Diagnostic and Prognostic Implications of Caspase-1 and PD-L1 Co-Expression Patterns in Myelodysplastic Syndromes
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Study Design
2.2. Selection of Patient and Control Samples
2.3. Definition of Disease and Patient Characteristics
2.4. Definition of Clinical Endpoints
2.5. Reagents
2.6. Data Collection, Scoring and Analysis Plan
2.7. Statistical Analysis
3. Results
3.1. Baseline Characteristics of MDS Patients and Controls
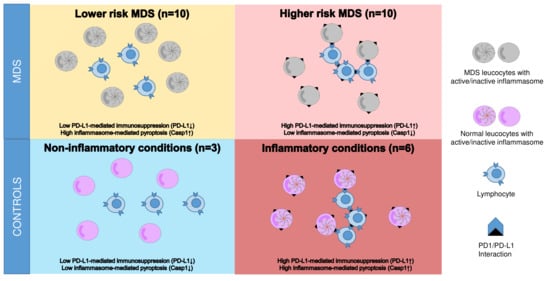
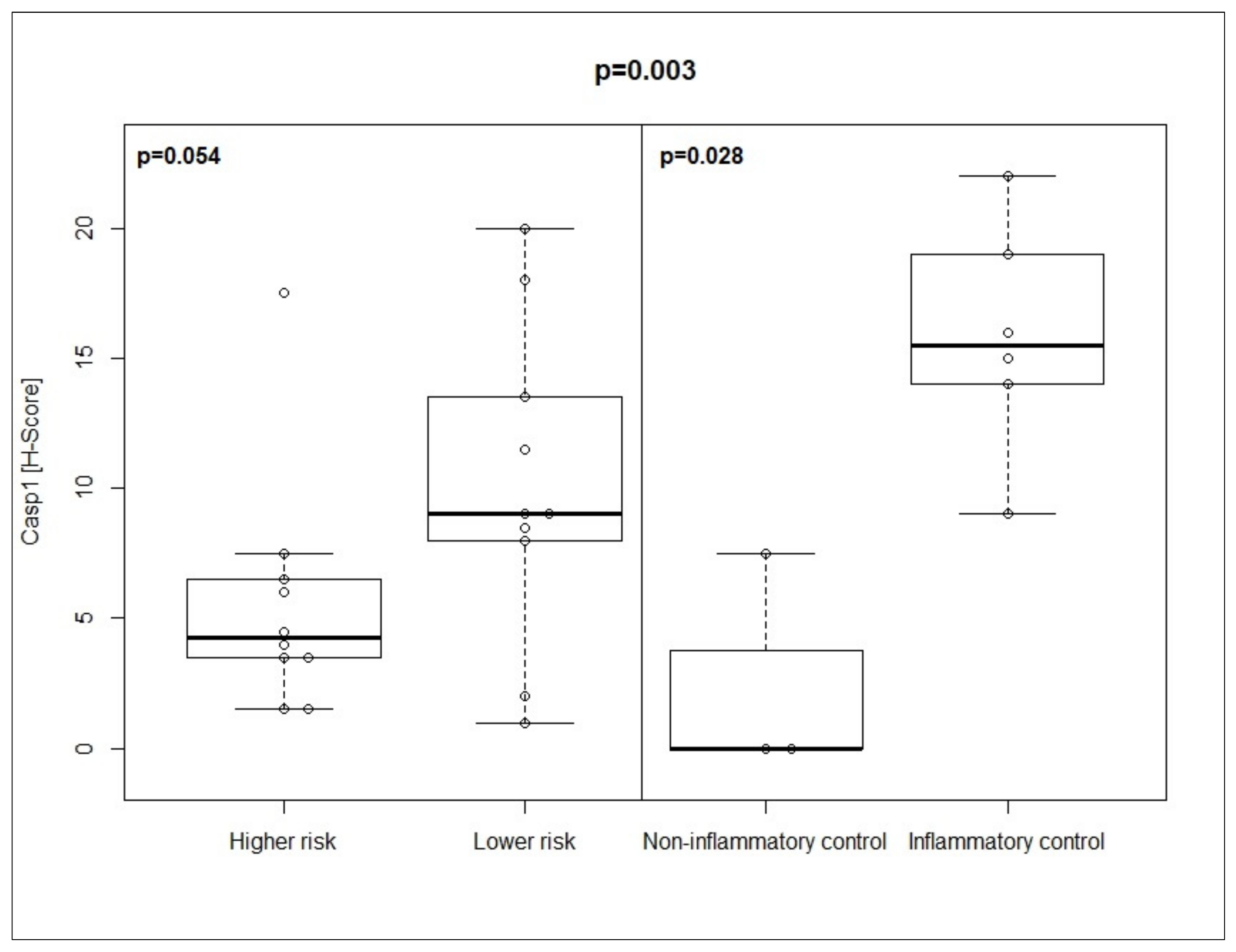
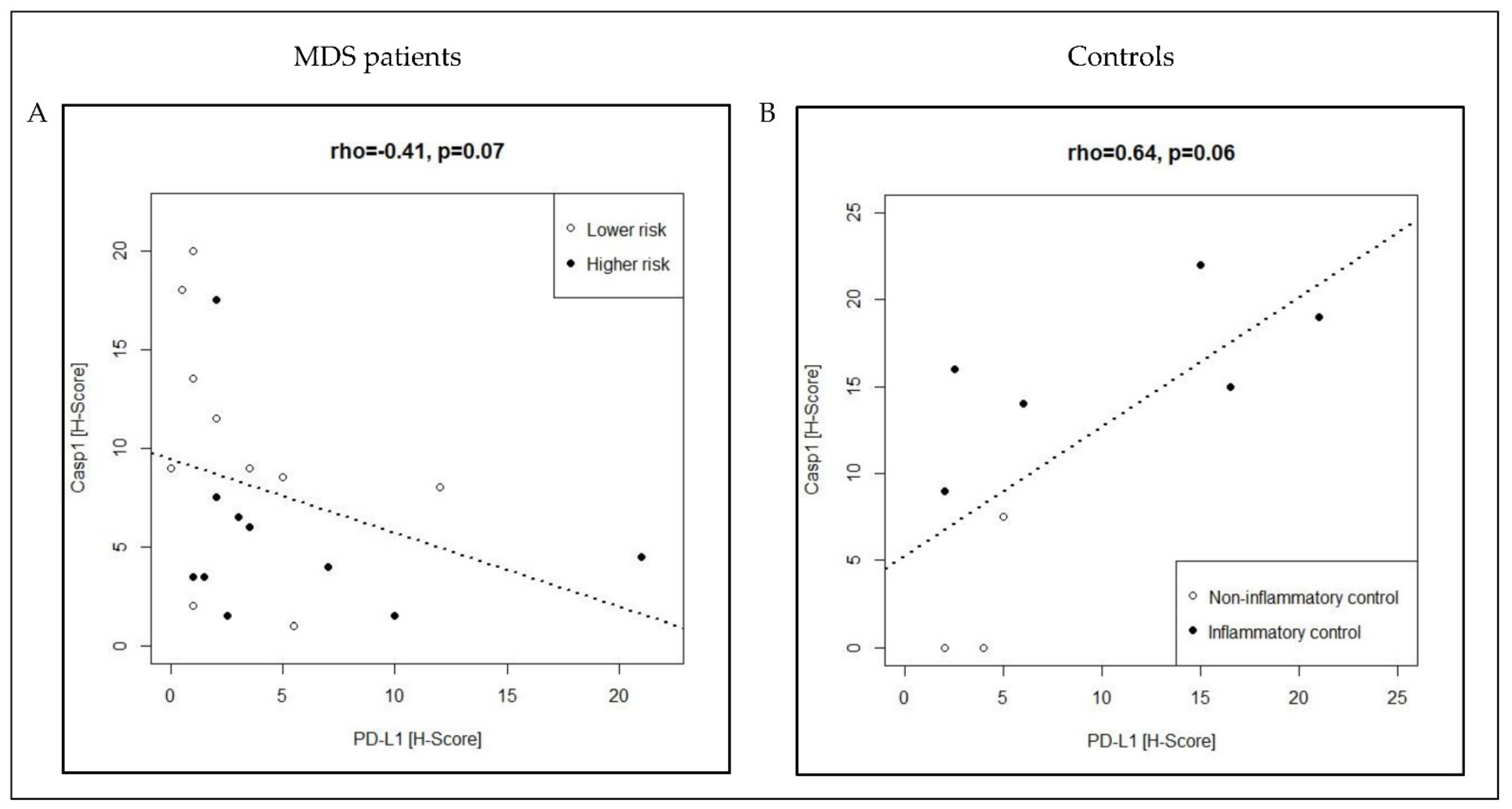
3.2. Co-Expression Pattern of Casp1/PD-L1 in MDS Patients and Controls
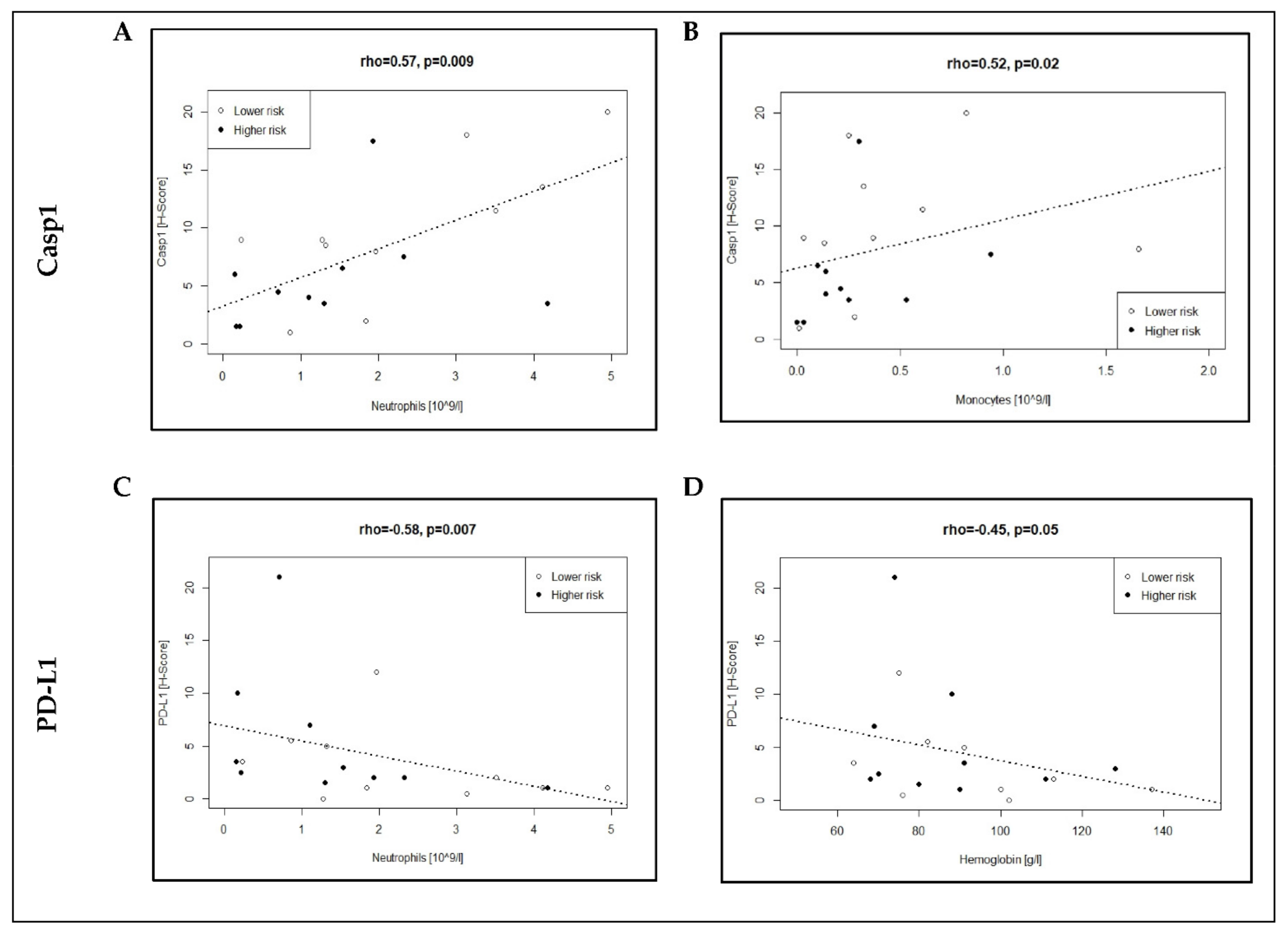
3.3. Association of Casp1/PD-L1 Expression with Neutrophil and Monocyte Counts in MDS Patients
3.4. Association of Immune-Related Biomarkers with Clinical Endpoints in MDS Patients
3.5. Casp1/PD-L1 Co-Expression Patterns in Individual Clinical Courses of MDS Patients
3.6. Co-Expression Pattern of Casp1/PD-L1 Might Be Useful as Classifier for MDS Disease State
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sallman, D.A.; Cluzeau, T.; Basiorka, A.A.; List, A. Unraveling the Pathogenesis of MDS: The NLRP3 Inflammasome and Pyroptosis Drive the MDS Phenotype. Front. Oncol. 2016, 6, 151. [Google Scholar] [CrossRef]
- Gañán-Gómez, I.; Wei, Y.; Starczynowski, D.T.; Colla, S.; Yang, H.; Cabrero-Calvo, M.; Bohannan, Z.S.; Verma, A.; Steidl, U.; Garcia-Manero, G. Deregulation of Innate Immune and Inflammatory Signaling in Myelodysplastic Syndromes. Leukemia 2015, 29, 1458–1469. [Google Scholar] [CrossRef]
