Next-Generation Sequencing Targeted Panel in Routine Care for Metastatic Colon Cancers
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Methodology Overview
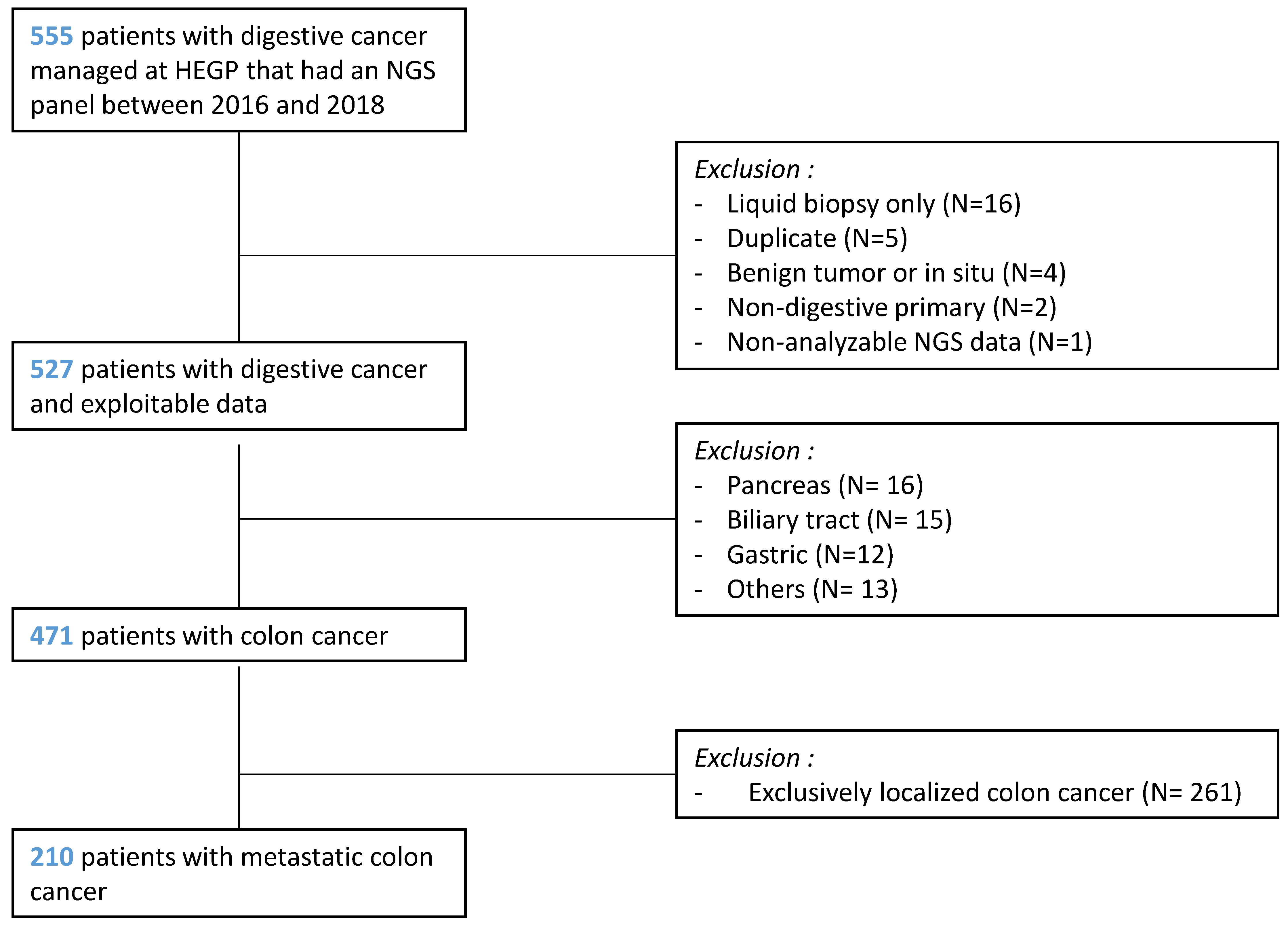
2.2. Inclusion Criteria
2.3. Exclusion Criteria
2.4. Patient Characteristics
2.5. TaqMan Probe Testing for KRAS and BRAF
2.6. Multigene Sequencing by NGS
- -
- NGS mutations with validated clinical impact: KRAS, BRAF, NRAS;
- -
- NGS mutations with potential clinical impact: PIK3CA, AKT1, ERBB2, PTEN, STK11, MAP2K1, ALK, MET, FGFR1, FGFR2, FGFR3, ERBB4, EGFR;
- -
- NGS mutations with unknown clinical impact: DDR2, CTNNB1, TP53, SMAD4, FBXW7, NOTCH1.
