MCF-7 Drug Resistant Cell Lines Switch Their Lipid Metabolism to Triple Negative Breast Cancer Signature
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Conditioned Media Obtainment
2.3. Oil Red Staining
2.4. RNA Extraction and Quantitative Real Time (qRT) PCR
2.5. Bodipy Fluorescent Staining
2.6. Enzyme-Linked Immunosorbent Assay (ELISA)
2.7. Lipid Uptake Assay: Nile Red Staining
2.8. C13 Palmitic Assay
2.9. Lipidomics
2.10. Statistical Analysis
3. Results
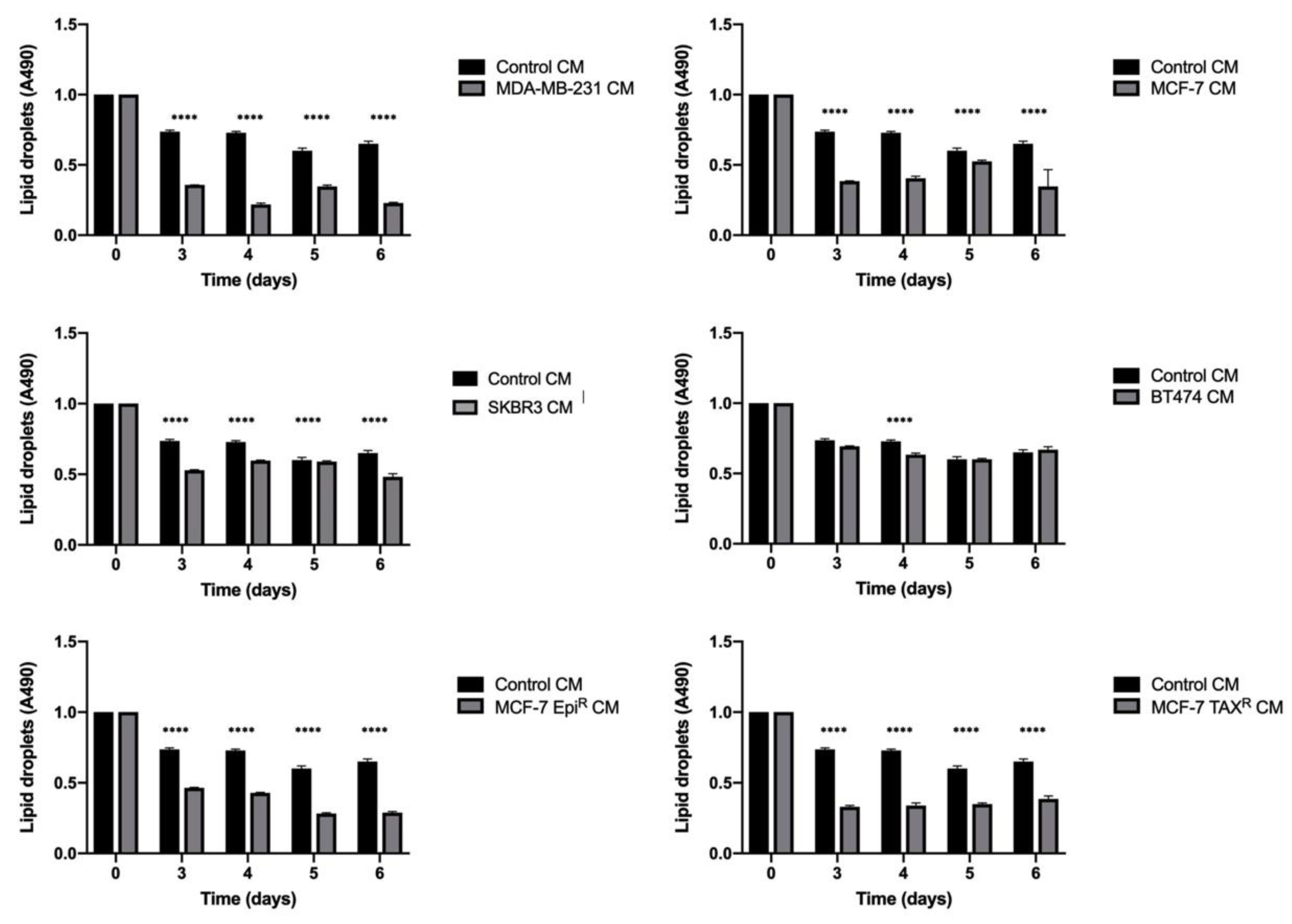
3.1. Breast Cancer Cell Conditioned Media Enhances Delipidation of Mature Adipocytes
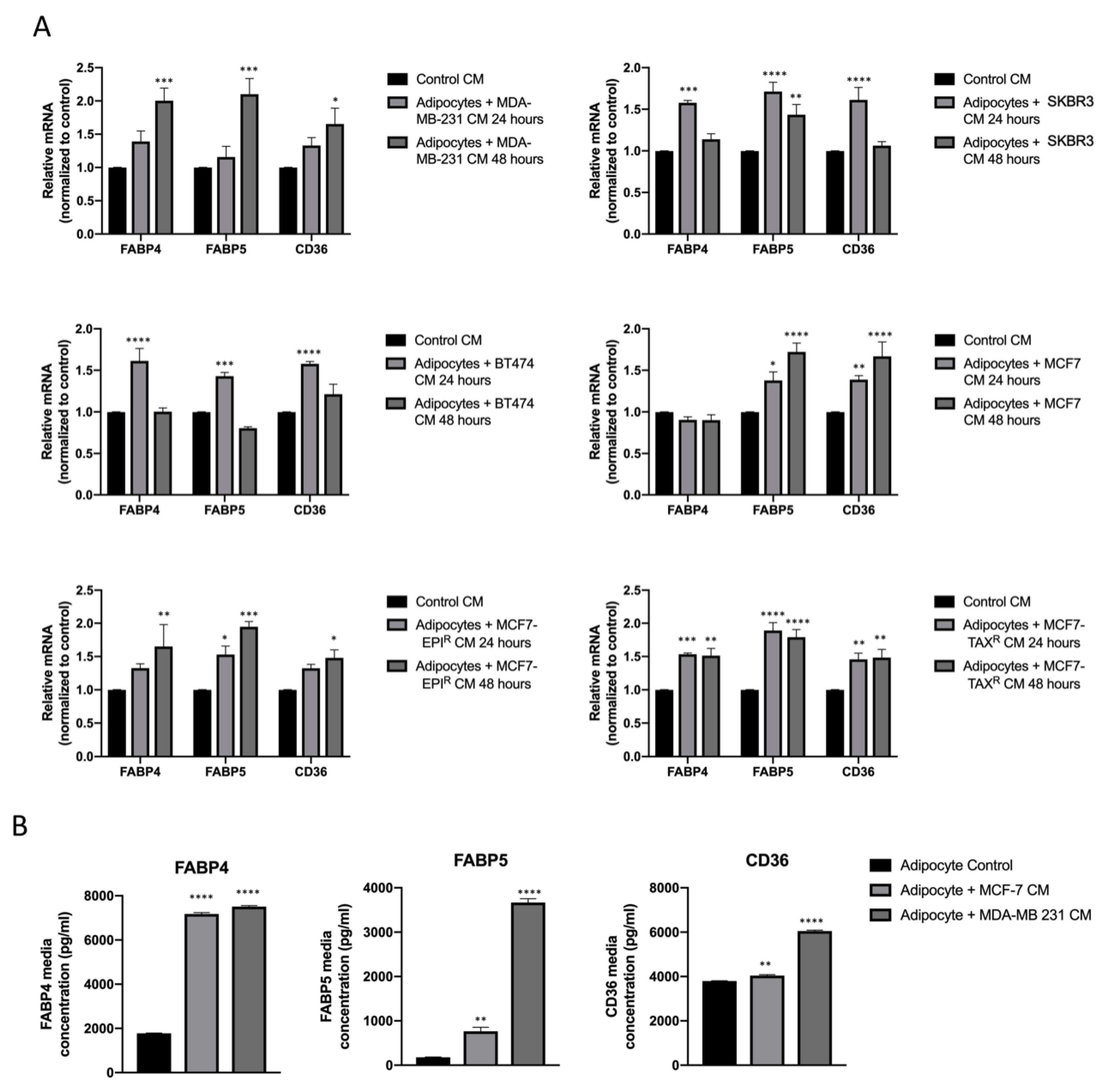
3.2. Breast Cancer Cell Conditioned Media Enhance FABP4, FABP5 and CD36 Release from Mature Adipocytes
3.3. Mature Adipocytes Conditioned Media Increases Fatty Acid Uptake in Breast Cancer Cell Lines
3.4. Mature Adipocytes Internalize Palmitic Acid during Differentiation and Export it as Lipids to Breast Cancer Cells
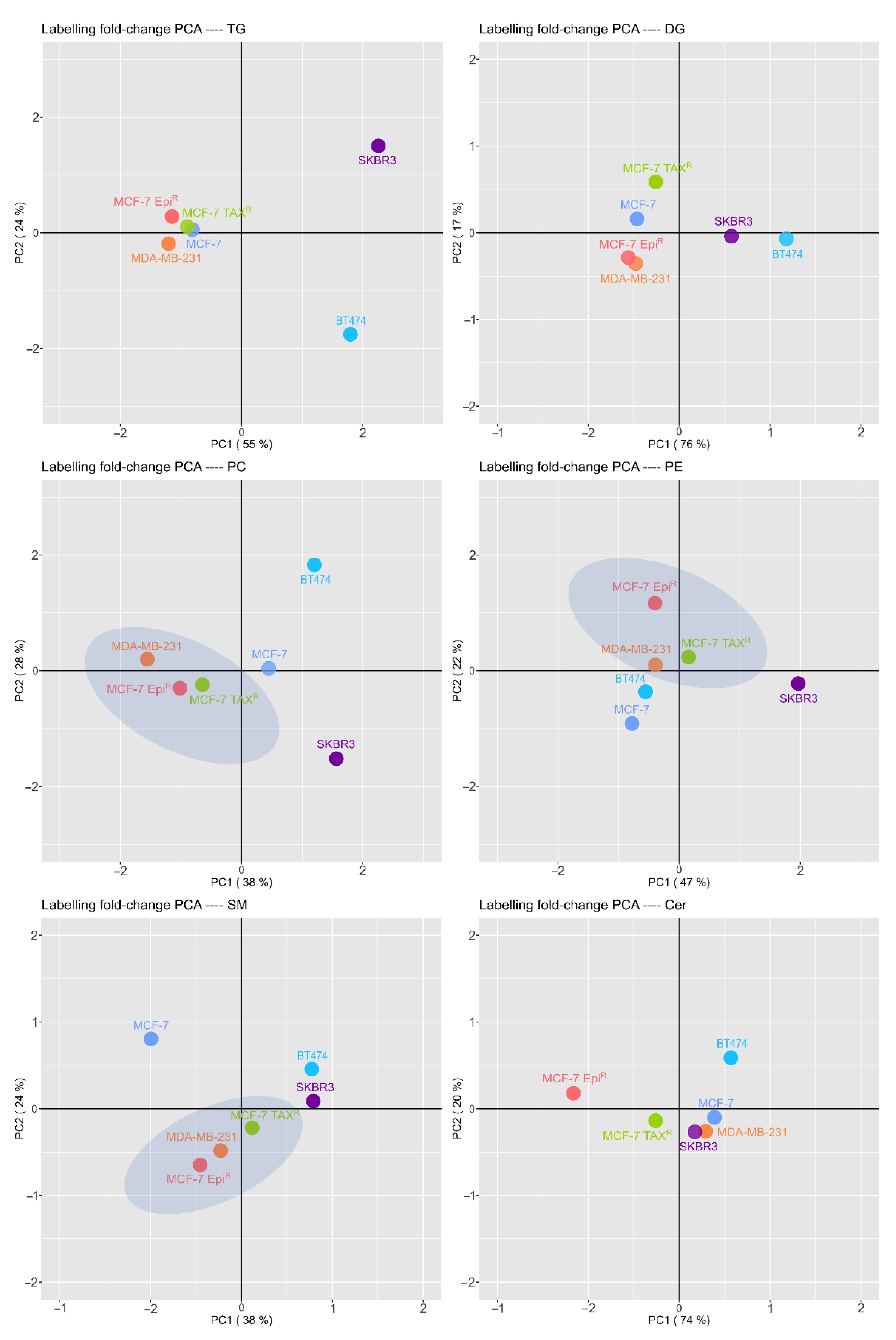
3.5. Breast Cancer Cells have a Different and Specific Lipid Signature in the Palmitic Transformation and Resistant Cells Lipid Pattern Differs from Their Sensitive Cell Line Being Closely to TNBC Cell Line Pattern
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Polednak, A.P. Estimating the number of U.S. incident cancers attributable to obesity and the impact on temporal trends in incidence rates for obesity-related cancers. Cancer Detect. Prev. 2008, 32, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Coelho, M.; Oliveira, T.; Fernandes, R. Biochemistry of adipose tissue: An endocrine organ. Arch. Med. Sci. 2013, 9, 191–200. [Google Scholar] [CrossRef] [Green Version]
- Muller, C. Tumour-surrounding adipocytes are active players in breast cancer progression. Ann. Endocrinol. 2013, 74, 108–110. [Google Scholar] [CrossRef]