- Greenberg, P.; Cox, C.; LeBeau, M.M.; Fenaux, P.; Morel, P.; Sanz, G.; Sanz, M.; Vallespi, T.; Hamblin, T.; Oscier, D.; et al. International Scoring System for Evaluating Prognosis in Myelodysplastic Syndromes. Blood 1997, 89, 2079–2088. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, P.L.; Tuechler, H.; Schanz, J.; Sanz, G.; Garcia-Manero, G.; Solé, F.; Bennett, J.M.; Bowen, D.; Fenaux, P.; Dreyfus, F.; et al. Revised International Prognostic Scoring System for Myelodysplastic Syndromes. Blood 2012, 120, 2454–2465. [Google Scholar] [CrossRef] [PubMed]
- Bonadies, N.; Bacher, V.U. What Role Can Next-Generation Sequencing Play in Myelodysplastic Syndrome Care? Expert. Rev. Hematol. 2019, 12, 379–382. [Google Scholar] [CrossRef] [Green Version]
- Sallman, D.A.; List, A. The Central Role of Inflammatory Signaling in the Pathogenesis of Myelodysplastic Syndromes. Blood 2019, 133, 1039–1048. [Google Scholar] [CrossRef] [Green Version]
- Chokr, N.; Patel, R.; Wattamwar, K.; Chokr, S. The Rising Era of Immune Checkpoint Inhibitors in Myelodysplastic Syndromes. Adv. Hematol. 2018, 2018, 2458679. [Google Scholar] [CrossRef]
- Cogle, C.R. Incidence and Burden of the Myelodysplastic Syndromes. Curr. Hematol. Malig. Rep. 2015, 10, 272–281. [Google Scholar] [CrossRef] [Green Version]
- Zeidan, A.M.; Shallis, R.M.; Wang, R.; Davidoff, A.; Ma, X. Epidemiology of Myelodysplastic Syndromes: Why Characterizing the Beast Is a Prerequisite to Taming It. Blood Rev. 2019, 34, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Bonadies, N.; Feller, A.; Rovo, A.; Ruefer, A.; Blum, S.; Gerber, B.; Stuessi, G.; Benz, R.; Cantoni, N.; Holbro, A.; et al. Trends of Classification, Incidence, Mortality, and Survival of MDS Patients in Switzerland between 2001 and 2012. Cancer Epidemiol. 2017, 46, 85–92. [Google Scholar] [CrossRef]