2.7. MSI Status
2.7.1. Immunohistochemistry
2.7.2. Molecular Test
2.8. Statistical Analysis
3. Results
3.1. Patients
3.2. Mutational Profile
3.3. Treatments Characteristics
3.4. Univariate Analysis
3.5. Multivariate Analysis
3.6. Cost and Turnaround Time
3.6.1. TaqMan
3.6.2. NGS
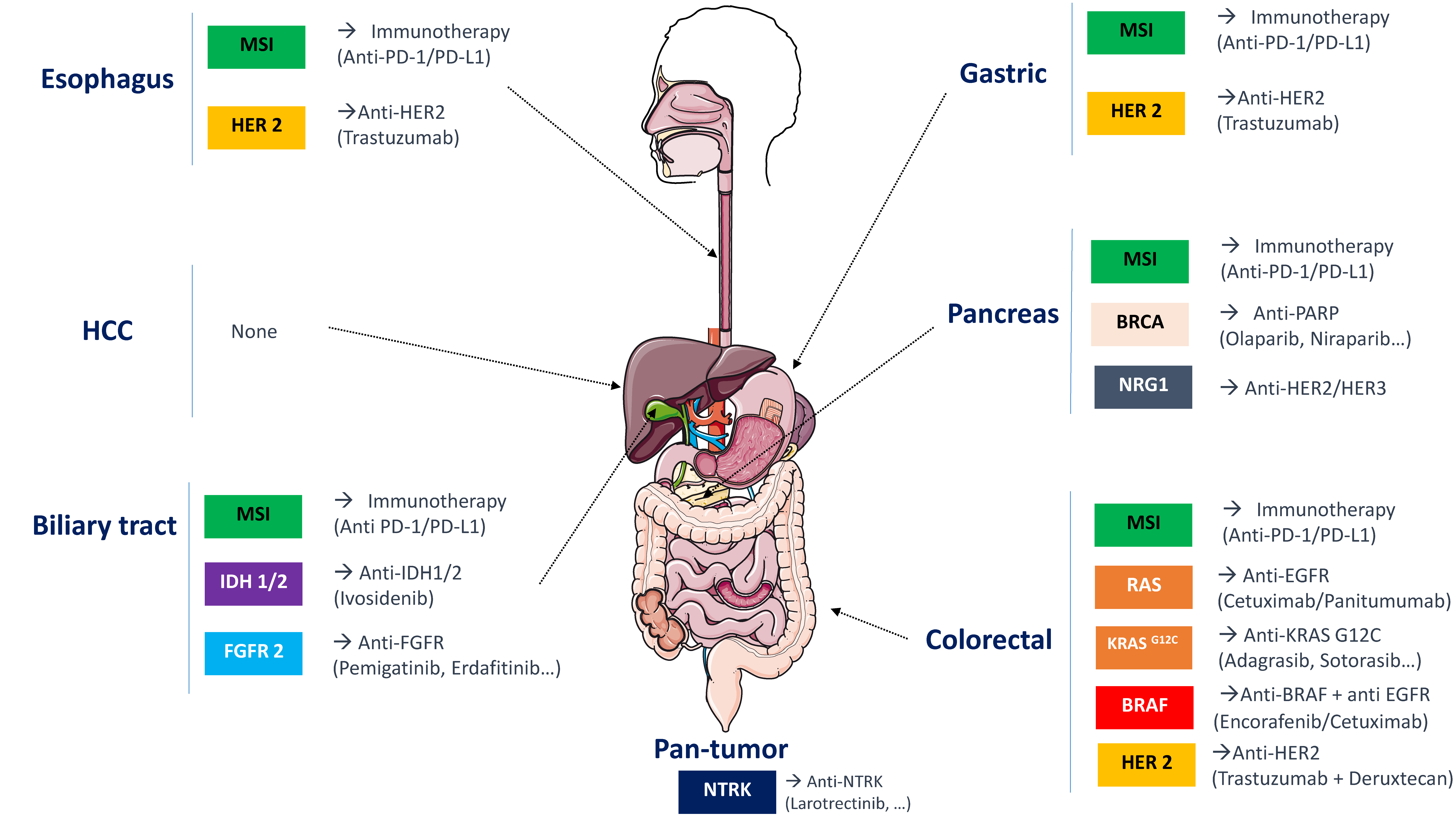
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Slatko, B.E.; Gardner, A.F.; Ausubel, F.M. Overview of Next Generation Sequencing Technologies. Curr. Protoc. Mol. Biol. 2018, 122, e59. [Google Scholar] [CrossRef]
- Colomer, R.; Mondejar, R.; Romero-Laorden, N.; Alfranca, A.; Sanchez-Madrid, F.; Quintela-Fandino, M. When Should We Order a next Generation Sequencing Test in a Patient with Cancer? EClinicalMedicine 2020, 25, 100487. [Google Scholar] [CrossRef] [PubMed]
- Yohe, S.; Thyagarajan, B. Review of Clinical Next-Generation Sequencing. Arch. Pathol. Lab. Med. 2017, 141, 1544–1557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulski, J.K. Next-Generation Sequencing—An Overview of the History, Tools, and “Omic” Applications; IntechOpen: London, UK, 2016; ISBN 978-953-51-2240-1. [Google Scholar]
- Global Cancer Observatory. Available online: https://gco.iarc.fr/ (accessed on 11 September 2020).
- Lièvre, A.; Bachet, J.-B.; Le Corre, D.; Boige, V.; Landi, B.; Emile, J.-F.; Côté, J.-F.; Tomasic, G.; Penna, C.; Ducreux, M.; et al. KRAS Mutation Status Is Predictive of Response to Cetuximab Therapy in Colorectal Cancer. Cancer Res. 2006, 66, 3992–3995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douillard, J.-Y.; Oliner, K.S.; Siena, S.; Tabernero, J.; Burkes, R.; Barugel, M.; Humblet, Y.; Bodoky, G.; Cunningham, D.; Jassem, J.; et al. Panitumumab-FOLFOX4 Treatment and RAS Mutations in Colorectal Cancer. N. Engl. J. Med. 2013, 369, 1023–1034. [Google Scholar] [CrossRef] [Green Version]
- Piton, N.; Lonchamp, E.; Nowak, F.; Sabourin, J.-C. Real-Life Distribution of KRAS and NRAS Mutations in Metastatic Colorectal Carcinoma from French Routine Genotyping. Cancer Epidemiol. Prev. Biomark. 2015, 24, 1416–1418. [Google Scholar] [CrossRef] [Green Version]
- Schirripa, M.; Cremolini, C.; Loupakis, F.; Morvillo, M.; Bergamo, F.; Zoratto, F.; Salvatore, L.; Antoniotti, C.; Marmorino, F.; Sensi, E.; et al. Role of NRAS Mutations as Prognostic and Predictive Markers in Metastatic Colorectal Cancer. Int. J. Cancer 2015, 136, 83–90. [Google Scholar] [CrossRef]