- Bochet, L.; Lehuédé, C.; Dauvillier, S.; Wang, Y.Y.; Dirat, B.; Laurent, V.; Dray, C.; Guiet, R.; Maridonneau-Parini, I.; Le Gonidec, S.; et al. Adipocyte-derived fibroblasts promote tumor progression and contribute to the desmoplastic reaction in breast cancer. Cancer Res. 2013, 73, 5657–5668. [Google Scholar] [CrossRef] [Green Version]
- Picon-Ruiz, M.; Pan, C.; Drews-Elger, K.; Jang, K.; Besser, A.H.; Zhao, D.; Morata-Tarifa, C.; Kim, M.; Ince, T.A.; Azzam, D.J.; et al. Interactions between adipocytes and breast cancer cells stimulate cytokine production and drive Src/Sox2/miR-302b-mediated malignant progression. Cancer Res. 2016, 76, 491–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishna Muppalla, J.N.; Muddana, K.; Reddy Dorankula, S.P.; Rao Thokala, M.; Prakash Pasupula, A. Microenvironment-A role in tumour progression and prognosis. J. Clin. Diagn. Res. 2013, 7, 2096–2099. [Google Scholar]
- Nieman, K.M.; Romero, I.L.; Van Houten, B.; Lengyel, E. Adipose tissue and adipocytes support tumorigenesis and metastasis. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2013, 1831, 1533–1541. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, N.; Escalona, R.; Leung, D.; Chan, E.; Kannourakis, G. Tumour microenvironment and metabolic plasticity in cancer and cancer stem cells: Perspectives on metabolic and immune regulatory signatures in chemoresistant ovarian cancer stem cells. Semin. Cancer Biol. 2018, 53, 265–281. [Google Scholar] [CrossRef]
- Rybinska, I.; Mangano, N.; Tagliabue, E.; Triulzi, T. Cancer-associated adipocytes in breast cancer: Causes and consequences. Int. J. Mol. Sci. 2021, 22, 3775. [Google Scholar] [CrossRef]
- Attané, C.; Muller, C. Drilling for Oil: Tumor-Surrounding Adipocytes Fueling Cancer. Trends Cancer 2020, 6, 593–604. [Google Scholar] [CrossRef]
