3.1. Single Cell Seeding and Viability
Figure 1 illustrates the steps used to to detect single cells secreted PSA protein during different treatments inside the microwells.
Figure 1a shows a graphical depiction of three of the 6400 wells of the microwell chip. A cell suspension is pipetted onto the top of the chip and a small negative pressure (−5 mbar) is applied across the microwell chip. The fluid enters the wells and exits through the pore in the bottom of the individual wells. The flow-through directs the cells towards the pore and, once a cell lands on the pore, the flow stops and no other cell will enter the same well (
Figure 1b). The microwell array functions as a sieve and captures cells based on their size [
13,
14,
15]. To measure secreted PSA, cells from the prostate cancer cell lines LNCaP and VCaP were loaded as single cells into the individual microwells. After the cells were loaded (
Figure 1b), the presence of cells in each well was visualized using fluorescence microscopy (
Figure 1c). The secreted PSA was captured at the bottom of the microwell array using a poly-vinylidenefluoride (PVDF) membrane that was coated with commercially available anti-PSA antibodies (
Figure 1d). The microwell chip with the PVDF membrane was incubated for 24 h to allow secreted PSA molecules to diffuse through the microwell pores towards the membrane (
Figure 1e). After incubation, the microwells and membranes were separated (
Figure 1f,g). The PSA on the membrane was fluorescently labeled to quantify the PSA at each position and correlated with the position of the microwell (
Figure 1h). The microwells were subsequently connected to a second anti-PSA-coated membrane and fresh medium supplemented with the anabolic steroid R1881 was added to stimulate PSA secretion over a period of 24 h (
Figure 1i). After 24 h, the membrane was replaced with a third membrane and the cells were supplemented with fresh medium containing an androgen inhibitor (enzalutamide or abiraterone,
Figure 1m–p). This procedure resulted in three membranes: one for the detection of PSA from unstimulated cells, one for the detection of PSA after R1881 stimulation, and one to measure the effect of either enzalutamide or abiraterone on PSA secretion. The PSA production and viability of the individual cells exposed to different conditions were monitored in this manner over a 72-h period.
After distributing single cells in the individual wells of the microwell array, images of the cells were acquired to determine their viability and morphological appearance. The design of the well in combination with the microfluidic forces only allow for a single cell per well; however, in cases where two cells are attached to one another, the microwell will contain two cells after the wells are filled [
11].
Figure 2a depicts typical images of one or two LNCaP cells inside the wells (blue and red circles, respectively). The cells show a spread morphology similar to that of cells grown in culture flasks. The captured single prostate cancer cells were viable and proliferated inside the microwell for 3 days of culture, indicating that the cell seeding process does not affect cell viability.
Figure 2b shows LNCaP and VCaP cells in a microwell at Day 1 and at Day 3 after cell division occurred. After filling the wells, the number of cells per well was quantified. The graph in
Figure 2c presents the number of cells per well found for each cell line. The majority of the wells contained a single cell (92%), 4% of the wells contained two cells, 3% of the wells contained three cells, and 2% of the wells contained no cells.
To determine the cell viability with or without drug stimulation, the number of viable cells after each treatment was counted and quantified using the fluorescence of calcein AM and dead cells were detected by the fluorescence of Ethidium homodimer-1.
Figure 2d–g show the viability of LNCaP and VCaP cells after different treatments. The viability was first measured for untreated cells left in the microwell chip (t = 24 h). Next, R1881 was added and cells were left for another 24 h before viability was measured (48 h). Finally, one of the drugs (enzalutamide or abiraterone) was added for 24 h and the cell viability was measured once again (t = 72 h). The data show no significant difference in viability between the no-drug group and the androgen-stimulated group (
Figure 2d,e). After 24 h, >85% of the cells were viable and >75% of the cells were viable after androgen stimulation (t = 48). However, after the addition of enzalutamide, the cell viability reduced significantly to 24% for LNCaP cells and to 29% for VCaP cells (
Figure 2d,f;
p < 0.0001) when compared with the no-drug group. After the addition of abiraterone, the cell viability was significantly reduced (
p < 0.0001) to 39% for LNCaP cells and to 46% for VCaP cells when compared with the no-drug group (
Figure 2e,g, respectively). In addition, there was also a significant difference between androgen stimulation and androgen inhibition (
p < 0.001 and
p < 0.0001). Comparing the same drug on different cell lines also showed a significant difference (
p < 0.001 for enzalutamide and abiraterone). Exposure to abiraterone resulted in a higher number of viable cells when compared with enzalutamide, with 39% for the LNCaP cells and 46% for the VCaP cells. These results indicate that both cell lines are more sensitive to enzalutamide than to abiraterone, which is in line with previous reports.
