Simple Summary
To produce the proteins needed for the cell to survive, the information in the DNA is converted to a mobile form known as messenger RNA, which exits the cell nucleus and binds to machines that convert RNAs into proteins in a process referred to as translation. Importantly, the RNA message can be altered at any point in its journey prior to translation. These alterations constitute changes to the chemical nature of the messenger RNA and modulate the ability of mRNAs to be converted into proteins and even lead to the production of proteins with different functionalities than those encoded by their original forms. Here we provide a conceptual framework for the integration of these mRNA maturation steps with translation with a focus on the proteins that escort these mRNAs through these steps and into the translation machines. We discuss the relevance to cancer and therapeutic strategies to target these in malignancy.
Abstract
The translation of RNA into protein is a dynamic process which is heavily regulated during normal cell physiology and can be dysregulated in human malignancies. Its dysregulation can impact selected groups of RNAs, modifying protein levels independently of transcription. Integral to their suitability for translation, RNAs undergo a series of maturation steps including the addition of the m7G cap on the 5′ end of RNAs, splicing, as well as cleavage and polyadenylation (CPA). Importantly, each of these steps can be coopted to modify the transcript signal. Factors that bind the m7G cap escort these RNAs through different steps of maturation and thus govern the physical nature of the final transcript product presented to the translation machinery. Here, we describe these steps and how the major m7G cap-binding factors in mammalian cells, the cap binding complex (CBC) and the eukaryotic translation initiation factor eIF4E, are positioned to chaperone transcripts through RNA maturation, nuclear export, and translation in a transcript-specific manner. To conceptualize a framework for the flow and integration of this genetic information, we discuss RNA maturation models and how these integrate with translation. Finally, we discuss how these processes can be coopted by cancer cells and means to target these in malignancy.
Keywords:
m7G cap; CBC; eIF4E; cap-chaperones; mRNA maturation; splicing; polyadenylation; nuclear export; cancer 1. Overview
The translation of RNAs into proteins is a fundamental step in the conversion of DNA signals (in the form of coding transcripts) into their active form (proteins). Most focus has been on the impact of modulating the translation of a given RNA on the ultimate levels of the corresponding protein produced. While this is clearly important, it is equally critical to consider the processes that chemically modify the transcripts which ultimately impact the composition of the final protein product, as well as the processes that regulate the availability of these transcripts to the ribosome. In this regard, many of the major RNA metabolism steps can physically alter the composition of transcripts, which can lead to altered abilities of RNAs to access the ribosome and/or the production of different functional forms of the resulting proteins. In this way, RNA metabolism provides a means to reprogram the transcriptional signals which are the ultimate substrates of the ribosomes. For instance, splice variants and alternatively polyadenylated transcript variants can produce proteins with opposing or modified functions [1,2]. Elevated RNA nuclear export can increase the availability of transcripts to the translation machinery, bypassing the need to increase transcription [3]. Understanding how RNA metabolism segues into translation is critical for the decryption of the subsequent functional proteome which in turn governs cell physiology and, when dysregulated, can underpin human malignancy.
A fundamental feature central to the control of RNA maturation, export, and translation is the m7G “cap” modification on the 5′end of coding RNAs [4]. Factors that bind this moiety can control the fate of the RNA. There are two major cap-binding factors in mammals [5]. The cap-binding complex (CBC) is comprised of the cap-binding protein NCBP2 (also known as CBP20) and its adaptor NCBP1 (also known as CBP80) which together escort RNAs through the different nuclear maturation steps including splicing, CPA, and nuclear RNA export [6,7,8,9,10]. CBC also acts in the pioneer-round of translation and even in the steady-state translation of some viral and cellular RNAs [11,12,13]. The eukaryotic translation initiation factor eIF4E, which is associated with the steady-state translation of RNAs [14], can also function in capping, splicing, CPA, and nuclear RNA export for selected transcripts [15,16,17,18,19,20,21]. Indeed, both CBC and eIF4E can be found in the nucleus and cytoplasm of mammalian cells, positioning them to act in all these functions simultaneously [7,8,9,10,18,22,23,24,25]. In this way, the proteins involved in these chemical alterations to the RNAs impact their translation. This suggests that RNA maturation and translation are more closely tied than previously considered. Indeed, the classic model is that CBC escorts RNAs in the nucleus as they undergo maturation steps that will define their function, transporting them to the cytoplasm where these RNAs are handed off to eIF4E for steady-state translation [14] (Figure 1). In this model, there is a clear separation-of-function based on subcellular location; however, more recent evidence suggests this needs to be revisited given the new cellular localization and functionalities discovered for CBC and eIF4E over the past 25 years.
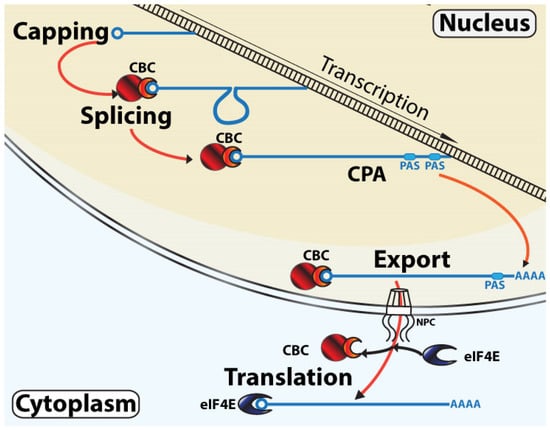
Figure 1.
Classical linear model of mRNA maturation. During transcription, the m7G cap is added to the RNA as it is transcribed. This chemical modification leads to CBC recruitment. While transcription elongation continues, CBC facilitates the recruitment of the spliceosome to the nascent RNA. At the end of transcription, CBC assists in the recruitment of the cleavage and polyadenylation (CPA) machinery at the PAS site. The CBC chaperones the mRNA to the NPC where the complex transits through the nuclear membrane and into the cytoplasm where eIF4E replaces the CBC as the predominant cap-chaperone. The transcript can now undergo steady-state translation. The m7G cap is depicted as a circle, CBC is in red with NCBP1 as the larger circle and NCBP2 as the semi-circle, and eIF4E is in blue. For simplicity, not all factors involved in these processes are shown.
Here we discuss the evidence supporting the roles of these cap-chaperones in RNA maturation and translation with the goal of understanding the flow of genetic information and its impact on the cell. These observations lead us to propose that there are coexistent, parallel RNA maturation paths characterized by CBC or eIF4E cap-chaperones. Moreover, we predict that RNAs can undergo cap-chaperone switching between CBC and eIF4E. In other words, transcripts can undergo certain RNA maturation events with one cap-chaperone and then switch the cap-chaperone for subsequent steps. This model provides a means by which groups of RNAs can be differentially matured and translated in response to a wide array of stimuli and collectively dysregulated in malignancy. This review is divided into three sections: (1) a description of RNA maturation, RNA export, and translation focusing on the role of cap-chaperones; (2) models for RNA maturation based on these observations and the interplay with translation; and (3) relevance to cancer.
2. RNA Maturation, Nuclear Export, and Translation Focusing on the Roles of Cap-Chaperones
One of the most highly held tenets in gene expression is that during transcription, coding as well as many non-coding RNAs, undergo a series of processing steps that physically modify the chemical structure of the RNA in order to generate mature transcripts that are substrates of the ribosome [26]. There are three major steps in this process for coding RNAs: m7G capping, splicing, and CPA [26]. This is followed by nuclear export to the cytoplasm which can amplify the signal by increasing cytoplasmic RNA availability to the ribosome without altering transcription levels [27,28]. In some cases, translation can also be used as a source for amplification of the transcriptional signal by increasing the number of ribosomes per transcript so that more protein is made from the same amount of transcript. The specific steps and relevant biochemical reactions are described in this section, followed by the relevance of cap-chaperones CBC and eIF4E to each step. These steps often occur co-transcriptionally, enabling a transcription–RNA maturation coupling. We also discuss recent findings which indicate that many of these steps can also occur post-transcriptionally in the nucleus and, in some cases, even in the cytoplasm.
2.1. The Process of m7G Capping
The first maturation step for most transcripts is referred to as m7G capping [4,29,30]. The function of the m7G cap is multifold. It enables the engagement of specific cap-binding factors such as CBC and eIF4E and, furthermore, it protects RNAs from degradation by 5′exonucleases [31,32]. Capping is key for the efficiency of subsequent RNA maturation steps including splicing, CPA, RNA export, and translation [9]. Generally, it is considered that RNAs are capped co-transcriptionally after generation of the first ~20–30 nucleotides. In mammals, capping is a three-step process involving RNGTT (RNA guanylyltransferase and 5′ phosphatase) and RNMT (RNA guanine-7-methyltransferase) [33,34,35], two proteins essential for capping and also for cell survival [33,34,36,37,38,39,40,41]. RNGTT removes the 5′ phosphate of the 5′ triphosphate on the pre-mRNA or non-coding RNA using its 5′ phosphatase activity [42]. The resulting 5′-diphosphate-RNA serves as an RNGTT substrate for the addition of guanosine monophosphate, resulting in a 5′-5′ pyrophosphate linkage. Then, RNMT uses its methyltransferase domain and the S-adenosyl methionine (SAM) methyl donor to methylate the cap guanylate [38,43,44]. RAM, a small protein co-factor, binds RNMT and increases its methylation activity and can also recruit some RNAs to the complex [43]. At steady state, the localization of RNMT and RNGTT is mainly nuclear; however, these factors are also found in the cytoplasm supporting the notion that capping can occur in both locals [36,45,46]. It also suggests that decapped RNAs are not always fated to be degraded but rather can be re-capped [47,48,49]. Further, while capping is generally considered to be co-transcriptional, the cytoplasmic localization of these enzymes and the observation of re-capping activity in that compartment suggest that re-capping could also occur independently of transcription [45,46,50]. Thus, as with splicing and CPA [51,52,53] (see below), it appears that capping can occur both co- and post-transcriptionally depending on the RNA and cellular context. The traditional view is that shortly after capping, RNAs directly bind to the NCBP2 component of the CBC [10]. At this stage, the RNA is available to be chaperoned through the splicing, CPA, and nuclear export steps by the CBC. The role of CBC in mRNA maturation has been reviewed extensively [9,13,54].
