S.; Zhang, S.; Luo, X.; Lee, J.; Li, L.; Sun, X.; Nguyen, L.H.; Chuang, J.-C.; et al. SWI/SNF component ARID1A restrains pancreatic neoplasia formation. Gut 2019, 68, 1259–1270. [Google Scholar] [CrossRef] [Green Version]
- Tomihara, H.; Carbone, F.; Perelli, L.; Huang, J.K.; Soeung, M.; Rose, J.L.; Robinson, F.S.; Deribe, Y.L.; Feng, N.; Takeda, M.; et al. Loss of ARID1A Promotes Epithelial–Mesenchymal Transition and Sensitizes Pancreatic Tumors to Proteotoxic Stress. Cancer Res. 2021, 81, 332–343. [Google Scholar] [CrossRef]
- Kim, H.S.; Lu, S.; Cao, Y.; Yi, Z.; Jeyarajan, S.; Zhao, L.; Bedi, K.; Narayanan, I.V.; Paulsen, M.T.; Ljungman, M.; et al. KMT2D mediates TGF-β-induced epithelial-to-mesenchymal transition to promote more aggressive pancreatic cancer. In Proceedings of the AACR Special Conference on Pancreatic Cancer: Advances in Science and Clinical Care, Boston, MA, USA, 6–9 September 2019; AACR; Cancer Res.: Philadelphia, PA, USA, 2019; Volume 79 (Suppl. 24). Abstract nr C28. [Google Scholar]
- Esquela-Kerscher, A.; Slack, F. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef] [Green Version]
- Rawat, M.; Kadian, K.; Gupta, Y.; Kumar, A.; Chain, P.S.; Kovbasnjuk, O.; Parasher, G. MicroRNA in Pancreatic Cancer: From Biology to Therapeutic Potential. Genes 2019, 10, 752. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Chen, L.; Li, C.; Huang, R.; Guo, M.; Ning, S.; Ji, J.; Guo, X.; Lou, G.; Jia, X.; et al. The multifaceted roles of long noncoding RNAs in pancreatic cancer: An update on what we know. Cancer Cell Int. 2020, 20, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Burk, U.; Schubert, J.; Wellner, U.; Schmalhofer, O.; Vincan, E.; Spaderna, S.; Brabletz, T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008, 9, 582–589. [Google Scholar] [CrossRef] [Green Version]
- Paterson, E.L.; Kolesnikoff, N.; Gregory, P.A.; Bert, A.G.; Khew-Goodall, Y.; Goodall, G.J. The microRNA-200 family regulates epithelial to mesenchymal transition. Sci. World J. 2008, 8, 901–904. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Cheng, H.; Shi, S.; Cui, X.; Yang, J.; Chen, L.; Cen, P.; Cai, X.; Lu, Y.; Wu, C.; et al. MicroRNA-34b inhibits pancreatic cancer metastasis through repressing Smad3. Curr. Mol. Med. 2013, 13, 467–478. [Google Scholar] [CrossRef]
- Frampton, A.E.; Krell, J.; Jacob, J.; Stebbing, J.; Castellano, L.; Jiao, L.R. Loss of miR-126 is crucial to pancreatic cancer progression. Expert Rev. Anticancer Ther. 2012, 12, 881–884. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Vandenboom, T.G., 2nd; Wang, Z.; Kong, D.; Ali, S.; Philip, P.A.; Sarkar, F.H. miR-146a suppresses invasion of pancreatic cancer cells. Cancer Res. 2010, 70, 1486–1495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, L.; Xiong, X.; Lin, Y.; Cheng, Y.; Lu, J.; Zhang, J.; Cheng, N. miR-203 inhibits tumor cell migration and invasion via caveolin-1 in pancreatic cancer cells. Oncol. Lett. 2014, 7, 658–662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, H.; Hao, S.J.; Yao, L.; Yang, F.; Di, Y.; Li, J.; Jiang, Y.J.; Jin, C.; Fu, D.L. MicroRNA-218 inhibits cell invasion and migration of pancreatic cancer via regulating ROBO1. Cancer Biol. Ther. 2014, 15, 1333–1339. [Google Scholar] [CrossRef] [Green Version]