3.2. Single Cell PSA Secretion Assay
To determine the PSA secretion levels of the cells, a fluorospot assay was developed using a capture membrane coated with antibodies directed against the cell-secreted PSA protein. The secreted PSA molecules captured on the membrane were visualized with fluorescent labeled antibodies directed against PSA. First, we verified that PSA secretion is affected by androgen stimulation (
Figure S1). The PSA secretion of prostate cell lines was measured on membranes in 12-well plates and showed the validity of the protocol. Next, we determined whether our microwell platform could be used to capture cell-secreted PSA on PVDF membranes. Cells were distributed into the microwells and their secreted PSA was captured on a PVDF membrane.
Figure 3 shows an array of calcein-AM-stained cells trapped in the wells, the corresponding membrane with the captured PSA, and a merged insert showing typical cells and the corresponding secreted PSA. The enlarged view shows high and low amounts of captured PSA corresponding to high-PSA-secreting and low-PSA-secreting cells. To quantify the amounts of PSA captured on the membrane from a well, an image analysis algorithm was developed. First, the images of the cells in the wells and the PSA imprint on the PVDF membrane were overlaid to match the well number with the captured PSA on the membrane. Next, the average fluorescence intensity of the PSA spots was measured. To quantify the PSA production, a calibration curve was plotted to provide the relation between the intensity and the amounts of PSA proteins in pg/cell/day (
Figure S2). The background levels were determined by assessment of the fluorescence intensity on the membrane of >50 empty wells across the microwell chip. The results in
Figure S3 shows that >90% of the single cells secreted PSA at significant higher concentration than the background (empty wells), (
p < 0.0001). The amount of captured PSA was determined for 1223 wells containing a single LNCaP cell and 329 wells containing two cells. Wells containing one LNCaP cell produced an average of 4.8 pg/cell/day of PSA, which was significantly less compared with wells containing two LNCaP cells with 7.5 pg/cell/day of PSA (
Figure S3), (
p < 0.001).
3.3. Drug Sensitivity of PC Cells
To detect the PSA secretion over a 72-h period from cells in wells, membranes were changed every 24 h.
Figure S4 illustrates the PSA microarray from LNCaP cells at different time intervals without any drug treatment. These results show a dynamic secretion profile and that the procedure for measuring protein over time is reproducible. Once the immunoassays were established, we performed anti-androgen treatments on the cells in the wells to identify their effect on the PSA secretion of PC cells. To do so, we distributed single cells into the microwells as described above. After that, the activated membrane was attached to the bottom of the microwell chip containing the cells.