While capping was widely considered to be a constitutive process by which all coding RNAs are 100% capped, recent findings suggest a more nuanced view should be adopted. Indeed, steady-state capping is dynamic, leading to the notion of cap homeostasis [4,47]. These findings are based on new methods developed to study capping which can be carried out on a per RNA or genome-wide basis [4,55]. These studies revealed that capping efficiency is positioned as an important control point for the protein-coding capacity of many mRNAs as well as the biochemical activity of non-coding transcripts [15,29,30,48,55,56]. Indeed, the capping of specific transcripts can be regulated during development and differentiation, and can be elevated in human malignancy [15,29,30,48,55,56]. The dynamic nature of capping is highlighted by the observation that RNAs can be de-capped and then re-capped [45,46,47,50]. The equilibrium between cap-chaperones and decapping enzymes could be modulated to transiently change the proportions of capped RNAs based on their recruitment affecting specific transcripts. It seems likely that since capping is dynamic, the cap-binding protein associated with RNAs varies depending on the cellular conditions, suggesting that transcripts could use different cap-chaperone proteins during disparate parts of their life cycle.
Recently, eIF4E was shown to play a role in steady-state capping [15]. eIF4E elevates the production of RNMT, RNGTT, and RAM through its RNA export and translation activities [15], and also physically interacts with RNMT in cells and in biophysical experiments [57]. Consistently, the overexpression of eIF4E in cell line models led to the increased capping of a subset of transcripts. Many of the eIF4E target RNAs encode proteins involved in oncogenesis, e.g., MDM2, CTNNB1, MYC, and CCND1, but not housekeeping transcripts. While eIF4E increased capping for some RNAs, it reduced the capping for others, suggesting there is a competition for limited resources (RNGTT, RAM, RNMT, SAM) so that eIF4E tips the balance to favor specific transcripts. These studies also revealed that in human cells the capping of specific RNAs at steady-state was lower than anticipated, positioning this as an important regulatory step in terms of the functionality of RNAs. eIF4E does not simply provide protection against decapping by directly binding to m7G caps of its target RNAs [15]. Indeed, eIF4E overexpression increases the steady-state capping of transcripts not found in eIF4E nuclear RIPs, indicating that eIF4E does not need to physically interact with these to enhance capping [15]. An RNA element, referred to as a cap sensitivity element (CapSE) conferred increased capping activity when fused to a LacZ reporter upon eIF4E overexpression [15] and could recruit relevant factors to the RNA. In all, eIF4E increases capping for a subset of RNAs, likely containing specific RNA elements that imbue these RNAs with sensitivity to this process by recruiting factors such as RNMT to the transcripts.
2.2. RNA Splicing
Splicing is the removal of introns and the joining of flanking exons in pre-messenger RNAs (pre-mRNAs) and some non-coding RNAs [58]. This process is catalyzed by the spliceosome, an intricate assembly of >150 proteins and 5 uridine-rich small nuclear UsnRNAs (U1, U2, U4, U5, and U6 snRNAs) [58]. This machine recognizes elements in the 5′ splice-site (5′SS), 3′ SS, and the branch site to catalyze the excision of targeted introns. Alternative splicing (AS) generates diversity in the proteome by producing multiple mRNAs from the same pre-mRNA [1]. About 95% of multi-exonic genes undergo AS [59]. AS events include altered selection of the 5′SS or 3′SS, skipped exons (SE), inclusion of mutually exclusive exons (MXE), or intron retention (IR) [59]. These events occur through competition between different splice sites in the pre-mRNA, where this balance is altered by cis-elements known as exon/intron splicing enhancers (ESE/ISE) or silencers (ESS/ISS) [59]. Strong ESS/ISS elements promote exon skipping whereas ESE/ISE sequences stimulate splicing. AS products can have opposing functions to their constitutive counterparts, lead to mis-localization, generate transcripts that are rapidly degraded, leading to protein loss, or other effects [1,60].
Recently, the location of splicing has been shown to be more diverse than first thought. Approximately 80% of splicing occurs co-transcriptionally and ~20% post-transcriptionally in nuclear speckles [52,61]. Features of intron location (e.g., near the 3′end, terminal introns) or intron composition (highly structured) favor post-transcriptional splicing [51,52,61,62]. Interestingly, in the same RNA, some introns can be removed co-transcriptionally with others post-transcriptionally [51,61]. In these cases, spliceosome assembly on the RNA can result from two independent assembly events (i.e., the spliceosome does not appear to travel with the RNA [61]). Additionally, cytoplasmic splicing was reported [63,64]. Indeed, ~0.5% of the transcriptome appears to undergo cytoplasmic splicing via the minor spliceosome. This complex shares many common components with the major spliceosome in the nucleus [63]. Cytoplasmic splicing has not been extensively studied and remains controversial, but the possibility of such post-transcriptional processing extends the capacity of mRNA maturation to modify transcripts and the resulting protein outputs.
The possibility that splicing occurs in different locations, co-transcriptionally and post-transcriptionally, leads to the question of whether different cap-chaperones would be used in different contexts. It is generally considered that RNAs are recruited to the spliceosome through interactions of their m7G cap with the CBC. CBC is most often associated with removal of the first intron in a co-transcriptional manner [65], where it triggers this event by promoting the recognition of the 5′ splice site by U1 snRNP [7]. Further, CBC binds the splicing machinery in an mRNA-independent manner [8]. For example, CBC promotes spliceosome assembly and interacts with UsnRNPs [8]. Additionally, CBC associates with U6 snRNP, which occurs concomitantly with the recruitment of the U4/U6-U5 tri-snRNP [65,66].
In addition to facilitating the constitutive splicing of first introns, CBC regulates AS through its co-factor, the positive transcription elongation factor P-TEFb, supporting a transcription-dependent regulation of AS by CBC [67]. Indeed, gene-specific recruitment of P-TEFb by the CBC [67] may explain how CBC affects AS for only a subset of transcripts. There is the proposed involvement of sequence-specific RNA-binding proteins such as SRSF1 [68], or structure-specific RNA-binding protein such as hnRNPF [69], suggesting that RNA cis-elements influence the CBC-dependent control of AS. Finally, CBC may have an indirect influence on AS through the regulation of snRNAs synthesis and export [70,71,72]. In all, while CBC generally associates with RNAs co-transcriptionally, it seems likely that is it not involved in all splicing events for all transcripts. This suggests that other cap-chaperones could escort pre-mRNAs through these steps. For example, in the nucleus, eIF4E can modulate the splicing of thousands of RNAs, increase the levels of components of the spliceosome, and physically interact with all the major UsnRNAs as well as protein components of the spliceosome [21]. This occurs in multiple human cell lines and in high-eIF4E acute myeloid leukemia (AML) patient specimens, suggesting that in parallel to CBC escorted RNAs, eIF4E can also chaperone a subset of RNAs to the spliceosome. It also raises the question of whether RNAs could be escorted through splicing by other cap-chaperones. However, further investigation is needed to confirm that eIF4E is associated directly with the mRNAs and not via interactions with splice factors or UsnRNA as seen for CBC. Interestingly, a recent genome-wide CRISPRi screen identified exon junction complex (EJC) and spliceosome components as lethal partners of eIF4E. The list of identified genetic eIF4E partners also includes more than twenty genes coding for proteins involved in ribonucleoprotein complex biogenesis, including ribosome biogenesis and other nuclear functions [73]. These findings clearly highlight the functional proximity of eIF4E with the nuclear pathways of RNA maturation.
2.3. Cleavage and Polyadenylation
Largely considered to be a co-transcriptional event, CPA is a two-step process required for maturation of the majority of coding RNAs and also for some non-coding transcripts [74]. This maturation is important for both nuclear RNA export and for ultimate translation for coding transcripts. The first step of CPA is the recognition of the polyadenylation signal (PAS) within the 3′ UTR of transcripts, which is then followed by cleavage of the polyadenylation site near to the PAS. Cleavage mainly occurs in the nucleus [74], and is followed by addition of the poly(A) tail via poly(A) polymerases which can occur in either the nucleus or cytoplasm [75,76,77]. Importantly, many transcripts have more than one PAS cleavage site, which can even occur within the coding region of the RNAs [75,76,77]. Alternative polyadenylation (APA) is defined as the cleavage of alternative PASs within the 3′UTR, introns, or coding region of transcripts; altered PAS cleavage efficiency; and/or alterations in poly(A) tail length [75,76,77]. APA can affect RNA stability, export, localization, translation, and in some cases, the functionality of the protein product by altering the coding sequence, such as by generating truncated proteins with alternative C-terminal domains [2]. The impact of APA on gene expression is highlighted by the observation that at least 70% of mammalian RNAs express APA isoforms [77].