- Pianko, M.J.; Goldberg, A.D.; Lesokhin, A.M. Clinical Development of PD-1 Blockade in Hematologic Malignancies. Cancer J. 2018, 24, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.S.; Mellman, I. Elements of Cancer Immunity and the Cancer-Immune Set Point. Nature 2017, 541, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Mekinian, A.; Grignano, E.; Braun, T.; Decaux, O.; Liozon, E.; Costedoat-Chalumeau, N.; Kahn, J.E.; Hamidou, M.; Park, S.; Puéchal, X.; et al. Systemic Inflammatory and Autoimmune Manifestations Associated with Myelodysplastic Syndromes and Chronic Myelomonocytic Leukaemia: A French Multicentre Retrospective Study. Rheumatology 2015, 55, 291–300. [Google Scholar] [CrossRef] [Green Version]
- De Hollanda, A.; Beucher, A.; Henrion, D.; Ghali, A.; Lavigne, C.; Lévesque, H.; Hamidou, M.; Subra, J.F.; Ifrah, N.; Belizna, C. Systemic and Immune Manifestations in Myelodysplasia: A Multicenter Retrospective Study. Arthritis Care Res. 2011, 63, 1188–1194. [Google Scholar] [CrossRef] [Green Version]
- Kipfer, B.; Daikeler, T.; Kuchen, S.; Hallal, M.; Andina, N.; Allam, R.; Bonadies, N. Increased Cardiovascular Comorbidities in Patients with Myelodysplastic Syndromes and Chronic Myelomonocytic Leukemia Presenting with Systemic Inflammatory and Autoimmune Manifestations. Semin. Hematol. 2018, 55, 242–247. [Google Scholar] [CrossRef]
- Kordasti, S.Y.; Afzali, B.; Lim, Z.; Ingram, W.; Hayden, J.; Barber, L.; Matthews, K.; Chelliah, R.; Guinn, B.; Lombardi, G.; et al. IL-17-Producing CD4+ T Cells, pro-Inflammatory Cytokines and Apoptosis Are Increased in Low Risk Myelodysplastic Syndrome. Br. J. Haematol. 2009, 145, 64–72. [Google Scholar] [CrossRef]
- Yang, H.; Bueso-Ramos, C.; Dinardo, C.; Estecio, M.R.; Davanlou, M.; Geng, Q.R.; Fang, Z.; Nguyen, M.; Pierce, S.; Wei, Y.; et al. Expression of PD-L1, PD-L2, PD-1 and CTLA4 in Myelodysplastic Syndromes Is Enhanced by Treatment with Hypomethylating Agents. Leukemia 2014, 28, 1280–1288. [Google Scholar] [CrossRef]