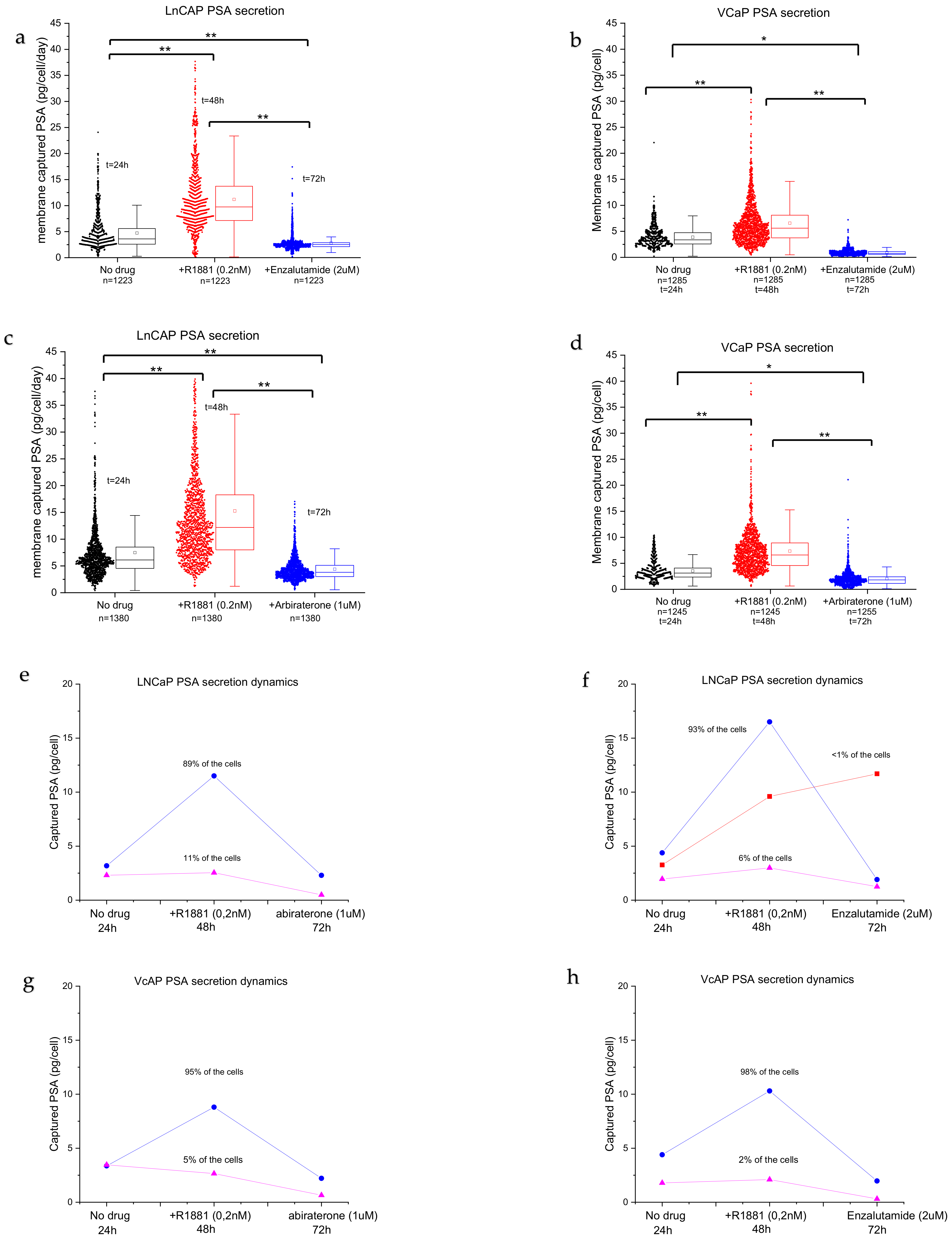
Figure 4 shows the captured PSA spots on three different membranes from the same cells in time. The membrane in
Figure 4a shows the PSA secretion from LNCaP cells after 24 h of incubation without the addition of any drug. A large spread in secreted amounts of PSA between individual cells was observed, indicating the existence of a high degree of heterogeneity between the individual cells.
Figure 4b shows the PSA secretion profile of the same cells after R1881 stimulation. The cells show an increase in PSA secretion (bigger spots) as well as an increase in the number of secreting cells (17%) with extensive heterogeneity between cells.
Figure 4c shows the PSA secretion of the same cells exposed to the anti-androgen drug enzalutamide for 24 h. After the cells were exposed to enzalutamide, the number of viable cells significantly decreased and the PSA secretion of the majority of cells also decreased. To visualize the PSA secretion of individual cells, the same area of the PVDF membrane was enlarged for the three subsequent time points and the secreted products of two cells are indicated with a 1 and a 2 in
Figure 4. These results show heterogeneous secretion of PSA by LNCaP cells. The treatment with R1881 induced cells to secrete higher levels of PSA and treatment with the anti-androgen drug enzalutamide showed a decrease in the PSA secretion of the LNCaP cells. Both PC cell lines LNCaP and VCaP show a similar pattern of heterogeneity in PSA secretion.