Interestingly, even though CPA is mainly a co-transcriptional nuclear process, transcripts can undergo additional CPA either post-transcriptionally in the nucleus or in the cytoplasm [53,78,79]. Analyses of the human transcriptome has revealed that thousands of cleaved and polyadenylated transcripts can be re-cleaved at a proximal internal site in the 3′-UTR, resulting in two stable, autonomous, RNA fragments: a coding sequence with a shorter 3′-UTR (body) and an uncapped 3′-UTR sequence downstream of the cleavage point (tail) [78]. This suggests a widespread post-transcriptional APA producing stable 3′-UTR RNA tails that co-exist alongside their transcripts of origin [78]. This study also suggested that shorter transcripts are more abundant in the cytoplasm and that these events are cell-type specific [78]. Observations of the CTN RNA (Cat2 transcribed nuclear RNA) showcased the notion of nuclear post-transcriptional APA [53]. Here, the CTN RNA undergoes a first round of CPA, likely co-transcriptionally, but then the polyadenylated mRNA remains stored in the nucleus until specific signaling events trigger the cleavage of another PAS within the CTN RNA to produce the CAT2 RNA, which is followed by poly(A) tail addition and is then exported to the cytoplasm and translated into protein [53]. Cytoplasmic, post-transcriptional APA in neurons provides a fascinating similar example [79]. Here, selected groups of transcripts are processed into mRNAs with long 3′UTRs in the nucleus. This selective APA seems to promote the nuclear export and the subsequent axonal transport of these mRNAs to be translated at specific sites within the neurons. In this case, longer 3′UTRs are necessary for the recruitment of RNA binding proteins (RBPs) and non-coding RNAs needed for their axonal transport and ultimate neuronal localization. Afterwards, efficient local translation requires, in most cases, shorter 3′UTRs so the cleavage of a proximal PAS is triggered at specific cytoplasmic locales [79]. One of the most interesting studies in this regard showed that hundreds of axonal transcripts with shorter 3′UTRs are produced in the cytoplasm. This cytoplasmic cleavage is mediated by a multi-protein complex centered around the endonuclease Ago2 [80].
Cap-chaperones are involved in PAS selection and can influence cleavage efficiency. CBC, for instance, specifically regulates the 3′end processing of structured transcripts from intronless genes such as replication-dependent histones (RDHs) and pre-snRNAs. CBC promotes the selection of proximal PAS through two mutually exclusive co-factors: the negative elongation factor (NELF) [81] and the arsenite resistance protein 2 (ARS2) [82,83,84]. Given its role in splicing by recruiting U1 snRNP to the 5′splice site of the first intron, CBC also inhibits the selection of some intronic PASs [83], providing a clear example of how cap-chaperones can be key contributors to the interplay between splicing and CPA. The alternative CBC component, the nuclear cap-binding protein NCBP3, also plays a role in suppressing premature CPA through its interaction with U1 snRNP [85]. Additionally, the other major cap-chaperone, eIF4E, is likely to be another regulator of CPA [19]. eIF4E physically associates with CPSF3 (the polyadenylation site cleavage enzyme) and its co-factor CPSF1 in cells and can increase the cleavage of model RNAs. It increased the cleavage efficiency of a few examined RNAs that harbored an eIF4E sensitivity element (4ESE) previously identified for eIF4E-dependent export [19]. In parallel, eIF4E drives the production of several cleavage co-factors by increasing both their RNA export and/or translation efficiency, including the cleavage enzyme CPSF3 and its co-factor CPSF1 [19]. Our unpublished results show that eIF4E overexpression or reduction alters the PAS site selection of ~1000 RNAs, suggesting a substantive role in the APA of selected transcripts.
APA contributes to disorders ranging from cancer to neurodegeneration [75,76,77]. In cancer, APA generally results in oncogenes coding mRNAs with shorter 3′UTRs, providing a mechanism to evade regulation by microRNAs or RBPs through the removal of key regulatory sequences [75,76,77]. These shorter RNAs are usually generated through the selection of PASs close to the stop codon (known as the proximal PAS). However, shorter 3′ UTRs are not always oncogenic, with effects often being context specific. APA can be dysregulated for many reasons [75]. However, the molecular mechanisms driving APA are only starting to be elucidated. Cap-chaperones, including CBC and eIF4E, are likely to be significant contributors to APA regulation as well as its dysregulation in malignancies.
2.4. Nuclear RNA Export
To reach the cytoplasm, mRNAs do not diffuse freely through the nuclear pore complex (NPC). Instead, mRNAs transit to the cytoplasm through the NPC escorted by NPC receptors and an array of export co-factors including cap-chaperones. Once in the cytoplasm, the mRNA cargoes are released from these factors so that they can be ultimately translated by the ribosome. NPC receptors and export factors are then reimported into the nucleus to act in future rounds of export. The addition of the cap enhanced the export of both snRNAs and mRNAs, highlighting the importance of cap-chaperones to this process [86]. Consistent with these observations, one of the protein co-factors facilitating the export of RNAs is the cap-chaperone CBC, which is associated with the m7G cap of mRNAs and also with the 2,2,7-trimethylguanosine cap on snRNAs during export, albeit in different complexes [87,88]. CBC-bound mRNAs associate with the bulk RNA export receptor NXF1/NXT1 [86,89] which in turn interacts with the nuclear basket of the NPC and then transits through the membrane via the central channel of NPC [90,91,92]. NXF1/NXT1 recruits mRNAs through the transcription export complex (TREX). TREX consists of UAP56, ALY/REF (ALY), CIP29, and the multi-subunit THO complex, which is comprised of THOC1/Hpr1, hTho2, THOC5, THOC6, THOC7, and Tex1 [89]. TREX is generally considered to recruit transcripts during splicing [93], and it interacts through ALY to 5′ and 3′ of transcripts [94,95]. In CBC, while NCBP2 is bound to the cap [96], NCBP1 interacts with different partners, including ALY, which in turn recruits TREX at the 5′ends of spliced mRNAs, leading to the export of the complex [97]. However, the inhibition of NCBP2 suggests that UsnRNA export needs CBC, while mRNA export does not [70], opening the possibility that other factors could play a cap-chaperone role here. The accessory CBC protein NCBP3, found to bind the m7G cap, albeit with less affinity than NCBP2 [6], was also identified as an interactor of TREX, suggesting a role in mRNA export [6]. Additionally, NCBP3 is associated with the exon-junction complex (EJC) and the stimulated export of polyadenylated RNAs through cooperation with TREX [98]. However, NCBP3 may not be a cap-binding protein in vivo, but rather, a protein associated with CBC, TREX, and the EJC to export spliced mRNAs [98,99,100]. Further investigation is required to dissect these possibilities.
An alternative nuclear export path for selected mRNAs employs CRM1/XPO1 to transit through the NPC rather than NXF1/NXT1 [27,28]. This pathway could utilize CBC and/or eIF4E. Indeed, eIF4E increases the export of many RNAs through the CRM1/XPO1 system. One well-defined means of selection of RNAs for this pathway is the presence of the 4ESE RNA element in the 3′UTR of transcripts which is comprised of a paired loop structure [17,18,20]. This element is recognized by LRPPRC (leucine-rich pentatricopeptide repeat C-terminus protein). The export complex formed with 4ESE-RNA is composed of eIF4E, LRPPRC, and the export receptor CRM1/XPO1 [20,101]. LRPPRC acts as an assembly platform where it interacts directly with the dorsal surface of eIF4E (bound to the m7G cap on the target RNA) and with CRM1/XPO1 [20,101]. The independence of eIF4E-dependent export from the bulk pathways was demonstrated by its sensitivity to leptomycin B treatment [18,102] which directly inhibits CRM1 [102] and its insensitivity to the genetic inhibition of NXF1/TAP1 [18]. Additionally, eIF4E remodels the NPC to promote 4ESE mRNA export [16]. Indeed, eIF4E overexpression leads to alterations to the NPC by reducing levels of RanBP2, the major cytoplasmic filament protein, and altering the localization of other NPC factors [16]. Moreover, eIF4E drives the expression of RanBP1, likely to compensate for the reduction in RanBP2, thereby allowing efficient RNA cargo release in the cytoplasm [16]. Intriguingly, eIF4E may also impact the bulk RNA export pathway through its ability to increase levels of Gle1 and DDX19 [16], which are required for the release of the RNA cargo into the cytoplasm in the NXF1/NXT1 pathway [16]. In this way, eIF4E is positioned to indirectly modulate the export of RNAs going through the CBC–NXF1/NXT1 pathway, highlighting an example of crosstalk between these cap-chaperone pathways. CBC uses the CRM1 pathway to export snRNAs [71], and thus it may also be able to export mRNAs similarly; this will require further studies to ascertain. The eIF4E pathway shares common elements but is distinct from, and parallel to, the CBC-directed RNA export pathway which usually employs NXF1/NXT1 [101].
2.5. Translation
Translation can be regulated at many steps and has been recently reviewed [103,104,105]; here, we focus on the initiation step. eIF4E-dependent translation initiation is considered as the “reference” pathway in that regard (see recent reviews [14,104,106]). In this pathway, once mRNAs are in the cytoplasm, the multi-protein translation initiation complex eIF4F assembles on the m7G cap of the RNA [107]. The eIF4F complex consists of eIF4E, eIF4A (an RNA helicase), and the scaffold protein eIF4G, which binds to both the 43S pre-initiation complex (PIC) and eIF4F, including eIF4E bound to the m7G-capped mRNA [107,108]. After assembly, the 43S PIC scans the 5′UTR until it reaches a start codon, and then recruits the 60S ribosome subunit to form the 80S ribosome, which can then start the elongation step [108]. In terms of translation, eIF4E overexpression often leads to enhanced translation efficiency, i.e., an increase of the number of ribosomes per transcript. Increases in levels of eIF4E increase the translation efficiency of specific RNAs in the cell while not altering others, such as GAPDH or ACTN [109]. In addition to its direct involvement in translation initiation, eIF4E could also impact translation via its ability to drive the expression of DDX19 and Gle1 [16], two export factors that also play independent roles in translation initiation and the termination of selected mRNAs [110,111].
Interestingly, CBC is also implicated in translation via its cap-binding activity [14]. The best described role for CBC here is in the pioneer round of translation which occurs as transcripts exit from the nuclear pore and is usually considered as a quality control mechanism to assess the readiness of RNAs for steady-state translation. The pioneer round of translation can occur using either cap-chaperone, CBC or eIF4E [95,112]. CBC-mediated translation is often associated with nonsense-mediated decay [113] or with a protein surveillance mechanism called the aggresome–autophagy pathway [114]. CBC also plays roles in steady-state translation during stress, including prolonged hypoxia, serum deprivation, or ionizing radiation [115,116,117,118,119]. For example, during hyperosmotic stress in yeast, CBC actively engages polysomes where it mediates translation of around 600 transcripts, about 10% of the transcriptome [120]. These conditions inhibit eIF4E-mediated translation [120]. During HIV infection, CBC plays a role in the translation of unspliced HIV transcripts under conditions where eIF4E-dependent translation is inhibited [11]. CBC also plays a role in the translation of specific types of RNAs, such as histone transcripts [81,121,122].
