MEOX2 Transcription Factor Is Involved in Survival and Adhesion of Glioma Stem-like Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. GSC-Lines and Cell Culture
2.2. Patient Cohort
2.3. Transfection
2.4. Cell Viability Assay
2.5. Pyrosequencing and Methylation
2.6. Subcellular Fractionation and Western Blotting
2.7. Immunofluorescence
2.8. Cell Cycle Analysis
2.9. Transcriptome Analysis
2.10. Statistical Analysis
3. Results
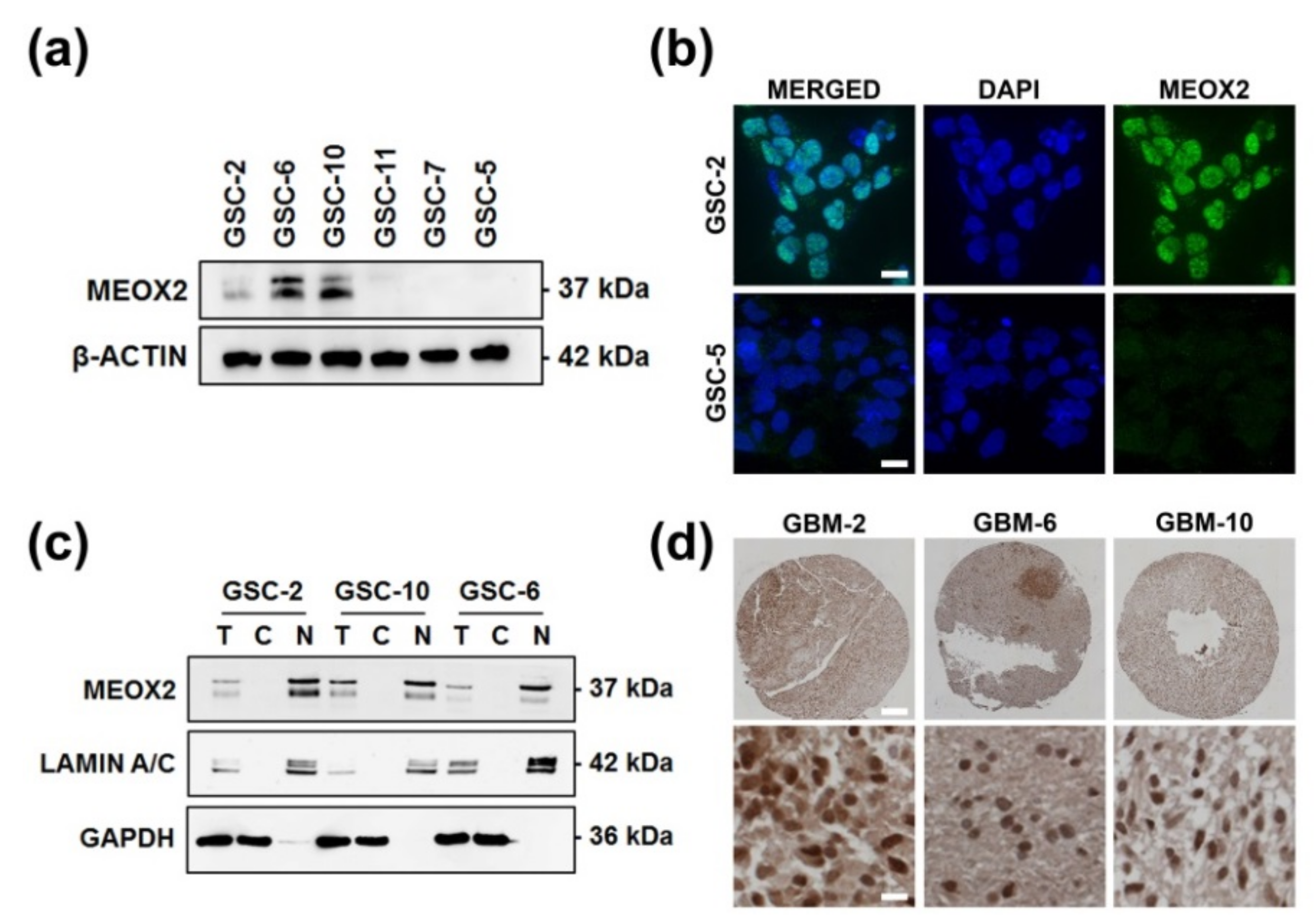
3.1. Nuclear and Heterogeneous MEOX2 Expression among GSC
3.2. MEOX2 Expression Is Partly Dependent on the Methylation Status of Its Promoter
3.3. MEOX2 Is Involved in Cell Survival through ERK/MAPK and PI3K/AKT Pathways
3.4. The Loss of MEOX2 Expression Correlates with the Differentiation of GSC and with Acquisition of Neuronal Lineage Features
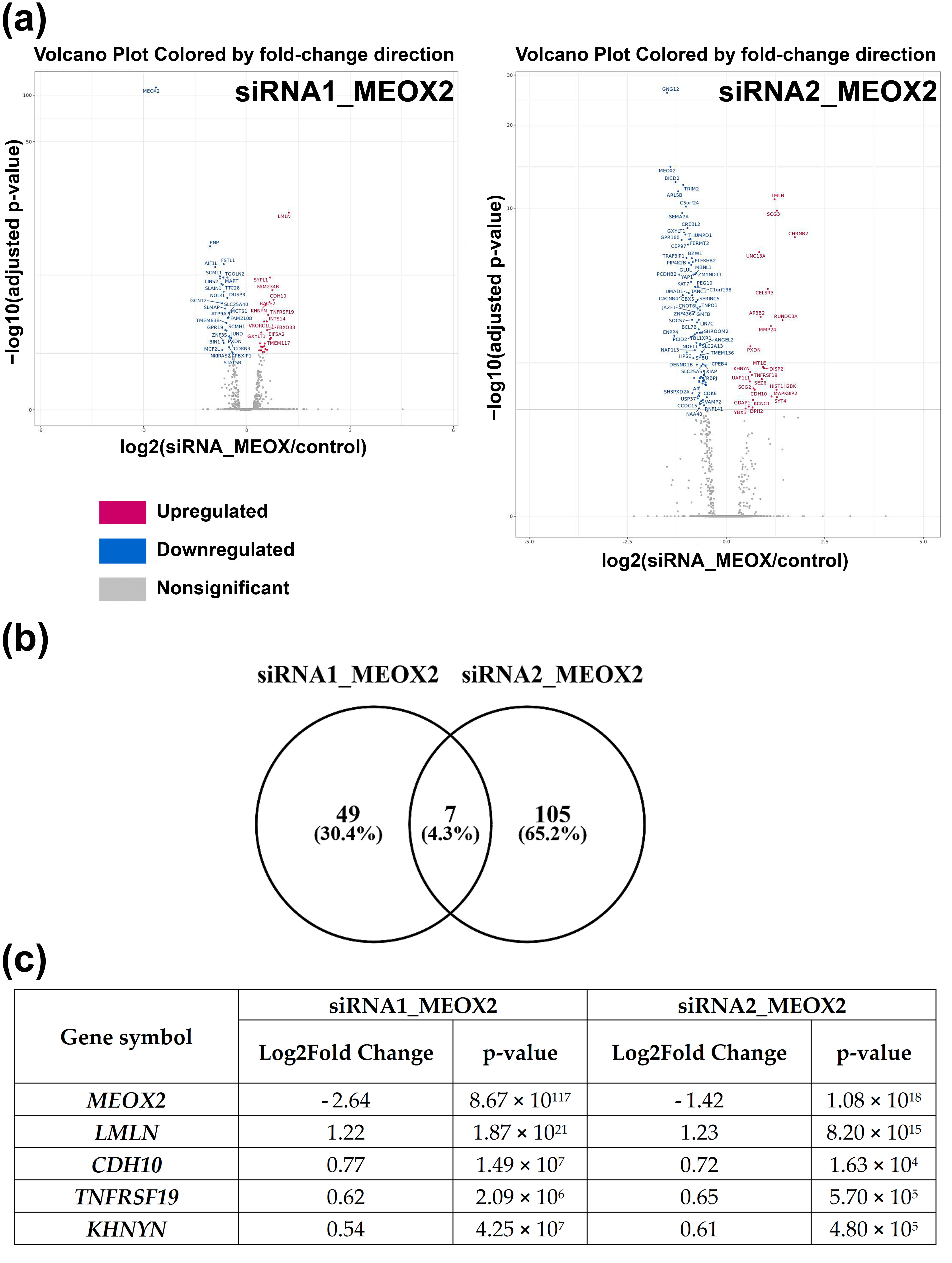
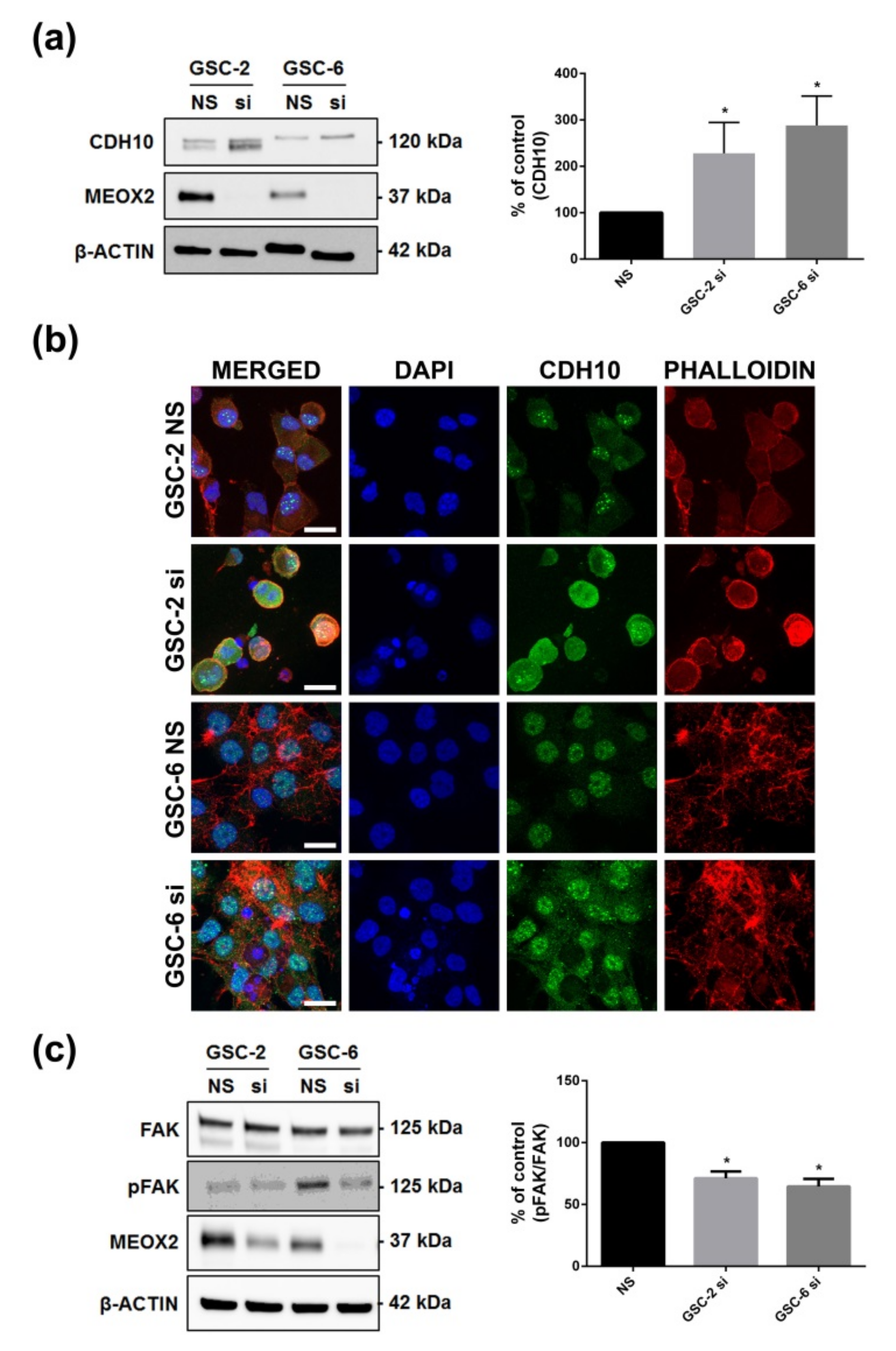
3.5. Inhibition of MEOX2 Regulates a Restricted Gene Network including Cadherin CDH10
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ignatova, T.N.; Kukekov, V.G.; Laywell, E.D.; Suslov, O.N.; Vrionis, F.D.; Steindler, D.A. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia 2002, 39, 193–206. [Google Scholar] [CrossRef]
- Singh, S.K.; Clarke, I.D.; Terasaki, M.; Bonn, V.E.; Hawkins, C.; Squire, J.; Dirks, P.B. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003, 63, 5821–5828. [Google Scholar]
- Galli, R.; Binda, E.; Orfanelli, U.; Cipelletti, B.; Gritti, A.; De Vitis, S.; Fiocco, R.; Foroni, C.; Dimeco, F.; Vescovi, A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004, 64, 7011–7021. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Curtin, J.; Xiong, Y.; Liu, G.; Waschsmann-Hogiu, S.; Farkas, D.L.; Black, K.L.; Yu, J.S. Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene 2004, 23, 9392–9400. [Google Scholar] [CrossRef] [PubMed]
- Desai, A.; Webb, B.; Gerson, S.L. CD133+ cells contribute to radioresistance via altered regulation of DNA repair genes in human lung cancer cells. Radiother. Oncol. J. Eur. Soc. Ther. Radiol. Oncol. 2014, 110, 538–545. [Google Scholar] [CrossRef] [PubMed]