- Yang, D.; Li, Y.; Xing, L.; Tan, Y.; Sun, J.; Zeng, B.; Xiang, T.; Tan, J.; Ren, G.; Wang, Y. Utilization of adipocyte-derived lipids and enhanced intracellular trafficking of fatty acids contribute to breast cancer progression. Cell Commun. Signal. 2018, 16, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harayama, T.; Riezman, H. Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol. 2018, 19, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Sunshine, H.; Iruela-Arispe, M.L. Membrane lipids and cell signaling. Curr. Opin. Lipidol. 2017, 28, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Fahy, E.; Subramaniam, S.; Murphy, R.C.; Nishijima, M.; Raetz, C.R.H.; Shimizu, T.; Spener, F.; Van Meer, G.; Wakelam, M.J.O.; Dennis, E.A. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 2009, 50, S9. [Google Scholar] [CrossRef] [Green Version]
- Beloribi-Djefaflia, S.; Vasseur, S.; Guillaumond, F. Lipid metabolic reprogramming in cancer cells. Oncogenesis 2016, 5, e189. [Google Scholar] [CrossRef]
- Orešič, M.; Hänninen, V.A.; Vidal-Puig, A. Lipidomics: A new window to biomedical frontiers. Trends Biotechnol. 2008, 26, 647–652. [Google Scholar] [CrossRef]
- Zeleznik, O.A.; Clish, C.B.; Kraft, P.; Avila-Pacheco, J.; Eliassen, A.H.; Tworoger, S.S. Circulating Lysophosphatidylcholines, Phosphatidylcholines, Ceramides, and Sphingomyelins and Ovarian Cancer Risk: A 23-Year Prospective Study. J. Natl. Cancer Inst. 2020, 112, 628–636. [Google Scholar] [CrossRef]
- Chen, X.; Chen, H.; Dai, M.; Ai, J.; Li, Y.; Mahon, B.; Dai, S.; Deng, Y. Plasma lipidomics profiling identified lipid biomarkers in distinguishing early-stage breast cancer from benign lesions. Oncotarget 2016, 7, 36622–36631. [Google Scholar] [CrossRef]
- Hammad, L.A.; Wu, G.; Saleh, M.M.; Klouckova, I.; Dobrolecki, L.E.; Hickey, R.J.; Schnaper, L.; Novotny, M.V.; Mechref, Y. Elevated levels of hydroxylated phosphocholine lipids in the blood serum of breast cancer patients. Rapid Commun. Mass Spectrom. 2009, 23, 863–876. [Google Scholar] [CrossRef]