CBC translation complexes are distinct form those employed by eIF4E. For example, CBC–RNA complexes recruit the CBC-dependent translation initiation factor (CTIF), which acts as an assembly platform protein similar to eIF4G. CBC–CTIF associates with eIF3 through eIF3g to recruit the small ribosomal subunit and to access the start codon [123]. In this situation, eIF4A3 is employed as a helicase component to unwind the RNA structure while scanning [122]. Recent work suggested that CBC-dependent translation could be restricted to perinuclear localization by the association of CTIF-DDX19, suggesting that the nuclear export factor is a co-regulator of CBC-dependent translation and/or that DDX19 also plays a role in the pioneer round of translation and/or during stress.
eIF3d plays a well-established role in ribosome recruitment during translation initiation [124]. However, recently it was shown that eIF3d is a cap-binding protein and is able to drive cap-dependent, eIF4E-independent translation [125,126]. eIF3d-capped RNAs associate with an assembly factor DAP5 rather than eIF4G or CTIF. eIF3d–DAP5 targets the translation of ~20–30% of the translatome [127]. Other factors are also involved in specific forms of cap-dependent translation, such as PARN and LARP1 [14,128]. As more cap-binding proteins continue to be identified, the spectrum of cap-dependent translation pathways will undoubtedly be enlarged. Indeed, these studies suggest that eIF4E does not monopolize the translation pathway for all RNAs or for all conditions.
3. Models for RNA Maturation Based on These Observations and Their Interplay with Translation
As described above, the combinatorial outcomes of RNA maturation define the functional proteome. Here, we describe classic notions for how these events are coordinated and discuss the possibility of parallel RNA maturation networks governed by cap-chaperones.
3.1. Classical Linear RNA Maturation Model
In this model, the CBC binds RNAs during the early stages of transcription, shortly after they have been capped [10,129] (Figure 1). The CBC then chaperones the RNAs through co-transcriptional splicing [7]. This occurs contemporaneously with, or just prior to, the selection of PAS cleavage sites followed by polyadenylation. In this model, the physical changes to the RNA are now complete (capping, intron removal, and PAS site cleavage), except for variation to poly(A) tail length [130]. Next, RNAs continue to be chaperoned by CBC to the nuclear pore complex where they associate with the bulk mRNA export receptors, NXF1/NXT1, and exit into the cytoplasm where they may undergo a pioneer round of translation to check mRNA quality using either CBC or eIF4E. These RNAs are then handed off to eIF4E for steady-state translation, which leads to protein generation. Eventually, RNAs are retired through decay. CBC is considered the sole nuclear cap-chaperone in this model.
3.2. Non-Linear RNA Maturation Model
This model is built upon the classical linear model but incorporates recent observations that capping, splicing, and CPA can sometimes occur post-transcriptionally as well as co-transcriptionally (Figure 2). Indeed, these events do not have to occur in the nucleus. For example, re-capping, CPA, and perhaps splicing can take place in the cytoplasm for specific transcripts (as described above). Furthermore, the same transcript can undergo both co- and post-transcriptional processing [53,79]. For example, intron-containing RNAs can be polyadenylated and then undergo splicing [51]. This indicates that RNA maturation is not always a process that is simply finished once RNAs leave the transcription sites, as predicted from the classical linear model. Further, it shows that the physical changes to the RNA can occur out-of-order i.e., not only in the order of capping-splicing-CPA-export-translation. We refer to this out-of-order processing as non-linear. This model incorporates temporal and geographical aspects of CBC-based RNA maturation that are not present in the classical, linear view of RNA maturation.
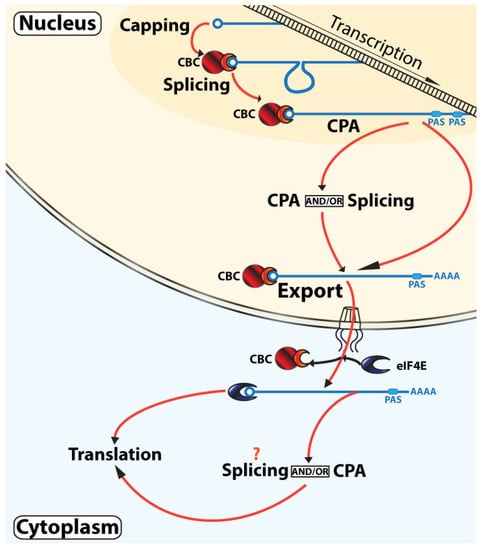
Figure 2.
Nonlinear model of RNA maturation. This model incorporates recent studies that demonstrate that splicing and CPA can take place both co-transcriptionally (darker orange highlighted region) and/or post-transcriptionally in the nucleus (lighter orange highlighted region) or in the cytoplasm (light blue). Once exported to the cytoplasm, the transcriptome can still be modified by capping, CPA, and perhaps splicing. As described in the text, CBC-mediated translation occurs but is not depicted here for simplicity. The m7G cap is depicted as a circle, CBC is in red with NCBP1 as the larger circle and NCBP2 as the semi-circle, and eIF4E is in blue. For simplicity, not all factors involved in these processes are shown.
3.3. Parallel, Non-Linear, Interleaved RNA Maturation Model
There are several observations that support the possibility that chaperoning capped RNAs through maturation steps is not limited to CBC but includes eIF4E, and by inference, other cap-binding proteins. These observations include (1) there are substantial amounts of both eIF4E and CBC found in the nucleus where most maturation occurs [9,22,23,24,131]; (2) both eIF4E and CBC bind to RNAs in the nucleus [9,16,17,18,101,132,133]; (3) both eIF4E and CBC physically associate with splicing [9,21], CPA [9,19], export [9,18,20,101], and translation machinery [14]; and (4) eIF4E and CBC have an impact on multiple steps of the maturation for specific subsets of RNAs [15,16,18,19,20,21,101]. Thus, we propose that multiple, parallel RNA maturation paths coexist, each employing different cap-chaperones (Figure 3). In this new model, CBC does not have a monopoly on flux through these nuclear RNA maturation steps, nor does eIF4E monopolize the cap-chaperone activity in the cytoplasm. RNAs can transit through the maturation steps in parallel, directed by either CBC or eIF4E cap-chaperones. Whether CBC or eIF4E both act in co-transcriptional maturation, and/or post-transcriptional maturation, remains an open question that needs to be addressed experimentally. In this way, eIF4E could provide a parallel path for selected RNAs to undergo maturation. Further, as in the non-linear model above, this model can incorporate aspects of both co- and post-transcriptional RNA maturation, i.e., it can account for out-of-order processing.
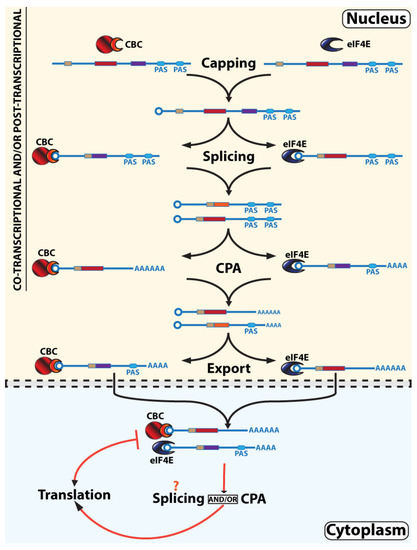
Figure 3.
Parallel, non-linear, interleaved RNA maturation model. We propose a model whereby multiple, parallel RNA maturation paths coexist, each employing different cap-chaperones. RNAs can interleave between eIF4E- or CBC-dependent pathways by a process we refer to as cap-chaperone switching. For simplicity, we focus here on demonstrating these aspects of the model but note that as for Figure 2, these processes can occur co- and/or post-transcriptionally and can be nuclear or cytoplasmic. The evidence for interactions of both CBC and eIF4E with these steps is described in the text. The different PAS sites (blue ellipses) and splicing events (exons in different coloured rectangles) are depicted to highlight our hypothesis that differing RNA compositions lead to different complements of USER codes, and this in turn will influence whether CBC or eIF4E is favored and under certain circumstances drive cap-chaperone switching as depicted in the figure. The m7G cap is depicted as a circle, CBC is in red with NCBP1 as the larger circle and NCBP2 as the semi-circle, and eIF4E is in blue. For simplicity, not all factors involved in these processes are shown.
It is likely that RNAs are not restricted to one cap-chaperone path. In this case, we propose that RNAs can undergo cap-chaperone switching during their maturation and translation in a process we refer to as interleaving (Figure 3). In this model, RNAs directed to CBC-mediated processing could generate better substrates for eIF4E in subsequent steps, or vice versa. In this way, RNAs may not be restricted to CBC- or eIF4E-dependent maturation paths. It is likely that there will be substantial heterogeneity in any given RNA population, whereby conditions would cause a population shift between predominant cap-chaperones rather than an “all-or-nothing” system. In this model, the predominant cap-chaperone can vary for the same RNA population at different steps of maturation, e.g., splicing vs. APA. In support of this, CCND1 and MYC transcripts are found to immunoprecipitate with both NCBP1 and eIF4E in the nucleus [101]. By contrast, GAPDH is restricted to associations with NCBP1 with no binding of this RNA with eIF4E detected in the nucleus [101]. Further, eIF4E overexpression drives more CCND1 and MYC transcripts to exit via eIF4E-dependent export rather than bulk export mediated by CBC, while GAPDH is not an eIF4E-dependent RNA export target [101]. These observations support a model that some RNA species can undergo cap-chaperone switching while others may be restricted to one cap-chaperone type. Ultimately, this transformed information flow will change the proteome functionality by impacting on the availability of the transcript to the translation machinery, translation efficiency, C-terminal definition of proteins, and protein structure based on differences derived from the splice isoform. A clear prediction from this model is that CBC and eIF4E should interact to hand off RNAs. Our lab and others had previously found it difficult to observe physical interactions between eIF4E and NCBP1 [95,101,134,135]. The recent availability of NCBP2 antibodies and more sensitive Western blot reagents prompted us to revisit this question. Indeed, these advances allowed us to now observe immunoprecipitations between NCBP2 and eIF4E (Mars et al., unpublished observations). In all, cap-chaperone switching can provide a molecular basis for RNAs to be exchanged back-and-forth between eIF4E- and CBC-parallel paths at different RNA maturation steps.