- Tachon, G.; Cortes, U.; Guichet, P.O.; Rivet, P.; Balbous, A.; Masliantsev, K.; Berger, A.; Boissonnade, O.; Wager, M.; Karayan-Tapon, L. Cell Cycle Changes after Glioblastoma Stem Cell Irradiation: The Major Role of RAD51. Int. J. Mol. Sci. 2018, 19, 18. [Google Scholar] [CrossRef]
- Fiscon, G.; Conte, F.; Licursi, V.; Nasi, S.; Paci, P. Computational identification of specific genes for glioblastoma stem-like cells identity. Sci. Rep. 2018, 8, 7769. [Google Scholar] [CrossRef]
- Suva, M.L.; Rheinbay, E.; Gillespie, S.M.; Patel, A.P.; Wakimoto, H.; Rabkin, S.D.; Riggi, N.; Chi, A.S.; Cahill, D.P.; Nahed, B.V.; et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell 2014, 157, 580–594. [Google Scholar] [CrossRef]
- Bhatlekar, S.; Fields, J.Z.; Boman, B.M. Role of HOX Genes in Stem Cell Differentiation and Cancer. Stem Cells Int. 2018, 2018, 3569493. [Google Scholar] [CrossRef]
- Ohshima, J.; Haruta, M.; Arai, Y.; Kasai, F.; Fujiwara, Y.; Ariga, T.; Okita, H.; Fukuzawa, M.; Hata, J.; Horie, H.; et al. Two candidate tumor suppressor genes, MEOX2 and SOSTDC1, identified in a 7p21 homozygous deletion region in a Wilms tumor. Genes Chromosomes Cancer 2009, 48, 1037–1050. [Google Scholar] [CrossRef]
- Avila-Moreno, F.; Armas-Lopez, L.; Alvarez-Moran, A.M.; Lopez-Bujanda, Z.; Ortiz-Quintero, B.; Hidalgo-Miranda, A.; Urrea-Ramirez, F.; Rivera-Rosales, R.M.; Vazquez-Manriquez, E.; Pena-Mirabal, E.; et al. Overexpression of MEOX2 and TWIST1 is associated with H3K27me3 levels and determines lung cancer chemoresistance and prognosis. PLoS ONE 2014, 9, e114104. [Google Scholar] [CrossRef]
- Tian, L.; Tao, Z.Z.; Ye, H.P.; Li, G.Y.; Zhan, Z.F.; Tuo, H.W. Over-expression of MEOX2 promotes apoptosis through inhibiting the PI3K/Akt pathway in laryngeal cancer cells. Neoplasma 2018, 65, 745–752. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.S.; Zhang, C.B.; Wang, H.J.; Yan, W.; Liu, Y.W.; Li, M.Y.; Zhang, W. Whole-genome mRNA expression profiling identifies functional and prognostic signatures in patients with mesenchymal glioblastoma multiforme. CNS Neurosci. Ther. 2013, 19, 714–720. [Google Scholar] [CrossRef] [PubMed]