- De Roock, W.; Claes, B.; Bernasconi, D.; De Schutter, J.; Biesmans, B.; Fountzilas, G.; Kalogeras, K.T.; Kotoula, V.; Papamichael, D.; Laurent-Puig, P.; et al. Effects of KRAS, BRAF, NRAS, and PIK3CA Mutations on the Efficacy of Cetuximab plus Chemotherapy in Chemotherapy-Refractory Metastatic Colorectal Cancer: A Retrospective Consortium Analysis. Lancet Oncol. 2010, 11, 753–762. [Google Scholar] [CrossRef]
- Karapetis, C.S.; Khambata-Ford, S.; Jonker, D.J.; O’Callaghan, C.J.; Tu, D.; Tebbutt, N.C.; Simes, R.J.; Chalchal, H.; Shapiro, J.D.; Robitaille, S.; et al. K-Ras Mutations and Benefit from Cetuximab in Advanced Colorectal Cancer. N. Engl. J. Med. 2008, 359, 1757–1765. [Google Scholar] [CrossRef] [Green Version]
- Taieb, J.; Jung, A.; Sartore-Bianchi, A.; Peeters, M.; Seligmann, J.; Zaanan, A.; Burdon, P.; Montagut, C.; Laurent-Puig, P. The Evolving Biomarker Landscape for Treatment Selection in Metastatic Colorectal Cancer. Drugs 2019, 79, 1375–1394. [Google Scholar] [CrossRef] [Green Version]
- Phelip, J.M.; Tougeron, D.; Léonard, D.; Benhaim, L.; Desolneux, G.; Dupré, A.; Michel, P.; Penna, C.; Tournigand, C.; Louvet, C.; et al. Metastatic colorectal cancer (mCRC): French intergroup clinical practice guidelines for diagnosis, treatments and follow-up (SNFGE, FFCD, GERCOR, UNICANCER, SFCD, SFED, SFRO, SFR). Dig. Liver Dis. 2019, 51, 1357–1363. [Google Scholar] [CrossRef]
- Van Cutsem, E.; Cervantes, A.; Nordlinger, B.; Arnold, D. Metastatic Colorectal Cancer: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2014, 25, iii1–iii9. [Google Scholar] [CrossRef]
- Overman, M.J.; McDermott, R.; Leach, J.L.; Lonardi, S.; Lenz, H.-J.; Morse, M.A.; Desai, J.; Hill, A.; Axelson, M.; Moss, R.A.; et al. Nivolumab in Patients with Metastatic DNA Mismatch Repair Deficient/Microsatellite Instability–High Colorectal Cancer (CheckMate 142): Results of an Open-Label, Multicentre, Phase 2 Study. Lancet Oncol. 2017, 18, 1182–1191. [Google Scholar] [CrossRef]
- Rey, J.-M.; Ducros, V.; Pujol, P.; Wang, Q.; Buisine, M.-P.; Aissaoui, H.; Maudelonde, T.; Olschwang, S. Improving Mutation Screening in Patients with Colorectal Cancer Predisposition Using Next-Generation Sequencing. J. Mol. Diagn. JMD 2017, 19, 589–601. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.H.; Yu, G.Y.; Hong, Y.G.; Lian, W.; Chouhan, H.; Xu, Y.; Liu, L.J.; Bai, C.G.; Zhang, W. Clinical Significance of Multiple Gene Detection with a 22-Gene Panel in Formalin-Fixed Paraffin-Embedded Specimens of 207 Colorectal Cancer Patients. Int. J. Clin. Oncol. 2019, 24, 141–152. [Google Scholar] [CrossRef]