- Ohuchida, K.; Mizumoto, K.; Lin, C.; Yamaguchi, H.; Ohtsuka, T.; Sato, N.; Toma, H.; Nakamura, M.; Nagai, E.; Hashizume, M.; et al. MicroRNA-10a is overexpressed in human pancreatic cancer and involved in its invasiveness partially via suppression of the HOXA1 gene. Ann. Surg. Oncol. 2012, 19, 2394–2402. [Google Scholar] [CrossRef]
- Weiss, F.U.; Marques, I.J.; Woltering, J.M.; Vlecken, D.H.; Aghdassi, A.; Partecke, L.I.; Heidecke, C.D.; Lerch, M.M.; Bagowski, C.P. Retinoic acid receptor antagonists inhibit miR-10a expression and block metastatic behavior of pancreatic cancer. Gastroenterology 2009, 137, 2136–2145.e7. [Google Scholar] [CrossRef]
- Liu, A.; Shao, C.; Jin, G.; Liu, R.; Hao, J.; Song, B.; Ouyang, L.; Hu, X. miR-208-induced epithelial to mesenchymal transition of pancreatic cancer cells promotes cell metastasis and invasion. Cell Biochem. Biophys. 2014, 69, 341–346. [Google Scholar] [CrossRef]
- Cui, L.; Dong, Y.; Wang, X.; Zhao, X.; Kong, C.; Liu, Y.; Jiang, X.; Zhang, X. Downregulation of long noncoding RNA SNHG1 inhibits cell proliferation, metastasis, and invasion by suppressing the Notch-1 signaling pathway in pancreatic cancer. J. Cell. Biochem. 2019, 120, 6106–6112. [Google Scholar] [CrossRef]
- Xu, B.; Gong, X.; Zi, L.; Li, G.; Dong, S.; Chen, X.; Li, Y. Silencing of DLEU 2 suppresses pancreatic cancer cell proliferation and invasion by upregulating microRNA-455. Cancer Sci. 2019, 110, 1676–1685. [Google Scholar] [CrossRef] [Green Version]
- Feng, H.; Wei, B.; Zhang, Y. Long non-coding RNA HULC promotes proliferation, migration and invasion of pancreatic cancer cells by down-regulating microRNA-15a. Int. J. Biol. Macromol. 2019, 126, 891–898. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, Y. LncRNA XIST enhanced TGF-β2 expression by targeting miR-141-3p to promote pancreatic cancer cells invasion. Biosci. Rep. 2019, 39, BSR20190332. [Google Scholar] [CrossRef] [Green Version]
- Zhou, D.-D.; Liu, X.-F.; Lu, C.-W.; Pant, O.P.; Liu, X.-D. Long non-coding RNA PVT1: Emerging biomarker in digestive system cancer. Cell Prolif. 2017, 50, e12398. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.-W.; Chen, Y.-F.; Li, J.; Huo, Y.-M.; Liu, D.-J.; Hua, R.; Zhang, J.-F.; Liu, W.; Yang, J.-Y.; Fu, X.-L.; et al. A novel long non-coding RNA ENST00000480739 suppresses tumour cell invasion by regulating OS-9 and HIF-1α in pancreatic ductal adenocarcinoma. Br. J. Cancer 2014, 111, 2131–2141. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Ding, X.; Hu, H.; He, Y.; Lu, Z.; Wu, P.; Tian, L.; Xia, T.; Yin, J.; Yuan, H.; et al. Long non-coding RNA lnc-PCTST predicts prognosis through inhibiting progression of pancreatic cancer by downregulation of TACC-3. Int. J. Cancer 2018, 143, 3143–3154. [Google Scholar] [CrossRef] [Green Version]
