Modeling Tumor: Lymphatic Interactions in Lymphatic Metastasis of Triple Negative Breast Cancer
Simple Summary
Abstract
1. Introduction
1.1. Lymphatic Metastasis of Breast Cancer (BC)
1.2. Lymphatic Metastasis of Triple Negative Breast Cancer (TNBC)
1.3. Avatars for Studying Lymphatic Metastasis
2. Materials and Methods
2.1. Reagents
2.2. Cells and Cell Maintenance
2.3. 3D Culture
2.4. Image Acquisition for Quantitative Analysis in 3D
2.5. LEC-Conditioned Media (CM)
2.6. 3D Invasion Assays
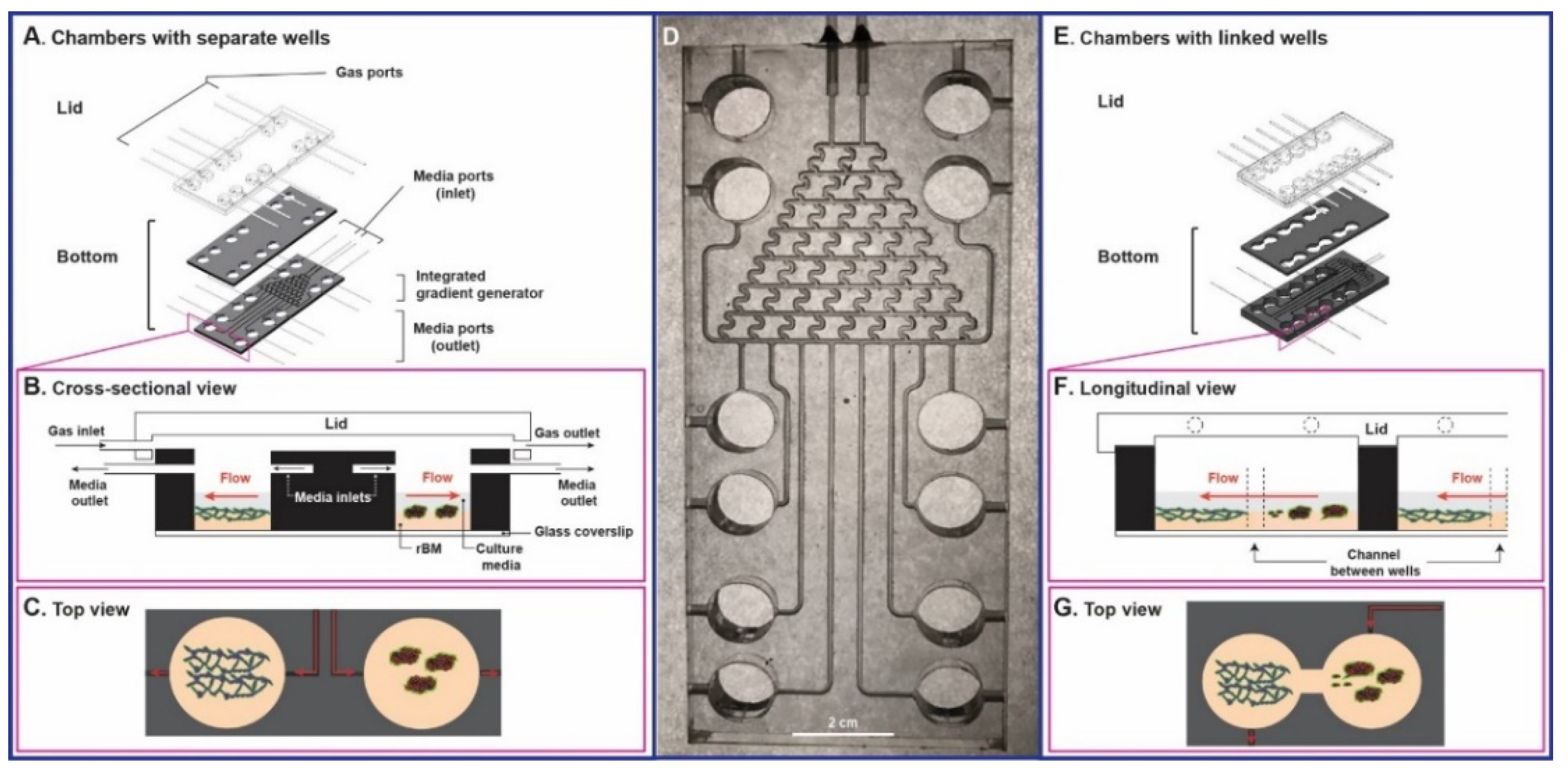
2.7. Fabrication of TAME Chambers
2.8. Statistical Analysis
3. Results
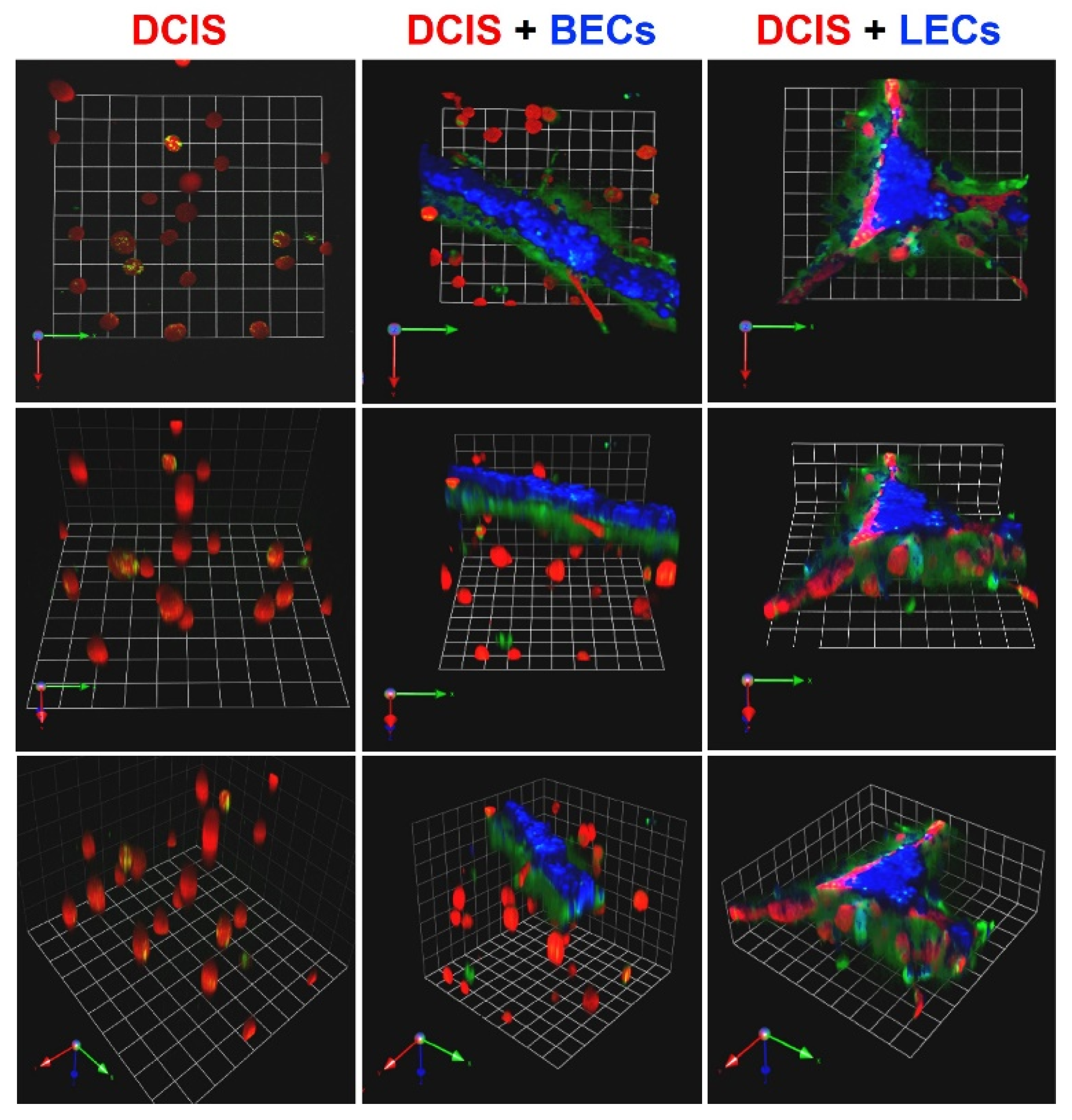
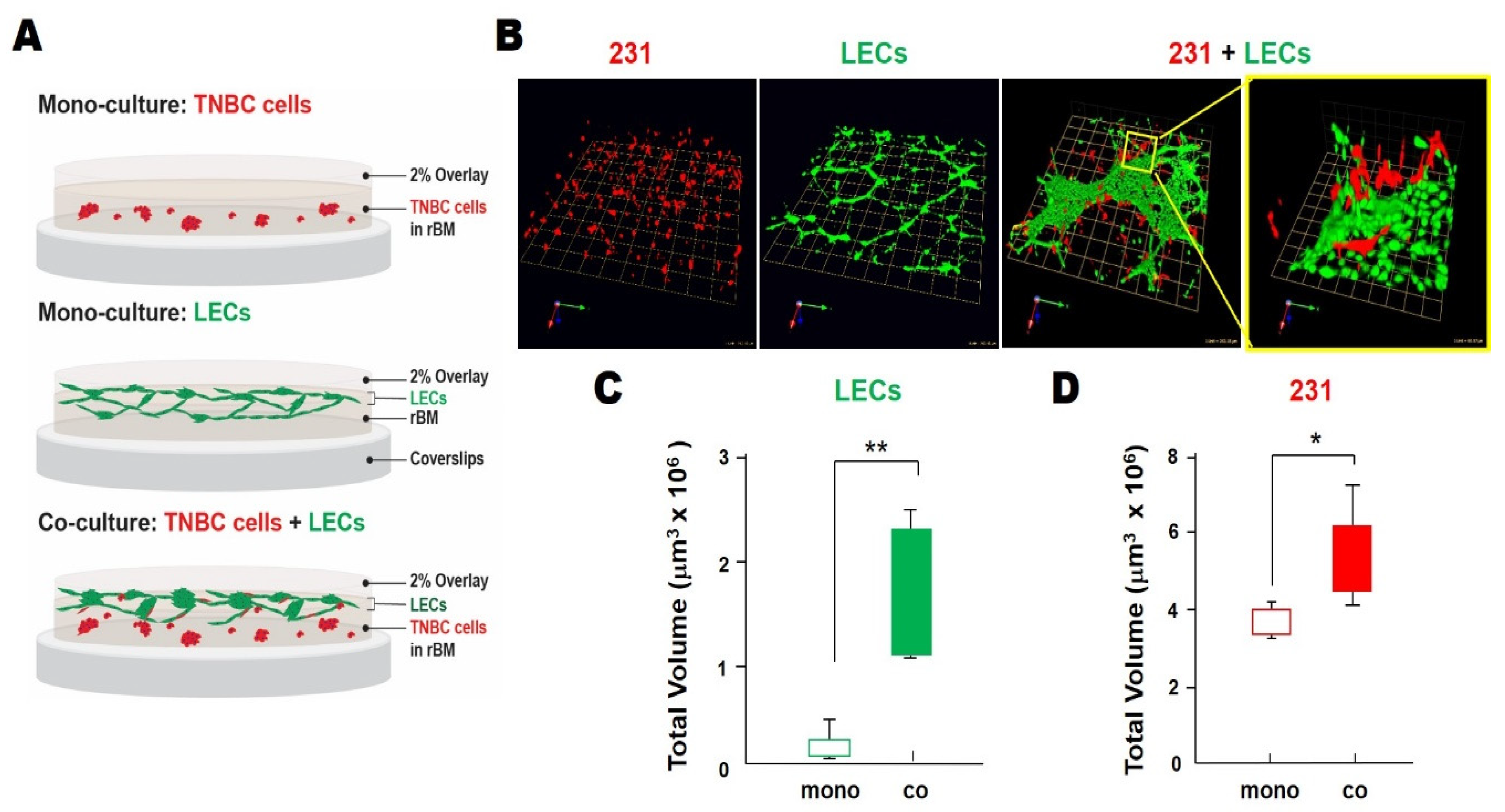
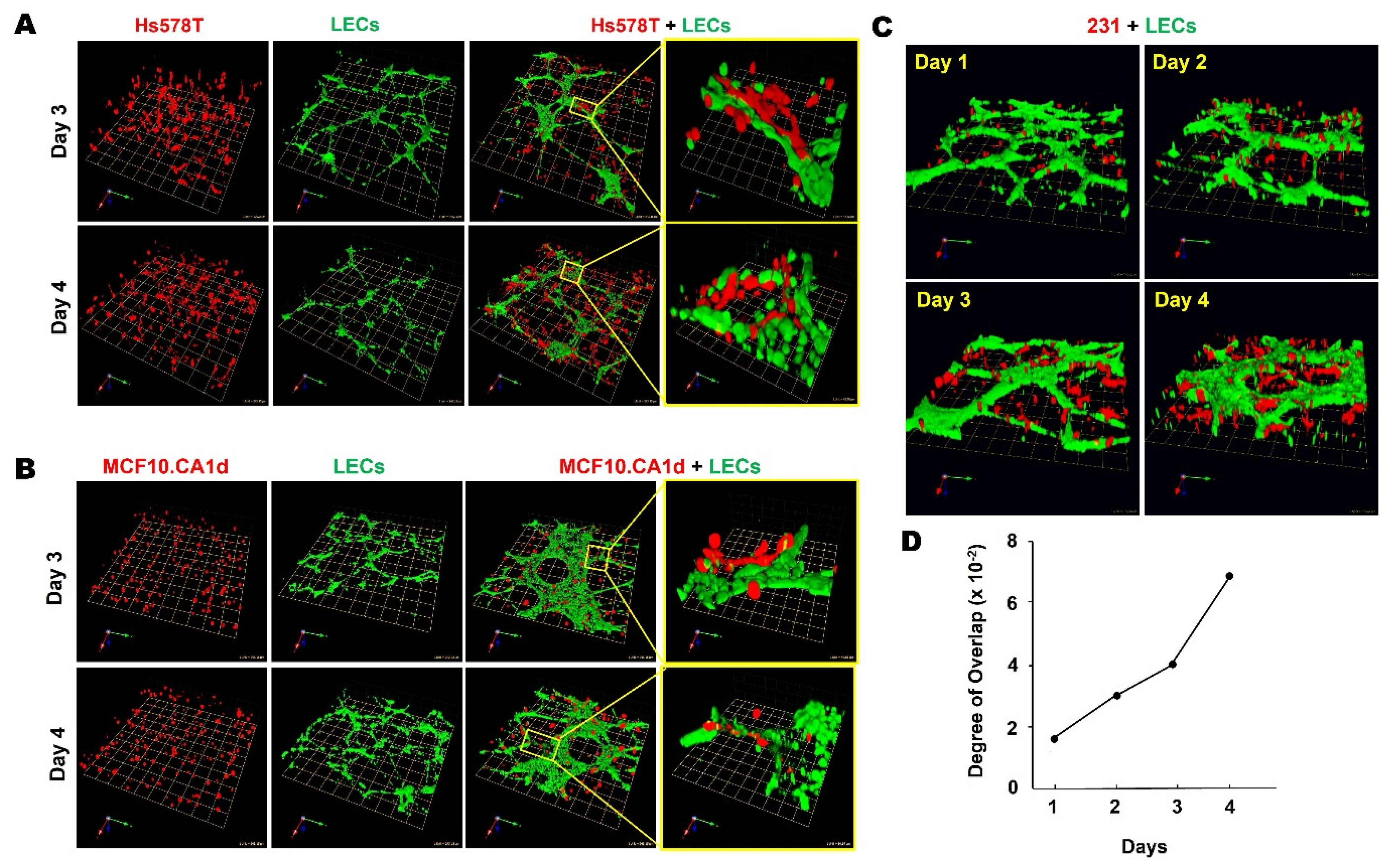
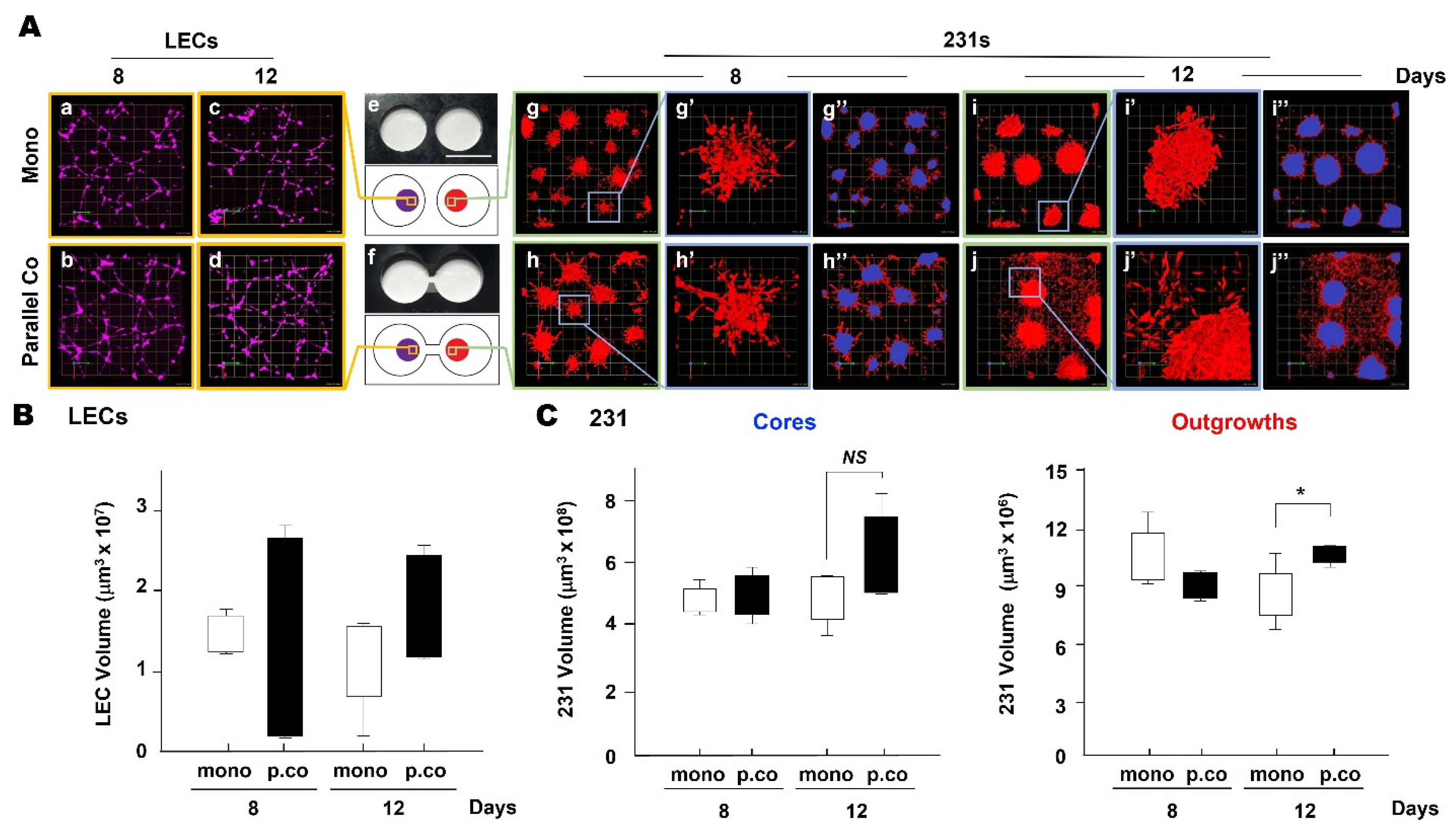
3.1. TNBC:LEC Interactions in 3D Cocultures
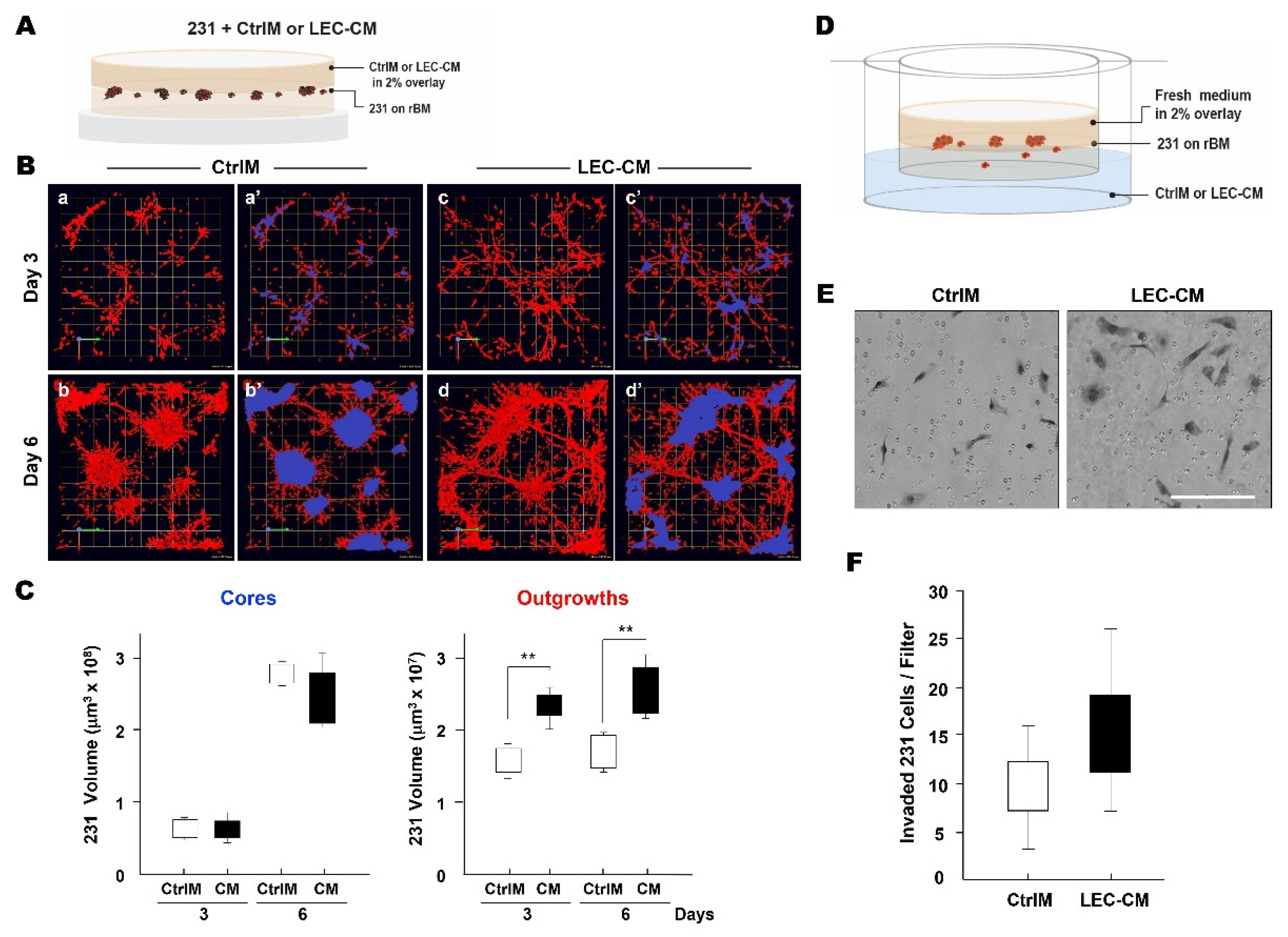
3.2. LEC Secretome and TNBC Invasion
3.3. Tissue Architecture and Microenvironment Engineering (TAME) Chambers
3.4. LEC Secretome-Induced TNBC Invasion in TAME Linked-Well Chambers
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Peart, O. Metastatic Breast Cancer. Radiol. Technol. 2017, 88, 519M–539M. [Google Scholar]
- Wang, H.; Zhang, C.; Zhang, J.; Kong, L.; Zhu, H.; Yu, J. The prognosis analysis of different metastasis pattern in patients with different breast cancer subtypes: A SEER based study. Oncotarget 2017, 8, 26368–26379. [Google Scholar] [CrossRef] [PubMed]
- Lauria, R.; Perrone, F.; Carlomagno, C.; De Laurentiis, M.; Morabito, A.; Gallo, C.; Varriale, E.; Pettinato, G.; Panico, L.; Petrella, G.; et al. The prognostic value of lymphatic and blood vessel invasion in operable breast cancer. Cancer 1995, 76, 1772–1778. [Google Scholar] [CrossRef]
- Woo, C.S.; Silberman, H.; Nakamura, S.K.; Ye, W.; Sposto, R.; Colburn, W.; Waisman, J.R.; Silverstein, M.J. Lymph node status combined with lymphovascular invasion creates a more powerful tool for predicting outcome in patients with invasive breast cancer. Am. J. Surg. 2002, 184, 337–340. [Google Scholar] [CrossRef]
- Schoppmann, S.F.; Bayer, G.; Aumayr, K.; Taucher, S.; Geleff, S.; Rudas, M.; Kubista, E.; Hausmaninger, H.; Samonigg, H.; Gnant, M.; et al. Prognostic value of lymphangiogenesis and lymphovascular invasion in invasive breast cancer. Ann. Surg. 2004, 240, 306–312. [Google Scholar] [CrossRef]
- Ran, S.; Volk, L.; Hall, K.; Flister, M.J. Lymphangiogenesis and lymphatic metastasis in breast cancer. Pathophysiology 2010, 17, 229–251. [Google Scholar] [CrossRef]
- Galimberti, V.; Cole, B.F.; Zurrida, S.; Viale, G.; Luini, A.; Veronesi, P.; Baratella, P.; Chifu, C.; Sargenti, M.; Intra, M.; et al. Axillary dissection versus no axillary dissection in patients with sentinel-node micrometastases (IBCSG 23-01): A phase 3 randomised controlled trial. Lancet Oncol. 2013, 14, 297–305. [Google Scholar] [CrossRef]