- Dienstmann, R.; Mason, M.J.; Sinicrope, F.A.; Phipps, A.I.; Tejpar, S.; Nesbakken, A.; Danielsen, S.A.; Sveen, A.; Buchanan, D.D.; Clendenning, M.; et al. Prediction of Overall Survival in Stage II and III Colon Cancer beyond TNM System: A Retrospective, Pooled Biomarker Study. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2017, 28, 1023–1031. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Li, W.; Huang, Y.; Huang, M.; Li, S.; Zhai, X.; Zhao, J.; Gao, C.; Xie, W.; Qin, H.; et al. A Next-Generation Sequencing-Based Strategy Combining Microsatellite Instability and Tumor Mutation Burden for Comprehensive Molecular Diagnosis of Advanced Colorectal Cancer. BMC Cancer 2021, 21, 282. [Google Scholar] [CrossRef] [PubMed]
- Almuzzaini, B.; Alghamdi, J.; Alomani, A.; AlGhamdi, S.; Alsharm, A.A.; Alshieban, S.; Sayed, A.; Alhejaily, A.G.; Aljaser, F.S.; Abudawood, M.; et al. Identification of Novel Mutations in Colorectal Cancer Patients Using AmpliSeq Comprehensive Cancer Panel. J. Pers. Med. 2021, 11, 535. [Google Scholar] [CrossRef] [PubMed]
- Mosele, F.; Remon, J.; Mateo, J.; Westphalen, C.B.; Barlesi, F.; Lolkema, M.P.; Normanno, N.; Scarpa, A.; Robson, M.; Meric-Bernstam, F.; et al. Recommendations for the Use of Next-Generation Sequencing (NGS) for Patients with Metastatic Cancers: A Report from the ESMO Precision Medicine Working Group. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2020. [Google Scholar] [CrossRef]
- Legras, A.; Barritault, M.; Tallet, A.; Fabre, E.; Guyard, A.; Rance, B.; Digan, W.; Pecuchet, N.; Giroux-Leprieur, E.; Julie, C.; et al. Validity of Targeted Next-Generation Sequencing in Routine Care for Identifying Clinically Relevant Molecular Profiles in Non-Small-Cell Lung Cancer: Results of a 2-Year Experience on 1343 Samples. J. Mol. Diagn. JMD 2018, 20, 550–564. [Google Scholar] [CrossRef] [Green Version]
- Umar, A.; Boland, C.R.; Terdiman, J.P.; Syngal, S.; de la Chapelle, A.; Rüschoff, J.; Fishel, R.; Lindor, N.M.; Burgart, L.J.; Hamelin, R.; et al. Revised Bethesda Guidelines for Hereditary Nonpolyposis Colorectal Cancer (Lynch syndrome) and Microsatellite Instability. J. Natl. Cancer Inst. 2004, 96, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Luchini, C.; Bibeau, F.; Ligtenberg, M.J.L.; Singh, N.; Nottegar, A.; Bosse, T.; Miller, R.; Riaz, N.; Douillard, J.-Y.; Andre, F.; et al. ESMO Recommendations on Microsatellite Instability Testing for Immunotherapy in Cancer, and Its Relationship with PD-1/PD-L1 Expression and Tumour Mutational Burden: A Systematic Review-Based Approach. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2019, 30, 1232–1243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, P.A.; Taylor, R.; Minor, B.L.; Elliott, V.; Fernandez, M.; O’Neal, L.; McLeod, L.; Delacqua, G.; Delacqua, F.; Kirby, J.; et al. The REDCap Consortium: Building an International Community of Software Platform Partners. J. Biomed. Inform. 2019, 95, 103208. [Google Scholar] [CrossRef] [PubMed]