- Turcan, S.; Makarov, V.; Taranda, J.; Wang, Y.; Fabius, A.W.M.; Wu, W.; Zheng, Y.; El-Amine, N.; Haddock, S.; Nanjangud, G.; et al. Mutant-IDH1-dependent chromatin state reprogramming, reversibility, and persistence. Nat. Genet. 2018, 50, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Tachon, G.; Masliantsev, K.; Rivet, P.; Petropoulos, C.; Godet, J.; Milin, S.; Wager, M.; Guichet, P.O.; Karayan-Tapon, L. Prognostic significance of MEOX2 in gliomas. Mod. Pathol. Off. J. U.S. Can. Acad. Pathol. Inc. 2019, 32, 774–786. [Google Scholar] [CrossRef] [PubMed]
- Villalva, C.; Martin-Lanneree, S.; Cortes, U.; Dkhissi, F.; Wager, M.; Le Corf, A.; Tourani, J.M.; Dusanter-Fourt, I.; Turhan, A.G.; Karayan-Tapon, L. STAT3 is essential for the maintenance of neurosphere-initiating tumor cells in patients with glioblastomas: A potential for targeted therapy? Int. J. Cancer 2011, 128, 826–838. [Google Scholar] [CrossRef] [PubMed]
- Villalva, C.; Cortes, U.; Wager, M.; Tourani, J.M.; Rivet, P.; Marquant, C.; Martin, S.; Turhan, A.G.; Karayan-Tapon, L. O6-Methylguanine-methyltransferase (MGMT) promoter methylation status in glioma stem-like cells is correlated to temozolomide sensitivity under differentiation-promoting conditions. Int. J. Mol. Sci. 2012, 13, 6983–6994. [Google Scholar] [CrossRef] [PubMed]
- Balbous, A.; Cortes, U.; Guilloteau, K.; Rivet, P.; Pinel, B.; Duchesne, M.; Godet, J.; Boissonnade, O.; Wager, M.; Bensadoun, R.J.; et al. A radiosensitizing effect of RAD51 inhibition in glioblastoma stem-like cells. BMC Cancer 2016, 16, 604. [Google Scholar] [CrossRef]
- Wouters, J.; Vizoso, M.; Martinez-Cardus, A.; Carmona, F.J.; Govaere, O.; Laguna, T.; Joseph, J.; Dynoodt, P.; Aura, C.; Foth, M.; et al. Comprehensive DNA methylation study identifies novel progression-related and prognostic markers for cutaneous melanoma. BMC Med. 2017, 15, 101. [Google Scholar] [CrossRef]
- Kruger, N.J. The Bradford method for protein quantitation. Methods Mol. Biol. 1994, 32, 9–15. [Google Scholar] [CrossRef]