- Donker, M.; van Tienhoven, G.; Straver, M.E.; Meijnen, P.; van de Velde, C.J.; Mansel, R.E.; Cataliotti, L.; Westenberg, A.H.; Klinkenbijl, J.H.; Orzalesi, L.; et al. Radiotherapy or surgery of the axilla after a positive sentinel node in breast cancer (EORTC 10981-22023 AMAROS): A randomised, multicentre, open-label, phase 3 non-inferiority trial. Lancet Oncol. 2014, 15, 1303–1310. [Google Scholar] [CrossRef]
- Poortmans, P.M.; Collette, S.; Kirkove, C.; Van Limbergen, E.; Budach, V.; Struikmans, H.; Collette, L.; Fourquet, A.; Maingon, P.; Valli, M.; et al. Internal Mammary and Medial Supraclavicular Irradiation in Breast Cancer. N. Engl. J. Med. 2015, 373, 317–327. [Google Scholar] [CrossRef]
- Whelan, T.J.; Olivotto, I.A.; Parulekar, W.R.; Ackerman, I.; Chua, B.H.; Nabid, A.; Vallis, K.A.; White, J.R.; Rousseau, P.; Fortin, A.; et al. Regional Nodal Irradiation in Early-Stage Breast Cancer. N. Engl. J. Med. 2015, 373, 307–316. [Google Scholar] [CrossRef]
- Brown, M.; Assen, F.P.; Leithner, A.; Abe, J.; Schachner, H.; Asfour, G.; Bago-Horvath, Z.; Stein, J.V.; Uhrin, P.; Sixt, M.; et al. Lymph node blood vessels provide exit routes for metastatic tumor cell dissemination in mice. Science 2018, 359, 1408–1411. [Google Scholar] [CrossRef]
- Pereira, E.R.; Kedrin, D.; Seano, G.; Gautier, O.; Meijer, E.F.J.; Jones, D.; Chin, S.M.; Kitahara, S.; Bouta, E.M.; Chang, J.; et al. Lymph node metastases can invade local blood vessels, exit the node, and colonize distant organs in mice. Science 2018, 359, 1403–1407. [Google Scholar] [CrossRef]
- Pereira, E.R.; Jones, D.; Jung, K.; Padera, T.P. The lymph node microenvironment and its role in the progression of metastatic cancer. Semin. Cell Dev. Biol. 2015, 38, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Stacker, S.A.; Baldwin, M.E.; Achen, M.G. The role of tumor lymphangiogenesis in metastatic spread. FASEB J. 2002, 16, 922–934. [Google Scholar] [CrossRef]
- Karaman, S.; Detmar, M. Mechanisms of lymphatic metastasis. J. Clin. Investig. 2014, 124, 922–928. [Google Scholar] [CrossRef] [PubMed]
- Stacker, S.A.; Williams, S.P.; Karnezis, T.; Shayan, R.; Fox, S.B.; Achen, M.G. Lymphangiogenesis and lymphatic vessel remodelling in cancer. Nat. Rev. Cancer 2014, 14, 159–172. [Google Scholar] [CrossRef] [PubMed]
- Dieterich, L.C.; Detmar, M. Tumor lymphangiogenesis and new drug development. Adv. Drug. Deliv. Rev. 2016, 99, 148–160. [Google Scholar] [CrossRef]
- Muller, A.; Homey, B.; Soto, H.; Ge, N.; Catron, D.; Buchanan, M.E.; McClanahan, T.; Murphy, E.; Yuan, W.; Wagner, S.N.; et al. Involvement of chemokine receptors in breast cancer metastasis. Nature 2001, 410, 50–56. [Google Scholar] [CrossRef]