- Gao, H.; Gong, N.; Ma, Z.; Miao, X.; Chen, J.; Cao, Y.; Zhang, G. LncRNA ZEB2-AS1 promotes pancreatic cancer cell growth and invasion through regulating the miR-204/HMGB1 axis. Int. J. Biol. Macromol. 2018, 116, 545–551. [Google Scholar] [CrossRef]
- Martinelli, P.; Carrillo-de Santa Pau, E.; Cox, T.; Sainz, B., Jr.; Dusetti, N.; Greenhalf, W.; Rinaldi, L.; Costello, E.; Ghaneh, P.; Malats, N.; et al. GATA6 regulates EMT and tumour dissemination, and is a marker of response to adjuvant chemotherapy in pancreatic cancer. Gut 2017, 66, 1665–1676. [Google Scholar] [CrossRef] [Green Version]
- Patil, S.; Steuber, B.; Kopp, W.; Kari, V.; Urbach, L.; Wang, X.; Küffer, S.; Bohnenberger, H.; Spyropoulou, D.; Zhang, Z.; et al. EZH2 Regulates Pancreatic Cancer Subtype Identity and Tumor Progression via Transcriptional Repression of GATA6. Cancer Res. 2020, 80, 4620–4632. [Google Scholar] [CrossRef]
- Russell, R.; Perkhofer, L.; Liebau, S.; Lin, Q.; Lechel, A.; Feld, F.M.; Hessmann, E.; Gaedcke, J.; Güthle, M.; Zenke, M.; et al. Loss of ATM accelerates pancreatic cancer formation and epithelial–mesenchymal transition. Nat. Commun. 2015, 6, 7677. [Google Scholar] [CrossRef] [Green Version]
- Mody, H.R.; Hung, S.W.; Naidu, K.; Lee, H.; Gilbert, C.A.; Hoang, T.T.; Pathak, R.K.; Manoharan, R.; Muruganandan, S.; Govindarajan, R. SET contributes to the epithelial-mesenchymal transition of pancreatic cancer. Oncotarget 2017, 8, 67966–67979. [Google Scholar] [CrossRef]
- Wang, L.; Gu, F.; Ma, N.; Zhang, L.; Bian, J.-M.; Cao, H.-Y. CIP2A expression is associated with altered expression of epithelial–mesenchymal transition markers and predictive of poor prognosis in pancreatic ductal adenocarcinoma. Tumor Biol. 2013, 34, 2309–2313. [Google Scholar] [CrossRef]
- Kyuno, T.; Kohno, T.; Konno, T.; Yamaguchi, H.; Kyuno, D.; Imamura, M.; Kimura, Y.; Kojima, T.; Takemasa, I. Glucose-Dependent FOXM1 Promotes Epithelial-to-Mesenchymal Transition Via Cellular Metabolism and Targeting Snail in Human Pancreatic Cancer. Pancreas 2020, 49, 273–280. [Google Scholar] [CrossRef]
- Recouvreux, M.V.; Moldenhauer, M.R.; Galenkamp, K.M.; Jung, M.; James, B.; Zhang, Y.; Lowy, A.; Bagchi, A.; Commisso, C. Glutamine depletion regulates Slug to promote EMT and metastasis in pancreatic cancer. J. Exp. Med. 2020, 217, e20200388. [Google Scholar] [CrossRef]
- Wang, M.; Estrella, J.S.; Katz, M.H.; Kim, M.; Rashid, A.; Lee, J.E.; Maitra, A.; Wistuba, I.I.; Wolff, R.A.; Varadhachary, G.R.; et al. Expression of Epithelial-Mesenchymal Transition Markers in Treated Pancreatic Ductal Adenocarcinoma. Pancreas 2019, 48, 1367–1372. [Google Scholar] [CrossRef]