Recent studies provide one possible example for how cap-chaperone switching might occur [119]. In this case, the studies were investigating the mechanisms underpinning the transition from CBC-mediated mRNA nuclear export to eIF4E-mediated translation initiation [119]. Here, the double-stranded RNA-binding protein Staufen1 interacts with the capped mRNA and facilitates the interaction between the CBC–importin-α/β complex, which leads to the dissociation of CBC from the cap and allows the recruitment of eIF4E to the RNA [119]. Interestingly, this switching process was inhibited in response to ionizing radiation to maintain CBC–mRNA association. Inhibiting the transition from CBC to eIF4E obviates eIF4E-dependent translation and therefore induces apoptosis under these conditions [119]. The regulation of cap-chaperone switching could be positioned to impact on the adaptation to stress triggers [115,116,117,118,119].
It is important to consider what underlies the preference of one cap-chaperone over another for a given transcript. Presumably, this depends on the availability of cap-chaperones and related co-factors as well as the physical nature of the RNA itself. In this latter case, the splice isoform, CPA isoform, and/or similar modifications will likely impact the recruitment or exclusion of co-factors that would then influence the subsequent processing steps available to that RNA (Figure 3). This specificity is likely informed as in the RNA regulon models by a complement of USER (untranslated sequence element of regulation) codes [136,137,138]. Since the m7G cap is a USER code for both CBC and eIF4E, other elements within the RNA are needed to confer further selectivity. In this model, USER codes in RNAs will engage certain steps such as splicing which can in turn modulate the USER code content to favor specific outcomes in terms of the functional protein (Figure 3).
There is evidence for cap-chaperone switching and the influence of USER codes in this process [16,17]. For example, examination of the physical associations LacZ-4ESE and LacZ RNAs with CBC and eIF4E in the nucleus revealed that both RNAs bound to CBC (but not eIF4E) in the nuclear matrix fraction; however, in the nucleoplasmic fraction, LacZ-4ESE only bound to eIF4E (and not CBC) while LacZ only bound to CBC (and not eIF4E) [101]. Thus, cap-chaperone switching occurred for LacZ-4ESE but not LacZ RNAs. Consistently, LacZ-4ESE was exclusively bound to eIF4E-dependent RNA export factors and was exported solely by that pathway [101]. By contrast, LacZ did not bind to eIF4E or its associated export co-factors in the nucleus and was exported via the CBC-dependent bulk pathway [101]. In the cytoplasm, eIF4E binds both RNAs while CBC does not, characteristic of a subsequent cap-chaperone switching step shortly after arrival in the cytoplasm [101]. In all, the 4ESE USER code was sufficient to select the eIF4E cap-chaperone path in the nucleus leading to eIF4E-dependent export [101]. In this way, the 4ESE confers a cap-chaperone preference which is also observed for some forms of eIF4E-dependent APA [19]. While the USER codes can impart selectivity for cap-chaperones, conversely the maturation steps could control whether those USER codes are present, which then influences the processing steps that are still available to that RNA (Figure 3).
Other examples of cap-chaperone switching have been observed as for the Staufen1 example above. Indeed, RNAs passed from the pioneer round of translation by CBC to steady-state translation by eIF4E in a step referred to as the “hand off” is a well characterized example of cap-chaperone switching [113,117]. Also, in support of cap-chaperone switching, nuclear eIF4E binds ~3500 RNAs but does not act in the RNA export for all of these. Thus, those bound to eIF4E at some point, must switch to CBC (or another cap-chaperone) to be exported. Indeed, it would be of interest to establish the relative fraction of RNAs bound to eIF4E and CBC to further develop this model. While eIF4E has been reported to bind to many transcripts in the nucleus, a direct assessment of the number of transcripts bound to CBC in parallel (from the same nuclear lysates) has not been conducted to our knowledge but would provide important information. While it may, at first glance, seem easy to determine the fractions of RNAs that are present in eIF4E or CBC RIPs in such an experiment, it is difficult to accurately quantitate the relative fractions bound, particularly as the relative avidity of the eIF4E and CBC antibodies would undoubtedly confound this issue. In all, experiments such as this will provide important insights into this process.
It is also important to consider that the status of the cap itself will be key for assigning which cap-chaperone(s) is favored. In this review, we focus on m7G caps. However, a small percentage of coding RNAs have FAD, NAD, UDP-glucuronic acid, UDP-glucosamine, m3G, or even potential protein caps such as the potyviral VPg protein [139,140]. While eIF4E is known to bind the viral VPg protein [139], the cap-chaperones associated with these other cap-types are not yet known. This suggests that for these more exotic caps, there will be specialized cap-chaperones which will engage other parallel paths for their maturation.
Notably, eIF4E has an additional impact on RNA maturation that has not been reported for CBC. eIF4E impacts the production of components of the capping, splicing, CPA, and export machinery [15,16,19,21]. This is driven by its ability to increase the RNA export and/or translation of many of these components. For example, eIF4E enhances the production of bulk RNA export/translation factors (e.g., Gle1, DDX19) through increased RNA export of their corresponding transcripts as well as by reducing levels of the NPC component RanBP2 indirectly [16]. eIF4E similarly stimulates the production of the capping (RNMT, RAM, and RNGTT), splicing, and CPA (CPSF3, CPSF1, and others) machineries [15,19,21]. This provides a basis for wide-scale reprogramming of the RNA machinery to impact “bulk processing” as well as the eIF4E maturation path. In contrast, CBC overexpression or knockdown is not reported to impact the production of the RNA maturation machinery. However, further experimentation to directly test this possibility needs to be carried out. In all, the flow of genetic information can be reprogrammed simultaneously at multiple levels by eIF4E through its cap-chaperone activity as well as its ability to modulate levels of multiple maturation factors simultaneously. Thus, eIF4E does not only co-opt information flow for eIF4E-bound transcripts but can impact other RNAs, that it is not bound to, through impacts on the production of this machinery. In all, the multipronged impacts of both CBC an eIF4E are positioned to influence the functional proteome through their direct roles in RNA maturation, nuclear export, and selective translation.
4. Dysregulation of Cap-Chaperones in Cancer
There is substantial evidence that RNA maturation, export, and translation can be dysregulated in many human malignancies. In cell lines, eIF4E overexpression promotes foci formation, growth in soft agar, invasion, migration, tumor formation, and apoptotic rescue from a variety of stimuli [141,142,143,144]. In xenograft mouse models, elevated eIF4E correlates with increased tumor numbers, invasion, and metastases [145,146,147,148], while pharmacological inhibition or reduction of eIF4E levels suppress tumor growth. In transgenic models of eIF4E overexpression, mice develop a variety of cancers [149,150]; while partial loss of eIF4E does not substantially impact transgenic mice, the complete deletion of eIF4E is lethal [144]. eIF4E-mediated transformation was thought to rely only on the increased translation of oncogenic mRNAs [151]. However, eIF4E’s nuclear functions are also critical for its oncogenic activities as shown by several studies. For example, an eIF4E mutant which completely impairs its translation activity but remains fully active in nuclear export (eIF4E-W73A) transforms cells as readily as wild-type eIF4E [16]. Conversely, the mutation of S53A in eIF4E impairs both RNA export and transformation but does not impede translation [23,24]. Furthermore, the nuclear localization of eIF4E is closely tied to its transformation properties. eIF4E is imported into the nucleus through a direct interaction with importin 8 via its cap-binding site [24]. Importin 8 overexpression led to increased foci formation which was lost in the case of the genetic reduction of eIF4E even though importin 8 levels remained elevated and, in turn, eIF4E lost its oncogenic activity upon the reduction of importin 8 [24]. Indeed, the addition of a nuclear localization signal (NLS) increased eIF4E’s nuclear levels and oncogenicity [24]. Further support of the relevance of the nuclear roles of eIF4E to its oncogenic potential arises from observations that eIF4E’s RNA export and oncogenic activity are inhibited by the nuclear protein PML through its direct interaction with eIF4E and its ability to impair eIF4E’s cap-binding activity in the nucleus [23,152]. Similarly, the proline rich homeodomain protein PRH (also known as Hex), but not PRH without an NLS, inhibits eIF4E-related mRNA export and oncogenic activity through direct nuclear interaction [133]. For many RNAs encoding oncogenes and related factors such as cyclin D1, GPI, hexokinase, and HAS3, eIF4E elevates their production via increased nuclear export, but not via increased translation [143,153], consistent with the relevance of nuclear eIF4E to its oncogenic activity [16,17,18,19,24,143,154,155]. Consistently, the ability of eIF4E to modify the nuclear pore is also central to its oncogenic activity [16,24]. Indeed, while eIF4E elevates NPC components such as RanBP1 and stimulates export factors such as Gle1 and DDX19, it reduces levels of the major component of the cytoplasmic fibrils of the NPC, RanBP2. The overexpression of RanBP2 inhibits both eIF4E-mediated mRNA nuclear export and eIF4E-induced cell transformation. The opposing roles of eIF4E and RanBP2 is highlighted by the observation that eIF4E overexpression reduces RanBP2 protein levels [16]. While the nuclear localization and nuclear export functions of eIF4E are drivers of its oncogenic potential, the roles of APA and AS are not yet known but are exciting areas of future study.