- Florea, L.D.; Salzberg, S.L. Genome-guided transcriptome assembly in the age of next-generation sequencing. IEEE/ACM Trans. Comput. Biol. Bioinform. 2013, 10, 1234–1240. [Google Scholar] [CrossRef] [PubMed]
- Guichet, P.O.; Masliantsev, K.; Tachon, G.; Petropoulos, C.; Godet, J.; Larrieu, D.; Milin, S.; Wager, M.; Karayan-Tapon, L. Fatal correlation between YAP1 expression and glioma aggressiveness: Clinical and molecular evidence. J. Pathol. 2018, 246, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Cao, G.; Huang, B.; Liu, Z.; Zhang, J.; Xu, H.; Xia, W.; Li, J.; Li, S.; Chen, L.; Ding, H.; et al. Intronic miR-301 feedback regulates its host gene, ska2, in A549 cells by targeting MEOX2 to affect ERK/CREB pathways. Biochem. Biophys. Res. Commun. 2010, 396, 978–982. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Kong, F.; Wang, J.; Lu, Q.; Xu, H.; Qi, T.; Meng, J. Involvement of IGF-1 and MEOX2 in PI3K/Akt1/2 and ERK1/2 pathways mediated proliferation and differentiation of perivascular adipocytes. Exp. Cell Res. 2015, 331, 82–96. [Google Scholar] [CrossRef]
- Gorski, D.H.; LePage, D.F.; Patel, C.V.; Copeland, N.G.; Jenkins, N.A.; Walsh, K. Molecular cloning of a diverged homeobox gene that is rapidly down-regulated during the G0/G1 transition in vascular smooth muscle cells. Mol. Cell. Biol. 1993, 13, 3722–3733. [Google Scholar] [CrossRef]
- Lee, J.; Kotliarova, S.; Kotliarov, Y.; Li, A.; Su, Q.; Donin, N.M.; Pastorino, S.; Purow, B.W.; Christopher, N.; Zhang, W.; et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 2006, 9, 391–403. [Google Scholar] [CrossRef]
- Gursel, D.B.; Shin, B.J.; Burkhardt, J.K.; Kesavabhotla, K.; Schlaff, C.D.; Boockvar, J.A. Glioblastoma stem-like cells-biology and therapeutic implications. Cancers 2011, 3, 2655–2666. [Google Scholar] [CrossRef]
- Ligon, K.L.; Huillard, E.; Mehta, S.; Kesari, S.; Liu, H.; Alberta, J.A.; Bachoo, R.M.; Kane, M.; Louis, D.N.; Depinho, R.A.; et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron 2007, 53, 503–517. [Google Scholar] [CrossRef]