- Li, X.; Liu, Z.L.; Wu, Y.T.; Wu, H.; Dai, W.; Arshad, B.; Xu, Z.; Li, H.; Wu, K.N.; Kong, L.Q. Status of lipid and lipoprotein in female breast cancer patients at initial diagnosis and during chemotherapy. Lipids Health Dis. 2018, 17, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Gumà, J.; Adriá-Cebrián, J.; Ruiz-Aguado, B.; Albacar, C.; Girona, J.; Rodríguez-Calvo, R.; Martínez-Micaelo, N.; Lam, E.W.F.; Masana, L.; Guaita-Esteruelas, S. Altered serum metabolic profile assessed by advanced 1h-nmr in breast cancer patients. Cancers 2021, 13, 4281. [Google Scholar] [CrossRef] [PubMed]
- Fahy, E.; Sud, M.; Cotter, D.; Subramaniam, S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007, 35 (Suppl. 2), W606–W612. [Google Scholar] [CrossRef] [Green Version]
- Kind, T.; Liu, K.H.; Lee, D.Y.; Defelice, B.; Meissen, J.K.; Fiehn, O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat. Methods 2013, 10, 755–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montenegro-Burke, J.R.; Guijas, C.; Siuzdak, G. METLIN: A Tandem Mass Spectral Library of Standards. Methods Mol. Biol. 2020, 2104, 149–163. [Google Scholar] [PubMed]
- Capellades, J.; Navarro, M.; Samino, S.; Garcia-Ramirez, M.; Hernandez, C.; Simo, R.; Vinaixa, M.; Yanes, O. GeoRge: A Computational Tool to Detect the Presence of Stable Isotope Labeling in LC/MS-Based Untargeted Metabolomics. Anal. Chem. 2016, 88, 621–628. [Google Scholar] [CrossRef]
- Aznar Benitah, S. Metastatic-initiating cells and lipid metabolism. Cell Stress 2017, 1, 110–114. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.; Narita, S.; Inoue, T.; Koizumi, A.; Saito, M.; Tsuruta, H.; Numakura, K.; Satoh, S.; Nanjo, H.; Sasaki, T.; et al. Fatty acid binding protein 4 enhances prostate cancer progression by upregulating matrix metalloproteinases and stromal cell cytokine production. Oncotarget 2017, 8, 111780–111794. [Google Scholar] [CrossRef] [Green Version]