eIF4E levels are increased in many cancers, where its elevation generally correlates with poor prognosis [132,141,142,145,148,154,155,156,157,158]. Some cancers characterized by dysregulated eIF4E include acute myeloid leukemia (AML), multiple myeloma, diffuse large B-cell lymphoma, breast cancer, prostate cancer, head and neck cancer, and others [132,142,145,154,155,156,157,159,160,161,162,163,164,165,166]. Several of eIF4E’s activities are found to be elevated in primary patient specimens, including capping, splicing, RNA export, and translation [15,132,154,155,157,159,160]. The genetic or pharmacological inhibition of eIF4E was shown to reduce oncogenicity and has been correlated with both its nuclear and cytoplasmic functions [154,155,156,157,159,160,161,162,163,164,165,166,167,168]. For example, 4EGI-1 inhibits the formation of the eIF4E–eIF4G complex in breast cancer stem cells [168] and induces apoptosis in malignant pleural mesothelioma [169]; presumably, 4EGI-1 also inhibits the nuclear functions of eIF4E, but this remains to be tested. The direct pharmacological inhibitor of eIF4E, ribavirin [167,170,171], competes for cap-binding and thereby impairs both its RNA export and translation functions [154,158,167]. Ribavirin impairs the binding of eIF4E to importin 8 and prevents eIF4E’s nuclear entry in cell culture and in patients treated with ribavirin [24,155]. In high-eIF4E AML patients prior to treatment, eIF4E is almost entirely nuclear [132,155]. During clinical response, ribavirin blocks eIF4E’s association with importin 8, leading to impaired nuclear entry and reduced eIF4E-dependent mRNA export, which correlates with clinical responses [24,155,160]. eIF4E is mainly cytoplasmic and at relapse eIF4E is once again highly nuclear, supporting that its oncogenic potential derives at least in part from its nuclear localization [155,160]. During disease progression in patients, ribavirin becomes chemically modified and no longer binds eIF4E, allowing eIF4E to associate with importin 8 and to re-enter the nucleus leading to enhanced eIF4E-dependent nuclear RNA export [24,155,160,172]. Here the assembly of nuclear export complexes appears slower than the export and/or release of cargoes, leading to the nuclear enrichment observed for eIF4E at steady-state in untreated patients or during disease progression [24]. To date, ribavirin is the only direct eIF4E inhibitor to be used in patients. Ribavirin treatment leads to objective responses including remissions in two completed and one ongoing clinical trial in AML as well as objective responses in head and neck cancer and castration-resistant prostate cancer clinical trials [155,160,162,173]. There are >15 ongoing trials using ribavirin to target eIF4E in cancer (see ClinicalTrials.gov (accessed on 25 November 2021)). Clearly, inhibition of eIF4E activity is likely to be potent because it impacts nuclear and cytoplasmic activities and also the capacity of eIF4E to produce relevant RNA maturation and export machinery.
eIF4E has also been targeted with antisense oligonucleotides (ASOs) in animal models and patients. Unfortunately, in patients, there were no responses better than short-term stable disease despite the promising results of ASOs in prostate mouse models [174]. This failure appears to be due to the low efficiency of ASO-mediated reduction of eIF4E in humans [174], whereas this was very potent in mice [156]. Increased levels and phosphorylation of eIF4E are associated with aggressiveness in patients [141]. Our lab was first to show that the phosphorylation of eIF4E is linked to its oncogenic potential as well as its RNA export activity [175]. There are ongoing studies using inhibitors of eIF4E phosphorylation (MNK inhibitors) (ClinicalTrials.gov NCT02040558 (accessed on 25 November 2021)), the results of which are not yet known. eIF4E phosphorylation by MNK impacts its nuclear RNA export function [175,176], but its role in translation remains controversial. Moreover, MNK phosphorylates other proteins as well as eIF4E [14,177,178,179], and thus MNK inhibitors are not specific to eIF4E or translation, but more broadly impact RNA metabolism, which could underpin their prospective potency. The eIF4E partner protein and RNA export target, CPSF3, has been shown to be a potential target candidate as well as a prognostic biomarker for a multitude of cancers, e.g., AML and Ewing’s sarcoma [180,181,182]. Simultaneous inhibition of these two factors would constitute a very interesting approach to investigate the impact on countering oncogenesis, especially in the case of myeloid malignancies, amongst others. In summary, the cap-chaperone eIF4E has been largely studied in the context of oncogenesis.
By contrast, CBC is relatively understudied in terms of its role in malignancies. The depletion of NCBP2 or NCBP1 is not lethal in mammalian cells [6]. However, a recent study pointed out a critical role in oncogenic processes such as cell proliferation, invasion, and migration [183]. NCBP1 is elevated in lung adenocarcinoma and a mechanism associating NCPB1 to NCBP3 increased the expression of CUL4B, leading to oncogenic phenotypes in this context [183]. The protein atlas also identified NCBP1 (https://www.proteinatlas.org/ENSG00000136937-NCBP1/pathology (access on 25 November 2021)) as a favorable prognostic marker for prostate cancer and unfavorable for pancreatic cancer, while NCBP2 (https://www.proteinatlas.org/ENSG00000114503-NCBP2/pathology (access on 25 November 2021)) has been identified as an unfavorable prognostic marker for liver, pancreatic, and prostate cancer. Further investigations are needed to more deeply investigate the involvement of the different CBC components in oncogenesis and whether they constitute biomarkers and/or targets.
5. Open Questions and Conclusions
Above, we discussed the information flow that generates the transcripts that will ultimately underpin the functional proteome. There remain several open questions to further probe and build on this model. For instance, does eIF4E or other cap-binding proteins aside from CBC interact with RNAs co-transcriptionally and act in maturation at that site? Does eIF4E act in RNA maturation in the cytoplasm beyond translation? Does CBC act in post-transcriptional CPA or splicing in the nucleus or cytoplasm? What are the USER code complements that provide enrichment for specific cap-chaperones? To date, the 4ESE, in combination with the m7G cap, is known for favoring eIF4E-dependent nuclear RNA export and CPA; however, further decoding is necessary to properly dissect the USER codes that will underpin cap-chaperone choice and the subsequent decision tree of information flow. This information flow can have substantive impacts on cell physiology, even leading to structural and functional changes to the cell by altering the functionality of the proteome.
Our model of non-linear, parallel, interleaved RNA maturation provides a molecular basis to subvert the flow of genetic information and thus the functional proteome (Figure 3). In principle, this model is equally applicable to non-coding RNAs, and their resulting functional transcriptome. The outcome of each maturation step will impact the possibilities for the subsequent steps. In other words, CBC and eIF4E can re-write the RNA message depending on the relative outcomes of each maturation step, thereby partially decoupling the original messages derived from the DNA from the ultimate translated product. Furthermore, it suggests that drugging cap-chaperones could lead to multiple simultaneous impacts on RNA maturation, export, and translation, potentially increasing their potency. Thus, understanding the complexity and organization of the information flow derived from parallel RNA maturation paths to translation will have important therapeutic implications. Further experimental investigation is ultimately required to test this model and will undoubtedly lead to a better understanding of how these processes are integrated to generate the functional proteome.
Author Contributions
J.-C.M., M.G., B.C.-K., K.L.B.B. have contributed to conception, writing, and reviewing of the article. All authors have read and agreed to the published version of the manuscript.
Funding
This worked was supported by NIH R01 CA80728, NIH R01 CA98571, Canadian Institutes of Health Research PJT159785, Leukemia & Lymphoma Society USA Translational Research Program R6513-20, and holds the Canada Research Chair in Molecular Biology of the Cell Nucleus. M.G. is supported by the Cole Foundation and Baumgartner fellowship.