We then used the fluorescence images to analyze the amounts of PSA protein secreted by PC cells during the three consecutive days. The software algorithm aligned the images and measured the intensities in the captured PSA spots across the three membranes. The calibration curves (
Figure S2) enabled the estimation of the amounts of PSA proteins for >1000 stimulated and non-stimulated cells. Using this method, we demonstrated that the cells display extensive heterogeneity (
Figure 5a–d). LNCaPs show a distribution of low producers and highly active PSA-producing cells with a large tail (
Figure 5a,b, no drug), with an average secretion of ~7 pg/cells/day of PSA protein. Androgen stimulation significantly induced PSA secretion (2-fold) when compared with non-stimulated cells (
Figure 5a,b, +R1881,
p < 0.001). Furthermore, the long tail indicates that more cells are actively secreting PSA. The cells can be grouped into low PSA secretors and a wide distribution of high-PSA-secreting cells. The number of secreting cells decreased when the anti-androgen drug enzalutamide was added (
Figure 5a,b). The distribution was more homogenous, the amount of high-PSA-secreting cells decreased, and the average amount of secreted PSA per cell decreased significantly when compared with no drug treatment and R1881 treatment. VCaP cells (
Figure 5c,d) show a more uniform secretion pattern when compared with LNCaP cells (
Figure 5a,b), with an average of 3.7 pg/cell/day of PSA protein for non-treated cells. The average PSA production level increased significantly (
p < 0.001) when the cells were stimulated with R1881 to ~4.8 pg/cell/day. Stimulation with anti-androgens significantly reduced the number of cells producing PSA, resulting in a reduction in the median PSA production to <1 pg/cell in VCaP cells (
Figure 5c,d, abiraterone or enzalutamide,
p < 0.001). In addition, there was a difference in the expression of PSA when comparing the effect of the same drug on each cell line (for both drugs,
p < 0.001).
The percentage of viable LNCaP cells with a PSA secretion level above the background was 53% after 24 h (no drug treatment), 67% after 48 h (R1881 stimulation for 24 h), and 21% after 72 h (abiraterone treatment for 24 h) or 33% after 72 h (enzalutamide treatment for 24 h). The percentage of viable VCaP cells with a PSA secretion level above the background was 30% after 24 h (no drug treatment), 50% after 48 h (R1881 stimulation for 24 h), and 26% after 72 h (abiraterone treatment for 24 h) or 17% after 72 h (enzalutamide treatment for 24 h).
Although we observed a large distribution in secretion activity, clear patterns could be observed. Typical representative cases of the two cell lines are plotted in
Figure 5e,h. The majority of the cells responded to stimulation with androgen steroids and increased the PSA secretion levels. In the abiraterone group, LNCaP cells show two populations: androgen-sensitive cells (93–89%, blue lines) and insensitive/low-producing cells (6–11%, purple lines) (
Figure 5e,f), while the enzalutamide group shows an added population that is sensitive to androgens but not sensitive to enzalutamide stimulation (<1%) (
Figure 5d, red line). The androgen-sensitive cells display a sharp spike in PSA secretion and a decrease following anti-androgen stimulation. The population of androgen-insensitive LNCaP cells, however, did not respond to androgen stimulation and the PSA secretion levels remained low (
Figure 5e,f, pink lines), with low variation between assay points.
After the strong increase in PSA production, the PC cells showed a sharp drop in PSA secretion after the addition of abiraterone (
Figure 5e,g, blue lines) or enzalutamide (
Figure 5f,h, blue lines). Interestingly, a small number of LNCaP cells (<1%) did not show a drop after the addition of the anti-androgen drug enzalutamide and the PSA production remained high (
Figure 5f, red line), indicating that the cells required a longer stimulation duration.