- Skobe, M.; Hawighorst, T.; Jackson, D.G.; Prevo, R.; Janes, L.; Velasco, P.; Riccardi, L.; Alitalo, K.; Claffey, K.; Detmar, M. Induction of tumor lymphangiogenesis by VEGF-C promotes breast cancer metastasis. Nat. Med. 2001, 7, 192–198. [Google Scholar] [CrossRef]
- Li, Y.M.; Pan, Y.; Wei, Y.; Cheng, X.; Zhou, B.P.; Tan, M.; Zhou, X.; Xia, W.; Hortobagyi, G.N.; Yu, D.; et al. Upregulation of CXCR4 is essential for HER2-mediated tumor metastasis. Cancer Cell 2004, 6, 459–469. [Google Scholar] [CrossRef]
- Liang, Z.; Wu, T.; Lou, H.; Yu, X.; Taichman, R.S.; Lau, S.K.; Nie, S.; Umbreit, J.; Shim, H. Inhibition of breast cancer metastasis by selective synthetic polypeptide against CXCR4. Cancer Res. 2004, 64, 4302–4308. [Google Scholar] [CrossRef]
- Cunningham, H.D.; Shannon, L.A.; Calloway, P.A.; Fassold, B.C.; Dunwiddie, I.; Vielhauer, G.; Zhang, M.; Vines, C.M. Expression of the C-C chemokine receptor 7 mediates metastasis of breast cancer to the lymph nodes in mice. Transl. Oncol. 2010, 3, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Foulkes, W.D.; Smith, I.E.; Reis-Filho, J.S. Triple-negative breast cancer. N. Engl. J. Med. 2010, 363, 1938–1948. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Castro, A.C.; Lin, N.U.; Polyak, K. Insights into Molecular Classifications of Triple-Negative Breast Cancer: Improving Patient Selection for Treatment. Cancer Discov. 2019, 9, 176–198. [Google Scholar] [CrossRef]
- Dent, R.; Trudeau, M.; Pritchard, K.I.; Hanna, W.M.; Kahn, H.K.; Sawka, C.A.; Lickley, L.A.; Rawlinson, E.; Sun, P.; Narod, S.A. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin. Cancer Res. 2007, 13, 4429–4434. [Google Scholar] [CrossRef] [PubMed]
- Dent, R.; Hanna, W.M.; Trudeau, M.; Rawlinson, E.; Sun, P.; Narod, S.A. Pattern of metastatic spread in triple-negative breast cancer. Breast Cancer Res. Treat. 2009, 115, 423–428. [Google Scholar] [CrossRef]
- Trivers, K.F.; Lund, M.J.; Porter, P.L.; Liff, J.M.; Flagg, E.W.; Coates, R.J.; Eley, J.W. The epidemiology of triple-negative breast cancer, including race. Cancer Causes Control. 2009, 20, 1071–1082. [Google Scholar] [CrossRef]
- Howland, N.K.; Driver, T.D.; Sedrak, M.P.; Wen, X.; Dong, W.; Hatch, S.; Eltorky, M.A.; Chao, C. Lymph node involvement in immunohistochemistry-based molecular classifications of breast cancer. J. Surg. Res. 2013, 185, 697–703. [Google Scholar] [CrossRef][Green Version]
- He, Z.Y.; Wu, S.G.; Yang, Q.; Sun, J.Y.; Li, F.Y.; Lin, Q.; Lin, H.X. Breast Cancer Subtype is Associated With Axillary Lymph Node Metastasis: A Retrospective Cohort Study. Medicine 2015, 94, e2213. [Google Scholar] [CrossRef]
- Liu, H.T.; Ma, R.; Yang, Q.F.; Du, G.; Zhang, C.J. Lymphangiogenic characteristics of triple negativity in node-negative breast cancer. Int. J. Surg. Pathol. 2009, 17, 426–431. [Google Scholar] [CrossRef]