- Pan, J.; Fan, Z.; Wang, Z.; Dai, Q.; Xiang, Z.; Yuan, F.; Yan, M.; Zhu, Z.; Liu, B.; Li, C. CD36 mediates palmitate acid-induced metastasis of gastric cancer via AKT/GSK-3β/β-catenin pathway. J. Exp. Clin. Cancer Res. 2019, 38, 52. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.Y.; Attané, C.; Milhas, D.; Dirat, B.; Dauvillier, S.; Guerard, A.; Gilhodes, J.; Lazar, I.; Alet, N.; Laurent, V.; et al. Mammary adipocytes stimulate breast cancer invasion through metabolic remodeling of tumor cells. J. Clin. Investig. 2017, 2, e87489. [Google Scholar] [CrossRef] [Green Version]
- Nieman, K.M.; Kenny, H.A.; Penicka, C.V.; Ladanyi, A.; Buell-Gutbrod, R.; Zillhardt, M.R.; Romero, I.L.; Carey, M.S.; Mills, G.B.; Hotamisligil, G.S.; et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med. 2011, 17, 1498–1503. [Google Scholar] [CrossRef] [Green Version]
- Fatima, S.; Hu, X.; Gong, R.-H.; Huang, C.; Chen, M.; Wong, H.L.X.; Bian, Z.; Kwan, H.Y. Palmitic acid is an intracellular signaling molecule involved in disease development. Cell. Mol. Life Sci. 2019, 76, 2547–2557. [Google Scholar] [CrossRef]
- Louie, S.M.; Roberts, L.S.; Mulvihill, M.M.; Luo, K.; Nomura, D.K. Cancer cells incorporate and remodel exogenous palmitate into structural and oncogenic signaling lipids. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2013, 1831, 1566–1572. [Google Scholar] [CrossRef] [Green Version]
- Hilvo, M.; Denkert, C.; Lehtinen, L.; Müller, B.; Brockmöller, S.; Seppänen-Laakso, T.; Budczies, J.; Bucher, E.; Yetukuri, L.; Castillo, S.; et al. Novel theranostic opportunities offered by characterization of altered membrane lipid metabolism in breast cancer progression. Cancer Res. 2011, 71, 3236–3245. [Google Scholar] [CrossRef] [Green Version]