- El Amrani, M.; Corfiotti, F.; Corvaisier, M.; Vasseur, R.; Fulbert, M.; Skrzypczyk, C.; Deshorgues, A.; Gnemmi, V.; Tulasne, D.; Lahdaoui, F.; et al. Gemcitabine-induced epithelial-mesenchymal transition-like changes sustain chemoresistance of pancreatic cancer cells of mesenchymal-like phenotype. Mol. Carcinog. 2019, 58, 1985–1997. [Google Scholar] [CrossRef]
- Shah, A.N.; Summy, J.M.; Zhang, J.; Park, S.I.; Parikh, N.U.; Gallick, G.E. Development and Characterization of Gemcitabine-Resistant Pancreatic Tumor Cells. Ann. Surg. Oncol. 2007, 14, 3629–3637. [Google Scholar] [CrossRef]
- Porter, R.L.; Magnus, N.K.C.; Thapar, V.; Morris, R.; Szabolcs, A.; Neyaz, A.; Kulkarni, A.S.; Tai, E.; Chougule, A.; Hillis, A.; et al. Epithelial to mesenchymal plasticity and differential response to therapies in pancreatic ductal adenocarcinoma. Proc. Natl. Acad. Sci. USA 2019, 116, 26835–26845. [Google Scholar] [CrossRef] [Green Version]
- Tiriac, H.; Belleau, P.; Engle, D.D.; Plenker, D.; Deschênes, A.; Somerville, T.D.D.; Froeling, F.E.M.; Burkhart, R.A.; Denroche, R.E.; Jang, G.H.; et al. Organoid Profiling Identifies Common Responders to Chemotherapy in Pancreatic Cancer. Cancer Discov. 2018, 8, 1112–1129. [Google Scholar] [CrossRef] [Green Version]
- Dagogo-Jack, I.; Shaw, A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef]
- Jing, N.; Gao, W.-Q.; Fang, Y.-X. Regulation of Formation, Stemness and Therapeutic Resistance of Cancer Stem Cells. Front. Cell Dev. Biol. 2021, 9, 641498. [Google Scholar] [CrossRef]
- Ercan, G.; Karlitepe, A.; Ozpolat, B. Pancreatic Cancer Stem Cells and Therapeutic Approaches. Anticancer Res. 2017, 37, 2761–2775. [Google Scholar] [CrossRef]
- Patil, K.; Khan, F.B.; Akhtar, S.; Ahmad, A.; Uddin, S. The plasticity of pancreatic cancer stem cells: Implications in therapeutic resistance. Cancer Metastasis Rev. 2021, 40, 691–720. [Google Scholar] [CrossRef] [PubMed]
- Purohit, V.; Wang, L.; Yang, H.; Li, J.; Ney, G.M.; Gumkowski, E.R.; Vaidya, A.J.; Wang, A.; Bhardwaj, A.; Zhao, E.; et al. ATDC binds to KEAP1 to drive NRF2-mediated tumorigenesis and chemoresistance in pancreatic cancer. Genes Dev. 2021, 35, 218–233. [Google Scholar] [CrossRef] [PubMed]
- Menegon, S.; Columbano, A.; Giordano, S. The Dual Roles of NRF2 in Cancer. Trends Mol. Med. 2016, 22, 578–593. [Google Scholar] [CrossRef] [PubMed]
- De La Vega, M.R.; Chapman, E.; Zhang, D.D. NRF2 and the Hallmarks of Cancer. Cancer Cell 2018, 34, 21–43. [Google Scholar] [CrossRef]
- Weadick, B.; Nayak, D.; Persaud, A.K.; Hung, S.W.; Raj, R.; Campbell, M.J.; Chen, W.; Li, J.; Williams, T.M.; Govindarajan, R. EMT-Induced Gemcitabine Resistance in Pancreatic Cancer Involves the Functional Loss of Equilibrative Nucleoside Transporter 1. Mol. Cancer Ther. 2021, 20, 410–422. [Google Scholar] [CrossRef]
- Dardare, J.; Witz, A.; Merlin, J.-L.; Bochnakian, A.; Toussaint, P.; Gilson, P.; Harlé, A. Epithelial to Mesenchymal Transition in Patients with Pancreatic Ductal Adenocarcinoma: State-of-the-Art and Therapeutic Opportunities. Pharmaceuticals 2021, 14, 740. [Google Scholar] [CrossRef]