Acknowledgments
We are grateful for discussions with Jack Keene.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Urbanski, L.M.; Leclair, N.; Anczukow, O. Alternative-splicing defects in cancer: Splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdiscip. Rev. RNA 2018, 9, e1476. [Google Scholar] [CrossRef] [PubMed]
- Mohanan, N.K.; Shaji, F.; Koshre, G.R.; Laishram, R.S. Alternative polyadenylation: An enigma of transcript length variation in health and disease. Wiley Interdiscip. Rev. RNA 2021, e1692. [Google Scholar] [CrossRef]
- Stewart, M. Polyadenylation and nuclear export of mRNAs. J. Biol. Chem. 2019, 294, 2977–2987. [Google Scholar] [CrossRef]
- Borden, K.; Culjkovic-Kraljacic, B.; Cowling, V.H. To cap it all off, again: Dynamic capping and recapping of coding and non-coding RNAs to control transcript fate and biological activity. Cell Cycle 2021, 20, 1347–1360. [Google Scholar] [CrossRef] [PubMed]
- Topisirovic, I.; Svitkin, Y.V.; Sonenberg, N.; Shatkin, A.J. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip. Rev. RNA 2011, 2, 277–298. [Google Scholar] [CrossRef]
- Gebhardt, A.; Habjan, M.; Benda, C.; Meiler, A.; Haas, D.A.; Hein, M.Y.; Mann, A.; Mann, M.; Habermann, B.; Pichlmair, A. mRNA export through an additional cap-binding complex consisting of NCBP1 and NCBP3. Nat. Commun. 2015, 6, 8192. [Google Scholar] [CrossRef]
- Lewis, J.D.; Izaurralde, E.; Jarmolowski, A.; McGuigan, C.; Mattaj, I.W. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5’ splice site. Genes Dev. 1996, 10, 1683–1698. [Google Scholar] [CrossRef] [PubMed]
- Pabis, M.; Neufeld, N.; Steiner, M.C.; Bojic, T.; Shav-Tal, Y.; Neugebauer, K.M. The nuclear cap-binding complex interacts with the U4/U6.U5 tri-snRNP and promotes spliceosome assembly in mammalian cells. RNA 2013, 19, 1054–1063. [Google Scholar] [CrossRef]
- Rambout, X.; Maquat, L.E. The nuclear cap-binding complex as choreographer of gene transcription and pre-mRNA processing. Genes Dev. 2020, 34, 1113–1127. [Google Scholar] [CrossRef]
- Visa, N.; Izaurralde, E.; Ferreira, J.; Daneholt, B.; Mattaj, I.W. A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J. Cell Biol. 1996, 133, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Yilmaz, A.; Marsh, K.; Cochrane, A.; Boris-Lawrie, K. Thriving under stress: Selective translation of HIV-1 structural protein mRNA during Vpr-mediated impairment of eIF4E translation activity. PLoS Pathog. 2012, 8, e1002612. [Google Scholar] [CrossRef]
- Toro-Ascuy, D.; Rojas-Araya, B.; Garcia-de-Gracia, F.; Rojas-Fuentes, C.; Pereira-Montecinos, C.; Gaete-Argel, A.; Valiente-Echeverria, F.; Ohlmann, T.; Soto-Rifo, R. A Rev-CBP80-eIF4AI complex drives Gag synthesis from the HIV-1 unspliced mRNA. Nucleic Acids Res. 2018, 46, 11539–11552. [Google Scholar] [CrossRef]
- Ryu, I.; Kim, Y.K. Translation initiation mediated by nuclear cap-binding protein complex. BMB Rep. 2017, 50, 186–193. [Google Scholar] [CrossRef]
- Borden, K.L.B.; Volpon, L. The diversity, plasticity, and adaptability of cap-dependent translation initiation and the associated machinery. RNA Biol. 2020, 17, 1239–1251. [Google Scholar] [CrossRef] [PubMed]
- Culjkovic-Kraljacic, B.; Skrabanek, L.; Revuelta, M.V.; Gasiorek, J.; Cowling, V.H.; Cerchietti, L.; Borden, K.L.B. The eukaryotic translation initiation factor eIF4E elevates steady-state m(7)G capping of coding and noncoding transcripts. Proc. Natl. Acad. Sci. USA 2020, 117, 26773–26783. [Google Scholar] [CrossRef]
- Culjkovic-Kraljacic, B.; Baguet, A.; Volpon, L.; Amri, A.; Borden, K.L.B. The oncogene eIF4E reprograms the nuclear pore complext to promote mRNA export and oncogenic transformation. Cell Rep. 2012, 2, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Culjkovic, B.; Topisirovic, I.; Skrabanek, L.; Ruiz-Gutierrez, M.; Borden, K.L. eIF4E promotes nuclear export of cyclin D1 mRNAs via an element in the 3’UTR. J. Cell Biol. 2005, 169, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Culjkovic, B.; Topisirovic, I.; Skrabanek, L.; Ruiz-Gutierrez, M.; Borden, K.L. eIF4E is a central node of an RNA regulon that governs cellular proliferation. J. Cell Biol. 2006, 175, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.R.; Delaleau, M.; Borden, K.L.B. Nuclear eIF4E Stimulates 3’-End Cleavage of Target RNAs. Cell Rep. 2019, 27, 1397–1408 e1394. [Google Scholar] [CrossRef] [PubMed]
- Volpon, L.; Culjkovic-Kraljacic, B.; Sohn, H.S.; Blanchet-Cohen, A.; Osborne, M.J.; Borden, K.L.B. A biochemical framework for eIF4E-dependent mRNA export and nuclear recycling of the export machinery. RNA 2017, 23, 927–937. [Google Scholar] [CrossRef]
- Ghram, M.; Morris, G.; Culjkovic-Kraljacic, B.; Gendron, P.; Skrabanek, L.; Revuelta, M.V.; Cercheitti, L.; Guzman, M.L.; Borden, K.L.B. The eukaryotic translation initiation factor eIF4E reprogrammes the splicing machinery and drives alternative splicing. biorXiv 2021. [Google Scholar] [CrossRef]
- Lejbkowicz, F.; Goyer, C.; Darveau, A.; Neron, S.; Lemieux, R.; Sonenberg, N. A fraction of the mRNA 5’ cap-binding protein, eukaryotic initiation factor 4E, localizes to the nucleus. Proc. Natl. Acad. Sci. USA 1992, 89, 9612–9616. [Google Scholar] [CrossRef] [PubMed]
- Cohen, N.; Sharma, M.; Kentsis, A.; Perez, J.M.; Strudwick, S.; Borden, K.L. PML RING suppresses oncogenic transformation by reducing the affinity of eIF4E for mRNA. EMBO J. 2001, 20, 4547–4559. [Google Scholar] [CrossRef] [PubMed]
- Volpon, L.; Culjkovic-Kraljacic, B.; Osborne, M.J.; Ramteke, A.; Sun, Q.; Niesman, A.; Chook, Y.M.; Borden, K.L. Importin 8 mediates m7G cap-sensitive nuclear import of the eukaryotic translation initiation factor eIF4E. Proc. Natl. Acad. Sci. USA 2016, 113, 5263–5268. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Park, J.; Hwang, H.J.; Kim, L.; Jeong, K.; Song, H.K.; Rufener, S.C.; Muhlemann, O.; Kim, Y.K. Translation mediated by the nuclear cap-binding complex is confined to the perinuclear region via a CTIF-DDX19B interaction. Nucleic Acids Res. 2021, 49, 8261–8276. [Google Scholar] [CrossRef]
- Proudfoot, N.J.; Furger, A.; Dye, M.J. Integrating mRNA processing with transcription. Cell 2002, 108, 501–512. [Google Scholar] [CrossRef]
- Culjkovic-Kraljacic, B.; Borden, K.L. Aiding and abetting cancer: mRNA export and the nuclear pore. Trends Cell Biol. 2013, 23, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Natalizio, B.J.; Wente, S.R. Postage for the messenger: Designating routes for nuclear mRNA export. Trends Cell Biol. 2013, 23, 365–373. [Google Scholar] [CrossRef]
- Grasso, L.; Suska, O.; Davidson, L.; Gonatopoulos-Pournatzis, T.; Williamson, R.; Wasmus, L.; Wiedlich, S.; Peggie, M.; Stavridis, M.P.; Cowling, V.H. mRNA Cap Methylation in Pluripotency and Differentiation. Cell Rep. 2016, 16, 1352–1365. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Galloway, A.; Cowling, V.H. mRNA cap regulation in mammalian cell function and fate. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 270–279. [Google Scholar] [CrossRef]
- Stevens, A. An exoribonuclease from Saccharomyces cerevisiae: Effect of modifications of 5’ end groups on the hydrolysis of substrates to 5’ mononucleotides. Biochem. Biophys. Res. Commun. 1978, 81, 656–661. [Google Scholar] [CrossRef]
- Ling, S.H.; Qamra, R.; Song, H. Structural and functional insights into eukaryotic mRNA decapping. Wiley Interdiscip. Rev. RNA 2011, 2, 193–208. [Google Scholar] [CrossRef]
- Pillutla, R.C.; Shimamoto, A.; Furuichi, Y.; Shatkin, A.J. Human mRNA capping enzyme (RNGTT) and cap methyltransferase (RNMT) map to 6q16 and 18p11.22-p11.23, respectively. Genomics 1998, 54, 351–353. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Shatkin, A.J. Apoptosis and autophagy induction in mammalian cells by small interfering RNA knockdown of mRNA capping enzymes. Mol. Cell. Biol. 2008, 28, 5829–5836. [Google Scholar] [CrossRef][Green Version]
- Mao, X.; Schwer, B.; Shuman, S. Yeast mRNA cap methyltransferase is a 50-kilodalton protein encoded by an essential gene. Mol. Cell. Biol. 1995, 15, 4167–4174. [Google Scholar] [CrossRef] [PubMed]
- Shafer, B.; Chu, C.; Shatkin, A.J. Human mRNA cap methyltransferase: Alternative nuclear localization signal motifs ensure nuclear localization required for viability. Mol. Cell. Biol. 2005, 25, 2644–2649. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Srinivasan, P.; Piano, F.; Shatkin, A.J. mRNA capping enzyme requirement for Caenorhabditis elegans viability. J. Biol. Chem. 2003, 278, 14168–14173. [Google Scholar] [CrossRef] [PubMed]
- Cowling, V.H. Regulation of mRNA cap methylation. Biochem. J. 2010, 425, 295–302. [Google Scholar] [CrossRef]