- Cheng, P.; Eksioglu, E.A.; Chen, X.; Kandell, W.; Le Trinh, T.; Cen, L.; Qi, J.; Sallman, D.A.; Zhang, Y.; Tu, N.; et al. S100A9-Induced Overexpression of PD-1/PD-L1 Contributes to Ineffective Hematopoiesis in Myelodysplastic Syndromes. Leukemia 2019, 33, 2034–2046. [Google Scholar] [CrossRef]
- Robert, C. A Decade of Immune-Checkpoint Inhibitors in Cancer Therapy. Nat. Commun. 2020, 11, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Drake, C.G.; Jaffee, E.; Pardoll, D.M. Mechanisms of Immune Evasion by Tumors. Adv. Immunol. 2006, 90, 51–81. [Google Scholar]
- Barreyro, L.; Chlon, T.M.; Starczynowski, D.T. Chronic Immune Response Dysregulation in MDS Pathogenesis. Blood 2018, 132, 1553–1560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perrichet, A.; Ghiringhelli, F.; Rébé, C. Understanding Inflammasomes and Pd-1/Pd-L1 Crosstalk to Improve Cancer Treatment Efficiency. Cancers 2020, 12, 3550. [Google Scholar] [CrossRef]
- Lu, F.; Zhao, Y.; Pang, Y.; Ji, M.; Sun, Y.; Wang, H.; Zou, J.; Wang, Y.; Li, G.; Sun, T.; et al. NLRP3 Inflammasome Upregulates PD-L1 Expression and Contributes to Immune Suppression in Lymphoma. Cancer Lett. 2021, 497, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI Gene Expression and Hybridization Array Data Repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-Based Map of the Human Proteome. Science 2015, 347. [Google Scholar] [CrossRef] [PubMed]
- Papatheodorou, I.; Moreno, P.; Manning, J.; Fuentes, A.M.P.; George, N.; Fexova, S.; Fonseca, N.A.; Füllgrabe, A.; Green, M.; Huang, N.; et al. Expression Atlas Update: From Tissues to Single Cells. Nucleic Acids Res. 2020, 48, D77–D83. [Google Scholar] [CrossRef] [Green Version]
- Swerdlow, S.H.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J.; Vardiman, J. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th ed.; International Agency for Research on Cancer: Lyon, France, 2017. [Google Scholar]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 Revision to the World Health Organization Classification of Myeloid Neoplasms and Acute Leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef]