- Melisi, D.; Garcia-Carbonero, R.; Macarulla, T.; Pezet, D.; Deplanque, G.; Fuchs, M.; Trojan, J.; Oettle, H.; Kozloff, M.; Cleverly, A.; et al. Galunisertib plus gemcitabine vs. gemcitabine for first-line treatment of patients with unresectable pancreatic cancer. Br. J. Cancer 2018, 119, 1208–1214. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, M.; Takahashi, H.; Kondo, S.; Lahn, M.M.F.; Ogasawara, K.; Benhadji, K.A.; Fujii, H.; Ueno, H. Phase 1b study of galunisertib in combination with gemcitabine in Japanese patients with metastatic or locally advanced pancreatic cancer. Cancer Chemother. Pharmacol. 2017, 79, 1169–1177. [Google Scholar] [CrossRef]
- Kim, B.-G.; Malek, E.; Choi, S.H.; Ignatz-Hoover, J.J.; Driscoll, J.J. Novel therapies emerging in oncology to target the TGF-β pathway. J. Hematol. Oncol. 2021, 14, 1–20. [Google Scholar] [CrossRef]
- Li, H.; Xiao, H.; Lin, L.; Jou, D.; Kumari, V.; Lin, J.; Li, C. Drug Design Targeting Protein–Protein Interactions (PPIs) Using Multiple Ligand Simultaneous Docking (MLSD) and Drug Repositioning: Discovery of Raloxifene and Bazedoxifene as Novel Inhibitors of IL-6/GP130 Interface. J. Med. Chem. 2014, 57, 632–641. [Google Scholar] [CrossRef]
- De Jesus-Acosta, A.; Sugar, E.A.; O’Dwyer, P.J.; Ramanathan, R.K.; Von Hoff, D.D.; Rasheed, Z.; Zheng, L.; Begum, A.; Anders, R.; Maitra, A.; et al. Phase 2 study of vismodegib, a hedgehog inhibitor, combined with gemcitabine and nab-paclitaxel in patients with untreated metastatic pancreatic adenocarcinoma. Br. J. Cancer 2020, 122, 498–505. [Google Scholar] [CrossRef]


| Stage of Tumorigenesis | Gene Affected | Type of Genetic Alteration [References] |
|---|---|---|
| Acinar to Ductal Metaplasia | KRAS | Constitutive activation (upregulation): gain-of-function mutation (KRAS 12GD) [15,16] |
| Low grade PanIN | CDKN2A (Ink4a/Arf) | Inactivation (downregulation): deletion, loss-of-function mutation [15,16] |
| High grade PanIN | TP53 | Inactivation (downregulation): loss-of-function mutation, deletion [15,16] |
| SMAD4 | Inactivation (downregulation): deletion, loss of function mutation [15,16] |
| Study (Reference) | Number of Patients | Overall Survival | Disease Free Survival |
|---|---|---|---|
| Chouat et al. [59] | 50 | HR = 6.09 (95% CI 1.11–33.28), p = 0.03 | HR = 2,87 (95% CI 1.41–17.51), p = 0.02 |
| Liu et al. [61] | 46 | p = 0.01 | p = 0.001 |
| Karamitopoulou et al. [63] | 117 | HR = 3.98 (95% CI 2.3–6.9), p < 0.0001 | p = 0.0005 |
| Lohnesi et al. [64] | 173 | HR = 1.040 (95% CI 1.019–1.061), p < 0.001 | HR = 1.037 (95% CI 1.017–1.058), p < 0.001 |
| O’Connor et al. [65] | 613 | RR = 1.46 (95% CI 1.13–1.88), p = 0.004 HR = 2.65 (95% CI 1.79–3.91), p < 0.0001 | RR = 1.61 (95% CI 1.05–2.47), p = 0.03 |
| Study [Reference] | Non-Coding RNA Molecule | Number of Patients | Results | OS | DFS |