- Trepant, A.L.; Bouchart, C.; Rorive, S.; Sauvage, S.; Decaestecker, C.; Demetter, P.; Salmon, I. Identification of OLIG2 as the most specific glioblastoma stem cell marker starting from comparative analysis of data from similar DNA chip microarray platforms. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2015, 36, 1943–1953. [Google Scholar] [CrossRef]
- Smith, K.R.; Jones, K.A.; Kopeikina, K.J.; Burette, A.C.; Copits, B.A.; Yoon, S.; Forrest, M.P.; Fawcett-Patel, J.M.; Hanley, J.G.; Weinberg, R.J.; et al. Cadherin-10 Maintains Excitatory/Inhibitory Ratio through Interactions with Synaptic Proteins. J. Neurosci. Off. J. Soc. Neurosci. 2017, 37, 11127–11139. [Google Scholar] [CrossRef]
- Marthiens, V.; Kazanis, I.; Moss, L.; Long, K.; Ffrench-Constant, C. Adhesion molecules in the stem cell niche--more than just staying in shape? J. Cell Sci. 2010, 123, 1613–1622. [Google Scholar] [CrossRef]
- Shimoyama, Y.; Tsujimoto, G.; Kitajima, M.; Natori, M. Identification of three human type-II classic cadherins and frequent heterophilic interactions between different subclasses of type-II classic cadherins. Biochem. J. 2000, 349, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Webb, D.J.; Donais, K.; Whitmore, L.A.; Thomas, S.M.; Turner, C.E.; Parsons, J.T.; Horwitz, A.F. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 2004, 6, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Huang, C.; Cao, H.; Lin, J.; Gong, X.; Li, J.; Chen, Y.; Tian, Z.; Fang, Z.; Huang, J. A Novel Prognostic Signature of Transcription Factors for the Prediction in Patients With GBM. Front. Genet. 2019, 10, 906. [Google Scholar] [CrossRef]
- Pollard, S.M.; Yoshikawa, K.; Clarke, I.D.; Danovi, D.; Stricker, S.; Russell, R.; Bayani, J.; Head, R.; Lee, M.; Bernstein, M.; et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell 2009, 4, 568–580. [Google Scholar] [CrossRef]
- Gallo, M.; Ho, J.; Coutinho, F.J.; Vanner, R.; Lee, L.; Head, R.; Ling, E.K.; Clarke, I.D.; Dirks, P.B. A tumorigenic MLL-homeobox network in human glioblastoma stem cells. Cancer Res. 2013, 73, 417–427. [Google Scholar] [CrossRef]