- Benton, C.B.; Khan, M.; Sallman, D.; Nazha, A.; Nogueras González, G.M.; Piao, J.; Ning, J.; Aung, F.; Al Ali, N.; Jabbour, E.; et al. Prognosis of Patients with Intermediate Risk IPSS-R Myelodysplastic Syndrome Indicates Variable Outcomes and Need for Models beyond IPSS-R. Am. J. Hematol. 2018, 93, 1245–1253. [Google Scholar] [CrossRef] [Green Version]
- della Porta, M.G.; Malcovati, L.; Strupp, C.; Ambaglio, I.; Kuendgen, A.; Zipperer, E.; Travaglino, E.; Invernizzi, R.; Pascutto, C.; Lazzarino, M.; et al. Risk Stratification Based on Both Disease Status and Extra-Hematologic Comorbidities in Patients with Myelodysplastic Syndrome. Haematologica 2011, 96, 441–449. [Google Scholar] [CrossRef] [Green Version]
- Hirsch, F.R.; Varella-Garcia, M.; Bunn, P.A.; Di Maria, M.V.; Veve, R.; Bremnes, R.M.; Barón, A.E.; Zeng, C.; Franklin, W.A. Epidermal Growth Factor Receptor in Non-Small-Cell Lung Carcinomas: Correlation between Gene Copy Number and Protein Expression and Impact on Prognosis. J. Clin. Oncol. 2003, 21, 3798–3807. [Google Scholar] [CrossRef]
- Choudhury, K.R.; Yagle, K.J.; Swanson, P.E.; Krohn, K.A.; Rajendran, J.G. A Robust Automated Measure of Average Antibody Staining in Immunohistochemistry Images. J. Histochem. Cytochem. 2010, 58, 95–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fedchenko, N.; Reifenrath, J. Different Approaches for Interpretation and Reporting of Immunohistochemistry Analysis Results in the Bone Tissue—A Review. Diagn. Pathol. 2014, 9, 221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2020; Available online: http://www.r-project-org/ (accessed on 23 September 2021).