- Harris, P.A.; Taylor, R.; Thielke, R.; Payne, J.; Gonzalez, N.; Conde, J.G. Research Electronic Data Capture (REDCap)--a Metadata-Driven Methodology and Workflow Process for Providing Translational Research Informatics Support. J. Biomed. Inform. 2009, 42, 377–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blons, H.; Rouleau, E.; Charrier, N.; Chatellier, G.; Côté, J.-F.; Pages, J.-C.; de Fraipont, F.; Boyer, J.-C.; Merlio, J.P.; Morel, A.; et al. Performance and Cost Efficiency of KRAS Mutation Testing for Metastatic Colorectal Cancer in Routine Diagnosis: The MOKAECM Study, a Nationwide Experience. PLoS ONE 2013, 8, e68945. [Google Scholar] [CrossRef] [Green Version]
- Yao, Z.; Yaeger, R.; Rodrik-Outmezguine, V.S.; Tao, A.; Torres, N.M.; Chang, M.T.; Drosten, M.; Zhao, H.; Cecchi, F.; Hembrough, T.; et al. Tumours with Class 3 BRAF Mutants Are Sensitive to the Inhibition of Activated RAS. Nature 2017, 548, 234–238. [Google Scholar] [CrossRef]
- Tutuka, C.S.A.; Andrews, M.C.; Mariadason, J.M.; Ioannidis, P.; Hudson, C.; Cebon, J.; Behren, A. PLX8394, a New Generation BRAF Inhibitor, Selectively Inhibits BRAF in Colonic Adenocarcinoma Cells and Prevents Paradoxical MAPK Pathway Activation. Mol. Cancer 2017, 16, 112. [Google Scholar] [CrossRef] [Green Version]
- Marabelle, A.; Le, D.T.; Ascierto, P.A.; Giacomo, A.M.D.; Jesus-Acosta, A.D.; Delord, J.-P.; Geva, R.; Gottfried, M.; Penel, N.; Hansen, A.R.; et al. Efficacy of Pembrolizumab in Patients With Noncolorectal High Microsatellite Instability/Mismatch Repair–Deficient Cancer: Results From the Phase II KEYNOTE-158 Study. J. Clin. Oncol. 2019, 38, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kopetz, S.; Grothey, A.; Yaeger, R.; Van Cutsem, E.; Desai, J.; Yoshino, T.; Wasan, H.; Ciardiello, F.; Loupakis, F.; Hong, Y.S.; et al. Encorafenib, Binimetinib, and Cetuximab in BRAF V600E–Mutated Colorectal Cancer. N. Engl. J. Med. 2019, 381, 1632–1643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meeting Library|A Phase II, Multicenter, Open-Label Study of Trastuzumab Deruxtecan (T-DXd; DS-8201) in Patients (Pts) with HER2-Expressing Metastatic Colorectal Cancer (MCRC): DESTINY-CRC01. Available online: https://meetinglibrary.asco.org/record/185482/abstract (accessed on 10 September 2020).