- Shaw, K.R.; Wrobel, C.N.; Brugge, J.S. Use of three-dimensional basement membrane cultures to model oncogene-induced changes in mammary epithelial morphogenesis. J. Mammary Gland Biol. Neoplasia 2004, 9, 297–310. [Google Scholar] [CrossRef] [PubMed]
- Nyga, A.; Cheema, U.; Loizidou, M. 3D tumour models: Novel in vitro approaches to cancer studies. J. Cell Commun. Signal. 2011, 5, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Weigelt, B.; Ghajar, C.M.; Bissell, M.J. The need for complex 3D culture models to unravel novel pathways and identify accurate biomarkers in breast cancer. Adv. Drug. Deliv. Rev. 2014, 69–70, 42–51. [Google Scholar] [CrossRef]
- Rijal, G.; Li, W. A versatile 3D tissue matrix scaffold system for tumor modeling and drug screening. Sci. Adv. 2017, 3, e1700764. [Google Scholar] [CrossRef] [PubMed]
- Regehr, K.J.; Domenech, M.; Koepsel, J.T.; Carver, K.C.; Ellison-Zelski, S.J.; Murphy, W.L.; Schuler, L.A.; Alarid, E.T.; Beebe, D.J. Biological implications of polydimethylsiloxane-based microfluidic cell culture. Lab Chip 2009, 9, 2132–2139. [Google Scholar] [CrossRef]
- Shamir, E.R.; Ewald, A.J. Three-dimensional organotypic culture: Experimental models of mammalian biology and disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 647–664. [Google Scholar] [CrossRef]
- Xu, X.; Farach-Carson, M.C.; Jia, X. Three-dimensional in vitro tumor models for cancer research and drug evaluation. Biotechnol. Adv. 2014, 32, 1256–1268. [Google Scholar] [CrossRef]
- Antoni, D.; Burckel, H.; Josset, E.; Noel, G. Three-dimensional cell culture: A breakthrough in vivo. Int. J. Mol. Sci. 2015, 16, 5517–5527. [Google Scholar] [CrossRef]
- Benam, K.H.; Dauth, S.; Hassell, B.; Herland, A.; Jain, A.; Jang, K.J.; Karalis, K.; Kim, H.J.; MacQueen, L.; Mahmoodian, R.; et al. Engineered in vitro disease models. Annu. Rev. Pathol. 2015, 10, 195–262. [Google Scholar] [CrossRef]
- Carneiro, K.; de Brito, J.M.; Rossi, M.I. Development by three-dimensional approaches and four-dimensional imaging: To the knowledge frontier and beyond. Birth Defects Res. C Embryo Today 2015, 105, 1–8. [Google Scholar] [CrossRef]
- Huch, M.; Koo, B.K. Modeling mouse and human development using organoid cultures. Development 2015, 142, 3113–3125. [Google Scholar] [CrossRef]
- Kim, J.; Tanner, K. Recapitulating the Tumor Ecosystem along the Metastatic Cascade Using 3D Culture Models. Front. Oncol. 2015, 5, 170. [Google Scholar] [CrossRef] [PubMed]
- Tanner, K.; Gottesman, M.M. Beyond 3D culture models of cancer. Sci. Transl. Med. 2015, 7, 283ps289. [Google Scholar] [CrossRef]
- Truong, D.; Puleo, J.; Llave, A.; Mouneimne, G.; Kamm, R.D.; Nikkhah, M. Breast Cancer Cell Invasion into a Three Dimensional Tumor-Stroma Microenvironment. Sci. Rep. 2016, 6, 34094. [Google Scholar] [CrossRef] [PubMed]