- Martin, G.; Martin, G. R Studio. In An Introduction to Programming with R; Boston MA, USA, 2021; Available online: http://www.rstudio.com/ (accessed on 23 September 2021).
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Müller, M. PROC: An Open-Source Package for R and S+ to Analyze and Compare ROC Curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Ggplot2; Use R! Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-24275-0. [Google Scholar]
- Basiorka, A.A.; McGraw, K.L.; Eksioglu, E.A.; Chen, X.; Johnson, J.; Zhang, L.; Zhang, Q.; Irvine, B.A.; Cluzeau, T.; Sallman, D.A.; et al. The NLRP3 Inflammasome Functions as a Driver of the Myelodysplastic Syndrome Phenotype. Blood 2016, 128, 2960–2975. [Google Scholar] [CrossRef] [PubMed]
- Hill, M.; Segovia, M.; Russo, S.; Girotti, M.R.; Rabinovich, G.A. The Paradoxical Roles of Inflammation during PD-1 Blockade in Cancer. Trends Immunol. 2020, 41, 982–993. [Google Scholar] [CrossRef] [PubMed]
- Schürch, C.M. Therapeutic Antibodies for Myeloid Neoplasms-Current Developments and Future Directions. Front. Oncol. 2018, 8, 152. [Google Scholar] [CrossRef] [Green Version]
- Boudard, D.; Sordet, O.; Vasselon, C.; Revol, V.; Berthéas, M.F.; Freyssenet, D.; Viallet, A.; Piselli, S.; Guyotat, D.; Campos, L. Expression and Activity of Caspases 1 and 3 in Myelodysplastic Syndromes. Leukemia 2000, 14, 2045–2051. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Liu, D.; Li, L. PD-1/PD-L1 Pathway: Current Researches in Cancer. Am. J. Cancer Res. 2020, 10, 727–742. [Google Scholar]
- Pellagatti, A.; Cazzola, M.; Giagounidis, A.A.N.; Malcovati, L.; Della Porta, M.G.; Killick, S.; Campbell, L.J.; Wang, L.; Langford, C.F.; Fidler, C.; et al. Gene Expression Profiles of CD34+ Cells in Myelodysplastic Syndromes: Involvement of Interferon-Stimulated Genes and Correlation to FAB Subtype and Karyotype. Blood 2006, 108, 337–345. [Google Scholar] [CrossRef] [Green Version]
- Pellagatti, A.; Cazzola, M.; Giagounidis, A.; Perry, J.; Malcovati, L.; Della Porta, M.G.; Jädersten, M.; Killick, S.; Verma, A.; Norbury, C.J.; et al. Deregulated Gene Expression Pathways in Myelodysplastic Syndrome Hematopoietic Stem Cells. Leukemia 2010, 24, 756–764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gry, M.; Rimini, R.; Strömberg, S.; Asplund, A.; Pontén, F.; Uhlén, M.; Nilsson, P. Correlations between RNA and Protein Expression Profiles in 23 Human Cell Lines. BMC Genom. 2009, 10, 365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koussounadis, A.; Langdon, S.P.; Um, I.H.; Harrison, D.J.; Smith, V.A. Relationship between Differentially Expressed MRNA and MRNA-Protein Correlations in a Xenograft Model System. Sci. Rep. 2015, 5, 10775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, E.A.; Rajan, J.V.; Aderem, A. Caspase-1-Induced Pyroptotic Cell Death. Immunol. Rev. 2011, 243, 206–214. [Google Scholar] [CrossRef]
- Ball, D.P.; Taabazuing, C.Y.; Griswold, A.R.; Orth, E.L.; Rao, S.D.; Kotliar, I.B.; Vostal, L.E.; Johnson, D.C.; Bachovchin, D.A. Caspase-1 Interdomain Linker Cleavage Is Required for Pyroptosis. Life Sci. Alliance 2020, 3, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Characteristics | MDS Patients | Controls | ||||