- Sartore-Bianchi, A.; Trusolino, L.; Martino, C.; Bencardino, K.; Lonardi, S.; Bergamo, F.; Zagonel, V.; Leone, F.; Depetris, I.; Martinelli, E.; et al. Dual-Targeted Therapy with Trastuzumab and Lapatinib in Treatment-Refractory, KRAS Codon 12/13 Wild-Type, HER2-Positive Metastatic Colorectal Cancer (HERACLES): A Proof-of-Concept, Multicentre, Open-Label, Phase 2 Trial. Lancet Oncol. 2016, 17, 738–746. [Google Scholar] [CrossRef]
- Johnson, M.L.; Ou, S.H.I.; Barve, M.; Rybkin, I.I.; Papadopoulos, K.P.; Leal, T.A.; Velastegui, K.; Christensen, J.G.; Kheoh, T.; Chao, R.C.; et al. KRYSTAL-1: Activity and Safety of Adagrasib (MRTX849) in Patients with Colorectal Cancer (CRC) and Other Solid Tumors Harboring a KRAS G12C Mutation. Eur. J. Cancer 2020, 138, S2. [Google Scholar] [CrossRef]
- Hong, D.S.; Fakih, M.G.; Strickler, J.H.; Desai, J.; Durm, G.A.; Shapiro, G.I.; Falchook, G.S.; Price, T.J.; Sacher, A.; Denlinger, C.S.; et al. KRASG12C Inhibition with Sotorasib in Advanced Solid Tumors. N. Engl. J. Med. 2020, 383, 1207–1217. [Google Scholar] [CrossRef] [PubMed]
- AACR Project GENIE Consortium AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017, 7, 818–831. [CrossRef] [Green Version]
- Massard, C.; Michiels, S.; Ferté, C.; Deley, M.-C.L.; Lacroix, L.; Hollebecque, A.; Verlingue, L.; Ileana, E.; Rosellini, S.; Ammari, S.; et al. High-Throughput Genomics and Clinical Outcome in Hard-to-Treat Advanced Cancers: Results of the MOSCATO 01 Trial. Cancer Discov. 2017, 7, 586–595. [Google Scholar] [CrossRef] [Green Version]
- Tourneau, C.L.; Delord, J.-P.; Gonçalves, A.; Gavoille, C.; Dubot, C.; Isambert, N.; Campone, M.; Trédan, O.; Massiani, M.-A.; Mauborgne, C.; et al. Molecularly Targeted Therapy Based on Tumour Molecular Profiling versus Conventional Therapy for Advanced Cancer (SHIVA): A Multicentre, Open-Label, Proof-of-Concept, Randomised, Controlled Phase 2 Trial. Lancet Oncol. 2015, 16, 1324–1334. [Google Scholar] [CrossRef]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The Consensus Molecular Subtypes of Colorectal Cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Schrock, A.B.; Ouyang, C.; Sandhu, J.; Sokol, E.; Jin, D.; Ross, J.S.; Miller, V.A.; Lim, D.; Amanam, I.; Chao, J.; et al. Tumor Mutational Burden Is Predictive of Response to Immune Checkpoint Inhibitors in MSI-High Metastatic Colorectal Cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2019, 30, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
| Variable (N=) | Median (IQR) (%) | ||
|---|---|---|---|
| Number of Patients | 210 | ||
| Median follow-up (months) | 25.4 | (14.9–39.5) | |
| Sex | Female | 101 | (48) |
| Male | 109 | (52) | |
| Age | 67.5 | (58.1–76.2) | |
| Location (N = 207) | Right Colon | 72 | (35) |
| Left colon | 102 | (49) | |
| Rectum | 33 | (16) | |
| Metastasis | Synchronous | 140 | (67) |
| Metachronous | 70 | (33) | |
| Systemic treatment | Yes | 186 | (89) |
| (N = 207) | No | 24 | (11) |