- Hirt, C.; Papadimitropoulos, A.; Mele, V.; Muraro, M.G.; Mengus, C.; Iezzi, G.; Terracciano, L.; Martin, I.; Spagnoli, G.C. “In vitro” 3D models of tumor-immune system interaction. Adv. Drug Deliv. Rev. 2014, 79–80, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, N.; Santiago, A.M.; Kessel, D.; Sloane, B.F. Photodynamic therapy as an effective therapeutic approach in MAME models of inflammatory breast cancer. Breast Cancer Res. Treat. 2015, 154, 251–262. [Google Scholar] [CrossRef]
- Unger, F.T.; Witte, I.; David, K.A. Prediction of individual response to anticancer therapy: Historical and future perspectives. Cell Mol. Life Sci. 2015, 72, 729–757. [Google Scholar] [CrossRef]
- Martin, K.J.; Patrick, D.R.; Bissell, M.J.; Fournier, M.V. Prognostic breast cancer signature identified from 3D culture model accurately predicts clinical outcome across independent datasets. PLoS ONE 2008, 3, e2994. [Google Scholar] [CrossRef]
- Li, Q.; Mullins, S.R.; Sloane, B.F.; Mattingly, R.R. p21-Activated kinase 1 coordinates aberrant cell survival and pericellular proteolysis in a three-dimensional culture model for premalignant progression of human breast cancer. Neoplasia 2008, 10, 314–329. [Google Scholar] [CrossRef]
- Li, Q.; Chow, A.B.; Mattingly, R.R. Three-dimensional overlay culture models of human breast cancer reveal a critical sensitivity to mitogen-activated protein kinase kinase inhibitors. J. Pharmacol. Exp. Ther. 2010, 332, 821–828. [Google Scholar] [CrossRef]
- Nam, J.M.; Onodera, Y.; Bissell, M.J.; Park, C.C. Breast cancer cells in three-dimensional culture display an enhanced radioresponse after coordinate targeting of integrin alpha5beta1 and fibronectin. Cancer. Res. 2010, 70, 5238–5248. [Google Scholar] [CrossRef]
- Maguire, S.L.; Peck, B.; Wai, P.T.; Campbell, J.; Barker, H.; Gulati, A.; Daley, F.; Vyse, S.; Huang, P.; Lord, C.J.; et al. Three-dimensional modelling identifies novel genetic dependencies associated with breast cancer progression in the isogenic MCF10 model. J. Pathol. 2016, 240, 315–328. [Google Scholar] [CrossRef]
- Breslin, S.; O’Driscoll, L. Three-dimensional cell culture: The missing link in drug discovery. Drug Discov. Today 2013, 18, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Langhans, S.A. Three-Dimensional in Vitro Cell Culture Models in Drug Discovery and Drug Repositioning. Front. Pharmacol. 2018, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Greenlee, J.D.; King, M.R. Engineered fluidic systems to understand lymphatic cancer metastasis. Biomicrofluidics 2020, 14, 011502. [Google Scholar] [CrossRef] [PubMed]
- Sigdel, I.; Gupta, N.; Faizee, F.; Khare, V.M.; Tiwari, A.K.; Tang, Y. Biomimetic Microfluidic Platforms for the Assessment of Breast Cancer Metastasis. Front. Bioeng. Biotechnol. 2021, 9, 633671. [Google Scholar] [CrossRef]