|---|---|---|---|---|---|---|
| Lower Risk (n = 10) | Higher Risk (n = 10) | Non-Inflammatory Controls (n = 3) | Inflammatory Controls (n = 6) | |||
| Gender | ||||||
| Male | 8 (80) | 6 (60) | ns | 2 (66) | 6 (100) | ns |
| Female | 2 (20) | 4 (40) | ns | 1 (33) | 0 (0) | ns |
| Age | 71 (28–79) | 69.5 (21–76) | ns | 59 (58–60) | 56 (18–67) | ns |
| Blood counts | ||||||
| hemoglobin [g/L] | 99.5 (28.8) | 86.9 (19.6) | ns | 131.5 (30.5) | 104 (25.6) | ns |
| thrombocytes [109/L] | 121 (132.7) | 76.4 (73.5) | ns | 250.5 (84.1) | 278 (172.3) | ns |
| neutrophils [109/L] | 2.32 (1.53) | 1.36 (1.24) | p = 0.14 | 4.61 (4.61) | 8.55 (7.75) | ns |
| monocytes [109/L] | 0.45 (0.49) | 0.26 (0.28) | ns | 0.37 (0.37) | 1.92 (1.92) | ns |
| Blood chemistry | ||||||
| CRP [mg/L] | 16.2 (19.7) | 26.3 (45) | ns | 3.5 (0.7) | 68.2 (47.1) | ns |
| creatinin [μmol/L] | 81.6 (14.5) | 85 (27.9) | ns | 73.0 (73.0) | 117.0 (77.3) | ns |
| ASAT [U/L] | 29.4 (16.9) | 20.4 (5.9) | ns | 44.0 (44.0) | 26.3 (7.5) | ns |
| LDH [U/L] | 431.6 (136.2) | 479.7 (107.3) | ns | 378.0 (378.0) | 486.2 (179.2) | ns |
| ferritin [μg/L] | 556.2 (444.6) | 395.4 (338.1) | ns | 484.0 (484.0) | 895.7 (568.9) | ns |
| Bone marrow | ||||||
| dysplastic myelopoiesis | 9 (90) | 10 (100) | ns | 0 (0) | 0 (0) | ns |
| dysplastic megakaryopoiesis | 8 (80) | 9 (90) | ns | 0 (0) | 0 (9) | ns |
| dysplastic erythropoiesis | 9 (90) | 8 (80) | ns | 0 (0) | 2 (33) | ns |
| fibrosis, grade 2 or higher | 0 (0) | 1 (10) | ns | 0 (0) | 0 (0) | ns |
| normocellularity | 3 (30) | 1 (10) | ns | 3 (100) | 5 (83) | ns |
| normal BM blast counts <5% | 10 (100) | 3 (10) | p = 0.003 | 3 (100) | 6 (100) | ns |
| cellularity [%] | 50 (30–90) | 45 (20–90) | ns | n. a. | n. a. | |
| blasts [%] | 3 (0–4) | 9 (2–15) | p = 0.003 | n. a. | n. a. | |
| Specifications of MDS patients | Specification of Controls | |||||
| Karyotype (IPSS-R) | Suspected transfusion reaction | Hodgkin lymphoma | ||||
| very good/good risk | 6 (60) | 5 (50) | ns | localized follicular lymphoma | diffuse large B-cell lymphoma | |
| intermediate risk | 4 (40) | 2 (20) | ns | Solitary plasmocytoma | post-transplant lympho- proliferative disorder | |
| poor/very poor risk | 0 (0) | 3 (30) | ns | Pulmonary adenocarcinoma | ||
| MDS-CI | AIDS with cerebral toxoplasmosis | |||||
| low risk | 5 (50) | 5 (50) | ns | multicentric Castleman disease | ||
| intermediate risk | 5 (50) | 2 (20) | ns | |||
| high risk | 0 (0) | 3 (30) | ns | |||
| Treatment received before assessment | ||||||
| red blood cell transfusion | 4 (40) | 7 (70) | ns | |||
| platelet transfusion | 0 (0) | 1 (10) | ns | |||
| lenalidomide | 0 (0) | 1 (10) 1 | ns | |||
| Biomarkers | MDS Patients | Controls | ||||
|---|---|---|---|---|---|---|
| Lower Risk (n = 10) | Higher Risk (n = 10) | Non-Inflammatory (n = 3) | Inflammatory (n = 6) | |||
| Univariate Analysis | ||||||