- Nagahashi, M.; Tsuchida, J.; Moro, K.; Hasegawa, M.; Tatsuda, K.; Woelfel, I.A.; Takabe, K.; Wakai, T. High levels of sphingolipids in human breast cancer. J. Surg. Res. 2016, 204, 435–444. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Y.; Zhou, B.; Su, M.; Baxter, S.; Zheng, X.; Zhao, X.; Yen, Y.; Jia, W. Mass spectrometry-based quantitative metabolomics revealed a distinct lipid profile in breast cancer patients. Int. J. Mol. Sci. 2013, 14, 8047–8061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Luo, Q.; Xiao, Y.; Li, H.; Kong, L.; Ren, G. The implication from RAS/RAF/ERK signaling pathway increased activation in epirubicin treated triple negative breast cancer. Oncotarget 2017, 8, 108249–108260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogretmen, B.; Hannun, Y.A. Biologically active sphingolipids in cancer pathogenesis and treatment. Nat. Rev. Cancer 2004, 4, 604–616. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Muñoz, A. Ceramide 1-phosphate/ceramide, a switch between life and death. Biochim. Biophys. Acta-Biomembr. 2006, 1758, 2049–2056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Regina Todeschini, A.; Hakomori, S. itiroh Functional role of glycosphingolipids and gangliosides in control of cell adhesion, motility, and growth, through glycosynaptic microdomains. Biochim. Biophys. Acta-Gen. Subj. 2008, 1780, 421–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiffmann, S.; Sandner, J.; Birod, K.; Wobst, I.; Angioni, C.; Ruckhäberle, E.; Kaufmann, M.; Ackermann, H.; Lötsch, J.; Schmidt, H.; et al. Ceramide synthases and ceramide levels are increased in breast cancer tissue. Carcinogenesis 2009, 30, 745–752. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Lipid Species | MCF-7 EpiR vs. MCF-7 | MCF-7 TAXR vs. MCF-7 | MDA-MB-231 vs. MCF-7 | ||||
|---|---|---|---|---|---|---|---|
| Fold Change | p Value | Fold Change | p Value | Fold Change | p Value | ||
| PC | PC(28:0)///PC(14:0/14:0) | 0.02990952 | 0.001 | 0.04615794 | 0.001 | 0.21447702 | 0.050 |