- Liu, P.; Zhang, C.; Feng, J.B.; Zhao, Y.X.; Wang, X.P.; Yang, J.M.; Zhang, M.X.; Wang, X.L.; Zhang, Y. Cross talk among Smad, MAPK, and integrin signaling pathways enhances adventitial fibroblast functions activated by transforming growth factor-beta1 and inhibited by Gax. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; He, H.; Cao, X.; Shen, X.; Han, S.; Cui, C.; Zhao, J.; Wei, Y.; Chen, Y.; Xia, L.; et al. MiR-148a-3p Regulates Skeletal Muscle Satellite Cell Differentiation and Apoptosis via the PI3K/AKT Signaling Pathway by Targeting Meox2. Front. Genet. 2020, 11, 512. [Google Scholar] [CrossRef]
- Berezovsky, A.D.; Poisson, L.M.; Cherba, D.; Webb, C.P.; Transou, A.D.; Lemke, N.W.; Hong, X.; Hasselbach, L.A.; Irtenkauf, S.M.; Mikkelsen, T.; et al. Sox2 promotes malignancy in glioblastoma by regulating plasticity and astrocytic differentiation. Neoplasia 2014, 16, 193–206. [Google Scholar] [CrossRef]
- Schulte, A.; Gunther, H.S.; Phillips, H.S.; Kemming, D.; Martens, T.; Kharbanda, S.; Soriano, R.H.; Modrusan, Z.; Zapf, S.; Westphal, M.; et al. A distinct subset of glioma cell lines with stem cell-like properties reflects the transcriptional phenotype of glioblastomas and overexpresses CXCR4 as therapeutic target. Glia 2011, 59, 590–602. [Google Scholar] [CrossRef]
- Pojo, M.; Goncalves, C.S.; Xavier-Magalhaes, A.; Oliveira, A.I.; Goncalves, T.; Correia, S.; Rodrigues, A.J.; Costa, S.; Pinto, L.; Pinto, A.A.; et al. A transcriptomic signature mediated by HOXA9 promotes human glioblastoma initiation, aggressiveness and resistance to temozolomide. Oncotarget 2015, 6, 7657–7674. [Google Scholar] [CrossRef] [PubMed]
- Raciti, M.; Granzotto, M.; Duc, M.D.; Fimiani, C.; Cellot, G.; Cherubini, E.; Mallamaci, A. Reprogramming fibroblasts to neural-precursor-like cells by structured overexpression of pallial patterning genes. Mol. Cell. Neurosci. 2013, 57, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, I.J.; Hofler, H.; Atkinson, M.J. Mapping of a cadherin gene cluster to a region of chromosome 5 subject to frequent allelic loss in carcinoma. Genomics 1999, 57, 160–163. [Google Scholar] [CrossRef]