| Casp1 | 9 (1–20) | 4.25 (1.5–17.5) | p = 0.054 | 0 (0–7.5) | 15.5 (9–22) | p = 0.028 |
| PD1 | 3 (0–10) | 2.5 (1–10) | p = 0.784 | 1 (1–10) | 4 (1–10) | p = 0.680 |
| PD-L1 | 1.5 (0–12) | 2.75 (1–21) | p = 0.238 | 4 (2–5) | 10.5 (2–21) | p = 0.300 |
| Multivariate Analysis | ||||||
| Casp1 | p = 0.003 | |||||
| X | X | p adj = 0.48 | ||||
| X | X | p adj = 0.23 | ||||
| X | X | p adj = 0.75 | ||||
| X | X | p adj = 1.00 | ||||
| X | X | p adj = 0.01 | ||||
| X | X | p adj = 0.01 | ||||
| PD1 | p = 0.895 | |||||
| X | X | p adj = 1.00 | ||||
| X | X | p adj = 1.00 | ||||
| X | X | p adj = 1.00 | ||||
| X | X | p adj = 1.00 | ||||
| X | X | p adj = 1.00 | ||||
| X | X | p adj = 1.00 | ||||
| PD-L1 | p = 0.133 | |||||
| X | X | p adj = 1.00 | ||||
| X | X | p adj = 1.00 | ||||
| X | X | p adj = 0.11 | ||||
| X | X | p adj = 1.00 | ||||
| X | X | p adj = 1.00 | ||||
| X | X | p adj = 1.00 | ||||
| Lower Risk (n = 10) | Higher Risk (n = 10) | ||
| Follow-Up (Days), Median (Range) | 648.5 (107–1799) | 471 (15–3576) | ns |
| Treatment, n (%) | |||
| Disease-Modifying Treatment (DMT) | 8 (80) | 9 (90) | ns |
| hypomethylating agents (HMA) | 4 (40) | 7 (70) | ns |
| allo-HSCT | 1 (10) | 3 (30) | ns |
| Other DMTs | 4 (40) | 5 (50) | ns |
| red blood cell transfusions | 8 (80) | 9 (90) | ns |
| platelet transfusions | 7 (70) | 6 (60) | ns |
| Fate, n (%) | |||
| alive | 2 (20) | 4 (40) | ns |
| Allo-HSCT survivor | 1 (50) | 3 (75) | ns |
| dead | 8 (80) | 6 (60) | ns |
| MDS-related death | 6 (75) | 6 (100) | ns |
| not MDS-related death | 1 (13) | 0 (0) | ns |
| unknown death | 1 (13) | 0 (0) | ns |
| Endpoints (days), median (range) | |||
| time to treatment (TTT) | 289 (54–1015) | 55.5 (8–2632) | p = 0.04 |
| progression-free survival (PFS) | 429 (88–1264) | 161.5 (15–3576) | ns |
| leukemia-free survival (LFS) | 951 (107–1799) | 143.5 (15–616) | ns |
| overall survival (OS) | 955 (107–1799) | 170.5 (15–625) | ns |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Graf, J.R.; Forster, S.; Bruehl, F.K.; Banz, Y.; Hallal, M.; Brodard, J.; Bacher, V.U.; Allam, R.; Schürch, C.M.; Bonadies, N., on behalf of the Swiss MDS Study Group. Diagnostic and Prognostic Implications of Caspase-1 and PD-L1 Co-Expression Patterns in Myelodysplastic Syndromes. Cancers 2021, 13, 5712. https://doi.org/10.3390/cancers13225712
Graf JR, Forster S, Bruehl FK, Banz Y, Hallal M, Brodard J, Bacher VU, Allam R, Schürch CM, Bonadies N on behalf of the Swiss MDS Study Group. Diagnostic and Prognostic Implications of Caspase-1 and PD-L1 Co-Expression Patterns in Myelodysplastic Syndromes. Cancers. 2021; 13(22):5712. https://doi.org/10.3390/cancers13225712
Chicago/Turabian StyleGraf, Johannes R., Stefan Forster, Frido K. Bruehl, Yara Banz, Mahmoud Hallal, Justine Brodard, Vera Ulrike Bacher, Ramanjaneyulu Allam, Christian M. Schürch, and Nicolas Bonadies on behalf of the Swiss MDS Study Group. 2021. "Diagnostic and Prognostic Implications of Caspase-1 and PD-L1 Co-Expression Patterns in Myelodysplastic Syndromes" Cancers 13, no. 22: 5712. https://doi.org/10.3390/cancers13225712