| PC(31:1)///PC(15:0/16:1) | 0.0689704 | 0.017 | 0.09403492 | 0.025 | 0.20846594 | 0.108 | |
| PC(30:1)///PC(16:1/14:0) | 0.01112429 | 0.002 | 0.00700047 | 0.000 | 0.01721311 | 0.002 | |
| PC(O-33:3) | 1.27651749 | 0.004 | 3.99971727 | 0.001 | 2.80090791 | <0.001 | |
| PC(38:3)///PC(18:0/20:3) | 0.27554774 | 0.044 | 0.26114733 | 0.007 | 0.65273596 | 0.738 | |
| PC(44:1)///PC(26:0/18:1) | 44.6634837 | 0.092 | 36.8772782 | 0.032 | 376.01122 | 0.001 | |
| PC(38:1)///PC(20:0/18:1) | 3.53236174 | 0.288 | 3.43871497 | 0.184 | 14.0520945 | 0.004 | |
| PE | PE(P-34:2)///PE(P-16:1/18:1) | 28.2972788 | <0.001 | 26.2359375 | <0.001 | 22.5278479 | <0.001 |
| PE(P-36:5)///PE(P-16:1/20:4) | 8.81558854 | 0.007 | 10.4244025 | 0.005 | 14.2692477 | 0.001 | |
| PE(P-36:2)///PE(P-18:1/18:1) | 25.9952287 | <0.001 | 19.896668 | <0.001 | 114.690505 | <0.001 | |
| PE(34:1)///PE(16:0/18:1) | 0.58971808 | 0.781 | 0.64063147 | 0.186 | 0.35193966 | 0.009 | |
| PE(O-34:2)///PE(O-15:1/19:1) | 29.4014166 | <0.001 | 23.5110141 | <0.001 | 22.0164115 | <0.001 | |
| PE(P-38:7)///PE(P-16:1/22:6) | 14.7720082 | 0.006 | 12.4773801 | 0.004 | 9.72151443 | 0.008 | |
| TG | Mix TG(44:1) | 0.03544991 | 0.003 | 0.04252924 | 0.001 | 0.04335925 | 0.001 |
| Mix TG(46:4) | 0.05574601 | <0.001 | 0.07728431 | <0.001 | 0.07396131 | <0.001 | |
| Mix TG(46:1) | 0.15810058 | 0.018 | 0.23540369 | 0.004 | 0.17216579 | 0.002 | |
| Mix TG(48:4) | 0.29777056 | 0.007 | 0.40895724 | 0.008 | 0.27500008 | 0.001 | |
| Mix TG(50:5) | 0.16080286 | <0.001 | 0.17598899 | <0.001 | 0.12880096 | <0.001 | |
| Mix TG(50:2) | 0.48675871 | 0.399 | 0.53214893 | 0.017 | 0.39973327 | 0.002 | |
| Mix TG(52:3) | 0.24951721 | 0.092 | 0.30606399 | 0.011 | 0.34407207 | 0.013 | |
| Mix TG(54:3) | 0.47035591 | 0.322 | 0.38418791 | 0.001 | 0.40637479 | 0.002 | |
| SM | SM(36:1) | 3.3947483 | 0.001 | 3.89343315 | 0.002 | 1.02089675 | 0.999 |
| SM(39:3) | 0.22395786 | 0.017 | 0.21373755 | 0.001 | 0.21085366 | 0.002 | |