- Velpula, K.K.; Rehman, A.A.; Chelluboina, B.; Dasari, V.R.; Gondi, C.S.; Rao, J.S.; Veeravalli, K.K. Glioma stem cell invasion through regulation of the interconnected ERK, integrin alpha6 and N-cadherin signaling pathway. Cell. Signal. 2012, 24, 2076–2084. [Google Scholar] [CrossRef]
- Kahlert, U.D.; Nikkhah, G.; Maciaczyk, J. Epithelial-to-mesenchymal(-like) transition as a relevant molecular event in malignant gliomas. Cancer Lett. 2013, 331, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Lewis-Tuffin, L.J.; Rodriguez, F.; Giannini, C.; Scheithauer, B.; Necela, B.M.; Sarkaria, J.N.; Anastasiadis, P.Z. Misregulated E-cadherin expression associated with an aggressive brain tumor phenotype. PLoS ONE 2010, 5, e13665. [Google Scholar] [CrossRef]
- Morizur, L.; Chicheportiche, A.; Gauthier, L.R.; Daynac, M.; Boussin, F.D.; Mouthon, M.A. Distinct Molecular Signatures of Quiescent and Activated Adult Neural Stem Cells Reveal Specific Interactions with Their Microenvironment. Stem Cell Rep. 2018, 11, 565–577. [Google Scholar] [CrossRef]
- Duan, R.; Han, L.; Wang, Q.; Wei, J.; Chen, L.; Zhang, J.; Kang, C.; Wang, L. HOXA13 is a potential GBM diagnostic marker and promotes glioma invasion by activating the Wnt and TGF-beta pathways. Oncotarget 2015, 6, 27778–27793. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tachon, G.; Masliantsev, K.; Rivet, P.; Desette, A.; Milin, S.; Gueret, E.; Wager, M.; Karayan-Tapon, L.; Guichet, P.-O. MEOX2 Transcription Factor Is Involved in Survival and Adhesion of Glioma Stem-like Cells. Cancers 2021, 13, 5943. https://doi.org/10.3390/cancers13235943
Tachon G, Masliantsev K, Rivet P, Desette A, Milin S, Gueret E, Wager M, Karayan-Tapon L, Guichet P-O. MEOX2 Transcription Factor Is Involved in Survival and Adhesion of Glioma Stem-like Cells. Cancers. 2021; 13(23):5943. https://doi.org/10.3390/cancers13235943
Chicago/Turabian StyleTachon, Gaëlle, Konstantin Masliantsev, Pierre Rivet, Amandine Desette, Serge Milin, Elise Gueret, Michel Wager, Lucie Karayan-Tapon, and Pierre-Olivier Guichet. 2021. "MEOX2 Transcription Factor Is Involved in Survival and Adhesion of Glioma Stem-like Cells" Cancers 13, no. 23: 5943. https://doi.org/10.3390/cancers13235943