- Chan, J.M.; Zervantonakis, I.K.; Rimchala, T.; Polacheck, W.J.; Whisler, J.; Kamm, R.D. Engineering of in vitro 3D capillary beds by self-directed angiogenic sprouting. PLoS ONE 2012, 7, e50582. [Google Scholar] [CrossRef]
- Yeon, J.H.; Ryu, H.R.; Chung, M.; Hu, Q.P.; Jeon, N.L. In vitro formation and characterization of a perfusable three-dimensional tubular capillary network in microfluidic devices. Lab Chip 2012, 12, 2815–2822. [Google Scholar] [CrossRef]
- Lee, H.; Park, W.; Ryu, H.; Jeon, N.L. A microfluidic platform for quantitative analysis of cancer angiogenesis and intravasation. Biomicrofluidics 2014, 8, 054102. [Google Scholar] [CrossRef]
- Jeon, J.S.; Bersini, S.; Gilardi, M.; Dubini, G.; Charest, J.L.; Moretti, M.; Kamm, R.D. Human 3D vascularized organotypic microfluidic assays to study breast cancer cell extravasation. Proc. Natl. Acad. Sci. USA 2015, 112, 214–219. [Google Scholar] [CrossRef]
- Truong, D.; Fiorelli, R.; Barrientos, E.S.; Melendez, E.L.; Sanai, N.; Mehta, S.; Nikkhah, M. A three-dimensional (3D) organotypic microfluidic model for glioma stem cells-Vascular interactions. Biomaterials 2019, 198, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Pisano, M.; Triacca, V.; Barbee, K.A.; Swartz, M.A. An in vitro model of the tumor-lymphatic microenvironment with simultaneous transendothelial and luminal flows reveals mechanisms of flow enhanced invasion. Integr. Biol. 2015, 7, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.Y.; Choi, J.H.; Kim, K.J.; Shin, M.; Choi, J.W. Microfluidic System to Analyze the Effects of Interleukin 6 on Lymphatic Breast Cancer Metastasis. Front. Bioeng. Biotechnol. 2020, 8, 611802. [Google Scholar] [CrossRef] [PubMed]
- Lugo-Cintron, K.M.; Ayuso, J.M.; White, B.R.; Harari, P.M.; Ponik, S.M.; Beebe, D.J.; Gong, M.M.; Virumbrales-Munoz, M. Matrix density drives 3D organotypic lymphatic vessel activation in a microfluidic model of the breast tumor microenvironment. Lab Chip 2020, 20, 1586–1600. [Google Scholar] [CrossRef] [PubMed]
- Sameni, M.; Anbalagan, A.; Olive, M.B.; Moin, K.; Mattingly, R.R.; Sloane, B.F. MAME models for 4D live-cell imaging of tumor: Microenvironment interactions that impact malignant progression. J. Vis. Exp. 2012, 17, 3661. [Google Scholar] [CrossRef]
- Sameni, M.; Dosescu, J.; Moin, K.; Sloane, B.F. Functional imaging of proteolysis: Stromal and inflammatory cells increase tumor proteolysis. Mol. Imaging 2003, 2, 159–175. [Google Scholar] [CrossRef]
- Jedeszko, C.; Victor, B.C.; Podgorski, I.; Sloane, B.F. Fibroblast hepatocyte growth factor promotes invasion of human mammary ductal carcinoma in situ. Cancer Res. 2009, 69, 9148–9155. [Google Scholar] [CrossRef]