| SM(42:2) | 0.55261516 | 0.632 | 0.37982867 | 0.013 | 0.46507858 | 0.086 | |
| SM(42:1) | 2.07385942 | 0.331 | 2.05459388 | 0.309 | 5.87791467 | 0.003 | |
| Cer | LacCer(18:1/16:0) | 57.8502342 | <0.001 | 48.320848 | <0.001 | 4.02257761 | 0.003 |
| DihydroCer(18:0(OH)/16:0) | 0.20022379 | 0.073 | 0.18622892 | 0.003 | 0.10548975 | 0.001 | |
| LacCer(18:1/24:0) | 103.862204 | <0.001 | 56.4934583 | <0.001 | 10.0750913 | <0.001 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adriá-Cebrián, J.; Guaita-Esteruelas, S.; Lam, E.W.-F.; Rodríguez-Balada, M.; Capellades, J.; Girona, J.; Jimenez-Santamaria, A.M.; Yanes, O.; Masana, L.; Gumà, J. MCF-7 Drug Resistant Cell Lines Switch Their Lipid Metabolism to Triple Negative Breast Cancer Signature. Cancers 2021, 13, 5871. https://doi.org/10.3390/cancers13235871
Adriá-Cebrián J, Guaita-Esteruelas S, Lam EW-F, Rodríguez-Balada M, Capellades J, Girona J, Jimenez-Santamaria AM, Yanes O, Masana L, Gumà J. MCF-7 Drug Resistant Cell Lines Switch Their Lipid Metabolism to Triple Negative Breast Cancer Signature. Cancers. 2021; 13(23):5871. https://doi.org/10.3390/cancers13235871
Chicago/Turabian StyleAdriá-Cebrián, Jose, Sandra Guaita-Esteruelas, Eric W.-F. Lam, Marta Rodríguez-Balada, Jordi Capellades, Josefa Girona, Ana Maria Jimenez-Santamaria, Oscar Yanes, Luís Masana, and Josep Gumà. 2021. "MCF-7 Drug Resistant Cell Lines Switch Their Lipid Metabolism to Triple Negative Breast Cancer Signature" Cancers 13, no. 23: 5871. https://doi.org/10.3390/cancers13235871
APA StyleAdriá-Cebrián, J., Guaita-Esteruelas, S., Lam, E. W.-F., Rodríguez-Balada, M., Capellades, J., Girona, J., Jimenez-Santamaria, A. M., Yanes, O., Masana, L., & Gumà, J. (2021). MCF-7 Drug Resistant Cell Lines Switch Their Lipid Metabolism to Triple Negative Breast Cancer Signature. Cancers, 13(23), 5871. https://doi.org/10.3390/cancers13235871