|---|---|---|---|---|---|
| Hamada et al. [162] | miR-126 | 5 | Reduced levels in PDAC compared to adjacent normal tissue. | NA | NA |
| Xu et al. [170] | lnc-RNA DLEU | 178 | Increased levels in PDAC compared to adjacent normal tissue. High levels correlate with worse OS. | p = 0.036 | |
| Feng et al. [171] | lnc-RNA HULC | 36 | Increased levels in PDAC compared to adjacent normal tissue. | NA | NA |
| Sun et al. [172] | Lnc-RNA XIST | 30 | Increased levels in PDAC compared to adjacent normal tissue. | NA | NA |
| Wang et al. [175] | Lnc-RNA PCTST | 48 | Reduced levels in PDAC compared to adjacent normal tissue. High levels correlate with longer OS. | HR = 0.11 (95% 0.02–0.49) p = 0.004 | NA |
| Gao et al. [176] | Lnc-RNA Zeb2-AS1 | 39 | Increased levels in PDAC compared to adjacent normal tissue. High levels correlate with worse OS and DFS. | p < 0.005 | p < 0.005 |
| Drug Name | Mechanism of Action | Clinical Trial Phase | NCT Registry Number |
|---|---|---|---|
| PF-06952229 | Inhibitor of TGF-b receptor (TGF-bRI) | I | NCT03685591 |
| BCA101 | Inhibitor of TGF-b/EGFR fusion | I | NCT04429542 |
| SHR-1701 | Inhibitor of TGF-b (ligand) | Ib/II | NCT04624217 |
| NIS793 | mAb anti-TGF-b (ligand) | I | NCT02947165 |
| II | NCT04390763 | ||
| Vactosertib (TEW-7197) | Inhibitor of TGF-bRI kinase | Ib | NCT03666832 |
| II | NCT04258072 | ||
| Galunisertib (LY2157299) | Inhibitor of TGF-bRI kinase | Ib | NCT02734160 |
| Ib | NCT02154646 | ||
| Ib/II | NCT01373164 | ||
| Trabedersen (AP 12009) | Antisense oligonucleotide specific for TGF-b | I | NCT00844064 |
| Tocilizumab | mAb anti-IL6 receptor | II | NCT02767557 |
| II | NCT04258150 | ||
| I/II | NCT03193190 | ||
| Siltuximab | mAb anti-IL6 | I/II | NCT00841191 |
| Ib/II | NCT04191421 | ||
| Bazedoxifene | Selective estrogen receptor modulator (SERM)-Inhibitor of IL-6/GP130 | - | NCT04812808 |
| Canakinumab (ACZ885) | mAb anti IL-1b | Ib | NCT04581343 |
| LDE225 | Hedgehog inhibition | I/II | NCT02358161 |
| Ib | NCT01485744 | ||
| NLM-001 | Hedgehog inhibition | Ib/IIa | NCT04827953 |
| IPI-926 | Hedgehog inhibition | Ib/II | NCT01130142 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palamaris, K.; Felekouras, E.; Sakellariou, S. Epithelial to Mesenchymal Transition: Key Regulator of Pancreatic Ductal Adenocarcinoma Progression and Chemoresistance. Cancers 2021, 13, 5532. https://doi.org/10.3390/cancers13215532
Palamaris K, Felekouras E, Sakellariou S. Epithelial to Mesenchymal Transition: Key Regulator of Pancreatic Ductal Adenocarcinoma Progression and Chemoresistance. Cancers. 2021; 13(21):5532. https://doi.org/10.3390/cancers13215532
Chicago/Turabian StylePalamaris, Kostas, Evangelos Felekouras, and Stratigoula Sakellariou. 2021. "Epithelial to Mesenchymal Transition: Key Regulator of Pancreatic Ductal Adenocarcinoma Progression and Chemoresistance" Cancers 13, no. 21: 5532. https://doi.org/10.3390/cancers13215532