| Performance status (at first-line treatment of metastatic disease) | 0 | 75 | (37) |
| (N = 203) | 1 | 93 | (46) |
| 2 | 31 | (15) | |
| 3 | 4 | (2) | |
| CEA 1 (N = 181) | 9.5 | (3–45) | |
| CA 19.9 1 (N = 170) | 39 | (13–246) | |
| Specific targeted therapy based on NGS results | Clinical trial | 2 | (40) |
| Off-label | 3 | (60) | |
| Variable | Median (Range)/N (%) | |||
|---|---|---|---|---|
| Molecular alteration on NGS panel (N = 199) | N (%) | |||
| Gene mutations | TP53 | 132 (63) | ERBB4 | 4 (2) |
| KRAS | 86 (41) | ERBB2 | 2 (1) | |
| PIK3CA | 39 (19) | FGFR2 | 2 (1) | |
| BRAF | 20 (10) | NOTCH1 | 2 (1) | |
| SMAD4 | 20 (10) | DDR2 | 2 (1) | |
| FBXW7 | 12 (6) | MAP2K1 | 2 (1) | |
| NRAS | 10 (5) | ALK | 1 (0.5) | |
| PTEN | 7 (3) | FGFR1 | 1 (0.5) | |
| AKT1 | 5 (2) | STK11 | 1 (0.5) | |
| CTNNB1 | 4 (2) | FGFR3 | 1 (0.5) | |
| Amplifications | ERBB2 | 5 (2.5) | MAP2K1 | 2(1) |
| FGFR 3 | 4 (2) | NOTCH 1 | 1 (0.5) | |
| MET | 3 (1.5) | PIK3CA | 1 (0.5) | |
| KRAS | 3 (1.5) | FGFR2 | 1(0.5) | |
| BRAF | 2(1) | |||
| Number of variants/patient | 0 | 16 (8) | ||
| 1 | 67 (34) | |||
| 2 | 72 (36) | |||
| 3 | 29 (15) | |||
| 4 et + | 15 (7) | |||
| Microsatellite instability status | ||||
| Immunohisto-chemistry | MMR proficiency | 164 (90) | ||
| (N = 183) | MMR deficiency | 19 (10) | ||
| Molecular biology | MSS | 146 (88) | ||
| (N = 165) | MSI | 19 (12) | ||
| Not evaluable | 4 (3) | |||
| Molecular alteration (TaqMan) (N = 196) | ||||
| KRAS | WT | 128 (65) | ||
| Mutated | 68 (35) | |||
| BRAF | WT | 181 (92) | ||
| Mutated | 15 (8) |
| Variable | OS Hazard Ratio [95% CI] p-Value | |
|---|---|---|
| Univariate Analysis | Multivariate Analysis | |
| Performance status (≥2 or <2) | 3.67 (2.10–6.42) <0.001 | 4.91 (1.84–13.1) 0.001 |
| Differentiation (poorly differentiated/undifferentiated vs. well or moderately differentiated) | 2.82 (1.25–6.35) 0.012 | 4.70 (1.51–14.6) 0.007 |
| T (T4 vs. T1/T2/T3) | 1.90 (1.05–3.44) 0.034 | 0.95 (0.51–2.63) 0.73 |
| Microsatellite instability status | 0.47 (0.22–1) 0.05 | 1.05 (0.23–4.70) 0.9 |
| BRAF (mutated vs. wild-type) | 2.44 (1.21–4.93) 0.013 | 1.75 (0.46–6.58) 0.41 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bayle, A.; Basile, D.; Garinet, S.; Rance, B.; Laurent-Puig, P.; Blons, H.; Taieb, J.; Perkins, G. Next-Generation Sequencing Targeted Panel in Routine Care for Metastatic Colon Cancers. Cancers 2021, 13, 5750. https://doi.org/10.3390/cancers13225750
Bayle A, Basile D, Garinet S, Rance B, Laurent-Puig P, Blons H, Taieb J, Perkins G. Next-Generation Sequencing Targeted Panel in Routine Care for Metastatic Colon Cancers. Cancers. 2021; 13(22):5750. https://doi.org/10.3390/cancers13225750
Chicago/Turabian StyleBayle, Arnaud, Debora Basile, Simon Garinet, Bastien Rance, Pierre Laurent-Puig, Hélène Blons, Julien Taieb, and Geraldine Perkins. 2021. "Next-Generation Sequencing Targeted Panel in Routine Care for Metastatic Colon Cancers" Cancers 13, no. 22: 5750. https://doi.org/10.3390/cancers13225750