- Osuala, K.O.; Sameni, M.; Shah, S.; Aggarwal, N.; Simonait, M.L.; Franco, O.E.; Hong, Y.; Hayward, S.W.; Behbod, F.; Mattingly, R.R.; et al. Il-6 signaling between ductal carcinoma in situ cells and carcinoma-associated fibroblasts mediates tumor cell growth and migration. BMC Cancer 2015, 15, 584. [Google Scholar] [CrossRef]
- Sameni, M.; Cavallo-Medved, D.; Franco, O.E.; Chalasani, A.; Ji, K.; Aggarwal, N.; Anbalagan, A.; Chen, X.; Mattingly, R.R.; Hayward, S.W.; et al. Pathomimetic avatars reveal divergent roles of microenvironment in invasive transition of ductal carcinoma in situ. Breast Cancer Res. 2017, 19, 56. [Google Scholar] [CrossRef]
- Sameni, M.; Tovar, E.A.; Essenburg, C.J.; Chalasani, A.; Linklater, E.S.; Borgman, A.; Cherba, D.M.; Anbalagan, A.; Winn, M.E.; Graveel, C.R.; et al. Cabozantinib (XL184) Inhibits Growth and Invasion of Preclinical TNBC Models. Clin. Cancer Res. 2016, 22, 923–934. [Google Scholar] [CrossRef]
- Brock, E.J.; Ji, K.; Shah, S.; Mattingly, R.R.; Sloane, B.F. In Vitro Models for Studying Invasive Transitions of Ductal Carcinoma In Situ. J. Mammary Gland Biol. Neoplasia 2019, 24, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Kriehuber, E.; Breiteneder-Geleff, S.; Groeger, M.; Soleiman, A.; Schoppmann, S.F.; Stingl, G.; Kerjaschki, D.; Maurer, D. Isolation and characterization of dermal lymphatic and blood endothelial cells reveal stable and functionally specialized cell lineages. J. Exp. Med. 2001, 194, 797–808. [Google Scholar] [CrossRef] [PubMed]
- Chalasani, A.; Ji, K.; Sameni, M.; Mazumder, S.H.; Xu, Y.; Moin, K.; Sloane, B.F. Live-Cell Imaging of Protease Activity: Assays to Screen Therapeutic Approaches. Methods Mol. Biol. 2017, 1574, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Sousa, C.M.; Biancur, D.E.; Wang, X.; Halbrook, C.J.; Sherman, M.H.; Zhang, L.; Kremer, D.; Hwang, R.F.; Witkiewicz, A.K.; Ying, H.; et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 2016, 536, 479–483. [Google Scholar] [CrossRef]
- Jeon, N.L.; Dertinger, S.K.W.; Chiu, D.T.; Choi, I.S.; Stroock, A.D.; Whitesides, G.M. Generation of Solution and Surface Gradients Using Microfluidic Systems. Langmuir 2000, 16, 8311–8316. [Google Scholar] [CrossRef]
- Lee, G.Y.; Kenny, P.A.; Lee, E.H.; Bissell, M.J. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat. Methods 2007, 4, 359–365. [Google Scholar] [CrossRef]
- Weigelt, B.; Bissell, M.J. Unraveling the microenvironmental influences on the normal mammary gland and breast cancer. Semin Cancer Biol. 2008, 18, 311–321. [Google Scholar] [CrossRef]
- Padera, T.P.; Kadambi, A.; di Tomaso, E.; Carreira, C.M.; Brown, E.B.; Boucher, Y.; Choi, N.C.; Mathisen, D.; Wain, J.; Mark, E.J.; et al. Lymphatic metastasis in the absence of functional intratumor lymphatics. Science 2002, 296, 1883–1886. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, L.; Chen, G. Fabrication, modification, and application of poly(methyl methacrylate) microfluidic chips. Electrophoresis 2008, 29, 1801–1814. [Google Scholar] [CrossRef]
- Zhao, J.; Dong, F.; Li, Y.; Kong, B.; Liu, Q. Effect of freeze–thaw cycles on the emulsion activity and structural characteristics of soy protein isolate. Process Biochem. 2015, 50, 1607–1613. [Google Scholar] [CrossRef]
- de Almeida Monteiro Melo Ferraz, M.; Henning, H.H.W.; Ferreira da Costa, P.; Malda, J.; Le Gac, S.; Bray, F.; van Duursen, M.B.M.; Brouwers, J.F.; van de Lest, C.H.A.; Bertijn, I.; et al. Potential Health and Environmental Risks of Three-Dimensional Engineered Polymers. Environ. Sci. Technol. Lett. 2018, 5, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Toepke, M.W.; Beebe, D.J. PDMS absorption of small molecules and consequences in microfluidic applications. Lab Chip 2006, 6, 1484–1486. [Google Scholar] [CrossRef] [PubMed]
- Moore, T.A.; Brodersen, P.; Young, E.W.K. Multiple Myeloma Cell Drug Responses Differ in Thermoplastic vs PDMS Microfluidic Devices. Anal. Chem. 2017, 89, 11391–11398. [Google Scholar] [CrossRef] [PubMed]
- Virumbrales-Munoz, M.; Ayuso, J.M.; Gong, M.M.; Humayun, M.; Livingston, M.K.; Lugo-Cintron, K.M.; McMinn, P.; Alvarez-Garcia, Y.R.; Beebe, D.J. Microfluidic lumen-based systems for advancing tubular organ modeling. Chem. Soc. Rev. 2020, 49, 6402–6442. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Lei, P.J.; Padera, T.P. Progression of Metastasis through Lymphatic System. Cells 2021, 10, 627. [Google Scholar] [CrossRef]
- Lee, E.; Fertig, E.J.; Jin, K.; Sukumar, S.; Pandey, N.B.; Popel, A.S. Breast cancer cells condition lymphatic endothelial cells within pre-metastatic niches to promote metastasis. Nat. Commun. 2014, 5, 4715. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, K.; Zhao, Z.; Sameni, M.; Moin, K.; Xu, Y.; Gillies, R.J.; Sloane, B.F.; Mattingly, R.R. Modeling Tumor: Lymphatic Interactions in Lymphatic Metastasis of Triple Negative Breast Cancer. Cancers 2021, 13, 6044. https://doi.org/10.3390/cancers13236044
Ji K, Zhao Z, Sameni M, Moin K, Xu Y, Gillies RJ, Sloane BF, Mattingly RR. Modeling Tumor: Lymphatic Interactions in Lymphatic Metastasis of Triple Negative Breast Cancer. Cancers. 2021; 13(23):6044. https://doi.org/10.3390/cancers13236044
Chicago/Turabian StyleJi, Kyungmin, Zhiguo Zhao, Mansoureh Sameni, Kamiar Moin, Yong Xu, Robert J. Gillies, Bonnie F. Sloane, and Raymond R. Mattingly. 2021. "Modeling Tumor: Lymphatic Interactions in Lymphatic Metastasis of Triple Negative Breast Cancer" Cancers 13, no. 23: 6044. https://doi.org/10.3390/cancers13236044
APA StyleJi, K., Zhao, Z., Sameni, M., Moin, K., Xu, Y., Gillies, R. J., Sloane, B. F., & Mattingly, R. R. (2021). Modeling Tumor: Lymphatic Interactions in Lymphatic Metastasis of Triple Negative Breast Cancer. Cancers, 13(23), 6044. https://doi.org/10.3390/cancers13236044