- Tsukamoto, T.; Shibagaki, Y.; Murakoshi, T.; Suzuki, M.; Nakamura, A.; Gotoh, H.; Mizumoto, K. Cloning and characterization of two human cDNAs encoding the mRNA capping enzyme. Biochem. Biophys. Res. Commun. 1998, 243, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Yamada-Okabe, T.; Doi, R.; Shimmi, O.; Arisawa, M.; Yamada-Okabe, H. Isolation and characterization of a human cDNA for mRNA 5’-capping enzyme. Nucleic Acids Res. 1998, 26, 1700–1706. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yue, Z.; Maldonado, E.; Pillutla, R.; Cho, H.; Reinberg, D.; Shatkin, A.J. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc. Natl. Acad. Sci. USA 1997, 94, 12898–12903. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Das, K.; Tyminski, J.R.; Bauman, J.D.; Guan, R.; Qiu, W.; Montelione, G.T.; Arnold, E.; Shatkin, A.J. Structure of the guanylyltransferase domain of human mRNA capping enzyme. Proc. Natl. Acad. Sci. USA 2011, 108, 10104–10108. [Google Scholar] [CrossRef] [PubMed]
- Gonatopoulos-Pournatzis, T.; Dunn, S.; Bounds, R.; Cowling, V.H. RAM/Fam103a1 is required for mRNA cap methylation. Mol. Cell 2011, 44, 585–596. [Google Scholar] [CrossRef]
- Fabrega, C.; Hausmann, S.; Shen, V.; Shuman, S.; Lima, C.D. Structure and mechanism of mRNA cap (guanine-N7) methyltransferase. Mol. Cell 2004, 13, 77–89. [Google Scholar] [CrossRef]
- Trotman, J.B.; Giltmier, A.J.; Mukherjee, C.; Schoenberg, D.R. RNA guanine-7 methyltransferase catalyzes the methylation of cytoplasmically recapped RNAs. Nucleic Acids Res. 2017, 45, 10726–10739. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, Y.; Kedersha, N.L.; Schoenberg, D.R. Identification of a cytoplasmic complex that adds a cap onto 5’-monophosphate RNA. Mol. Cell. Biol. 2009, 29, 2155–2167. [Google Scholar] [CrossRef] [PubMed]
- Schoenberg, D.R.; Maquat, L.E. Re-capping the message. Trends Biochem. Sci. 2009, 34, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Aregger, M.; Kaskar, A.; Varshney, D.; Fernandez-Sanchez, M.E.; Inesta-Vaquera, F.A.; Weidlich, S.; Cowling, V.H. CDK1-Cyclin B1 Activates RNMT, Coordinating mRNA Cap Methylation with G1 Phase Transcription. Mol. Cell 2016, 61, 734–746. [Google Scholar] [CrossRef] [PubMed]
- Grudzien-Nogalska, E.; Kiledjian, M. New insights into decapping enzymes and selective mRNA decay. Wiley Interdiscip. Rev. RNA 2017, 8, e1379. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, C.; Patil, D.P.; Kennedy, B.A.; Bakthavachalu, B.; Bundschuh, R.; Schoenberg, D.R. Identification of cytoplasmic capping targets reveals a role for cap homeostasis in translation and mRNA stability. Cell Rep. 2012, 2, 674–684. [Google Scholar] [CrossRef] [PubMed]
- Boutz, P.L.; Bhutkar, A.; Sharp, P.A. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 2015, 29, 63–80. [Google Scholar] [CrossRef]
- Girard, C.; Will, C.L.; Peng, J.; Makarov, E.M.; Kastner, B.; Lemm, I.; Urlaub, H.; Hartmuth, K.; Luhrmann, R. Post-transcriptional spliceosomes are retained in nuclear speckles until splicing completion. Nat. Commun. 2012, 3, 994. [Google Scholar] [CrossRef]
- Prasanth, K.V.; Prasanth, S.G.; Xuan, Z.; Hearn, S.; Freier, S.M.; Bennett, C.F.; Zhang, M.Q.; Spector, D.L. Regulating gene expression through RNA nuclear retention. Cell 2005, 123, 249–263. [Google Scholar] [CrossRef] [PubMed]
- Gonatopoulos-Pournatzis, T.; Cowling, V.H. Cap-binding complex (CBC). Biochem. J. 2014, 457, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Riechmann, J.L.; Meyerowitz, E.M. Transcriptome-wide analysis of uncapped mRNAs in Arabidopsis reveals regulation of mRNA degradation. Plant. Cell 2008, 20, 2571–2585. [Google Scholar] [CrossRef] [PubMed]
- Cowling, V.H. Enhanced mRNA cap methylation increases cyclin D1 expression and promotes cell transformation. Oncogene 2010, 29, 930–936. [Google Scholar] [CrossRef]
- Osborne, M.; Volpon, L.; Memarpooryazdi, M.; Thambipillai, A.; Czarnota, S.; Culjkovic-Kraljacic, B.; Tahran, C.; Oeffinger, M.; Cowling, V.H.; Borden, K.L.B. Identification and characterization of the interaction between the methyl-7-guanosine cap maturation enzyme RNMT and the cap-binding protein eIF4E. 2021; under review. [Google Scholar]
- Wahl, M.C.; Will, C.L.; Luhrmann, R. The spliceosome: Design principles of a dynamic RNP machine. Cell 2009, 136, 701–718. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef]
- Effenberger, K.A.; Urabe, V.K.; Jurica, M.S. Modulating splicing with small molecular inhibitors of the spliceosome. Wiley Interdiscip. Rev. RNA 2017, 8, e1381. [Google Scholar] [CrossRef]
- Vargas, D.Y.; Shah, K.; Batish, M.; Levandoski, M.; Sinha, S.; Marras, S.A.; Schedl, P.; Tyagi, S. Single-molecule imaging of transcriptionally coupled and uncoupled splicing. Cell 2011, 147, 1054–1065. [Google Scholar] [CrossRef]
- Pandya-Jones, A.; Black, D.L. Co-transcriptional splicing of constitutive and alternative exons. RNA 2009, 15, 1896–1908. [Google Scholar] [CrossRef] [PubMed]
- Turunen, J.J.; Niemela, E.H.; Verma, B.; Frilander, M.J. The significant other: Splicing by the minor spliceosome. Wiley Interdiscip. Rev. RNA 2013, 4, 61–76. [Google Scholar] [CrossRef]
- Bell, T.J.; Miyashiro, K.Y.; Sul, J.Y.; Buckley, P.T.; Lee, M.T.; McCullough, R.; Jochems, J.; Kim, J.; Cantor, C.R.; Parsons, T.D.; et al. Intron retention facilitates splice variant diversity in calcium-activated big potassium channel populations. Proc. Natl. Acad. Sci. USA 2010, 107, 21152–21157. [Google Scholar] [CrossRef]
- Gornemann, J.; Kotovic, K.M.; Hujer, K.; Neugebauer, K.M. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol. Cell 2005, 19, 53–63. [Google Scholar] [CrossRef]
- O’Mullane, L.; Eperon, I.C. The pre-mRNA 5’ cap determines whether U6 small nuclear RNA succeeds U1 small nuclear ribonucleoprotein particle at 5’ splice sites. Mol. Cell. Biol. 1998, 18, 7510–7520. [Google Scholar] [CrossRef] [PubMed]
- Lenasi, T.; Peterlin, B.M.; Barboric, M. Cap-binding protein complex links pre-mRNA capping to transcription elongation and alternative splicing through positive transcription elongation factor b (P-TEFb). J. Biol. Chem. 2011, 286, 22758–22768. [Google Scholar] [CrossRef]
- Anczukow, O.; Akerman, M.; Clery, A.; Wu, J.; Shen, C.; Shirole, N.H.; Raimer, A.; Sun, S.; Jensen, M.A.; Hua, Y.; et al. SRSF1-Regulated Alternative Splicing in Breast Cancer. Mol. Cell 2015, 60, 105–117. [Google Scholar] [CrossRef]
- Huang, H.; Zhang, J.; Harvey, S.E.; Hu, X.; Cheng, C. RNA G-quadruplex secondary structure promotes alternative splicing via the RNA-binding protein hnRNPF. Genes Dev. 2017, 31, 2296–2309. [Google Scholar] [CrossRef] [PubMed]
- Izaurralde, E.; Lewis, J.; Gamberi, C.; Jarmolowski, A.; McGuigan, C.; Mattaj, I.W. A cap-binding protein complex mediating U snRNA export. Nature 1995, 376, 709–712. [Google Scholar] [CrossRef] [PubMed]
- Ohno, M.; Segref, A.; Bachi, A.; Wilm, M.; Mattaj, I.W. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 2000, 101, 187–198. [Google Scholar] [CrossRef]
- Segref, A.; Mattaj, I.W.; Ohno, M. The evolutionarily conserved region of the U snRNA export mediator PHAX is a novel RNA-binding domain that is essential for U snRNA export. RNA 2001, 7, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Kuzuoglu-Ozturk, D.; Hu, Z.; Rama, M.; Devericks, E.; Weiss, J.; Chiang, G.G.; Worland, S.T.; Brenner, S.E.; Goodarzi, H.; Gilbert, L.A.; et al. Revealing molecular pathways for cancer cell fitness through a genetic screen of the cancer translatome. Cell Rep. 2021, 35, 109321. [Google Scholar] [CrossRef]
- Proudfoot, N.J. Ending the message: Poly(A) signals then and now. Genes Dev. 2011, 25, 1770–1782. [Google Scholar] [CrossRef]
- Curinha, A.; Oliveira Braz, S.; Pereira-Castro, I.; Cruz, A.; Moreira, A. Implications of polyadenylation in health and disease. Nucleus 2014, 5, 508–519. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.R.; Martin, G.; Keller, W.; Zavolan, M. Means to an end: Mechanisms of alternative polyadenylation of messenger RNA precursors. Wiley Interdiscip. Rev. RNA 2014, 5, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Manley, J.L. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 2017, 18, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Malka, Y.; Steiman-Shimony, A.; Rosenthal, E.; Argaman, L.; Cohen-Daniel, L.; Arbib, E.; Margalit, H.; Kaplan, T.; Berger, M. Post-transcriptional 3-UTR cleavage of mRNA transcripts generates thousands of stable uncapped autonomous RNA fragments. Nat. Commun. 2017, 8, 2029. [Google Scholar] [CrossRef] [PubMed]
- Bae, B.; Miura, P. Emerging Roles for 3’ UTRs in Neurons. Int. J. Mol. Sci. 2020, 21, 3413. [Google Scholar] [CrossRef] [PubMed]
- Andreassi, C.; Luisier, R.; Crerar, H.; Darsinou, M.; Blokzijl-Franke, S.; Lenn, T.; Luscombe, N.M.; Cuda, G.; Gaspari, M.; Saiardi, A.; et al. Cytoplasmic cleavage of IMPA1 3’ UTR is necessary for maintaining axon integrity. Cell Rep. 2021, 34, 108778. [Google Scholar] [CrossRef]
- Narita, T.; Yung, T.M.; Yamamoto, J.; Tsuboi, Y.; Tanabe, H.; Tanaka, K.; Yamaguchi, Y.; Handa, H. NELF interacts with CBC and participates in 3’ end processing of replication-dependent histone mRNAs. Mol. Cell 2007, 26, 349–365. [Google Scholar] [CrossRef]
- Gruber, J.J.; Olejniczak, S.H.; Yong, J.; La Rocca, G.; Dreyfuss, G.; Thompson, C.B. Ars2 promotes proper replication-dependent histone mRNA 3’ end formation. Mol. Cell 2012, 45, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Hallais, M.; Pontvianne, F.; Andersen, P.R.; Clerici, M.; Lener, D.; Benbahouche Nel, H.; Gostan, T.; Vandermoere, F.; Robert, M.C.; Cusack, S.; et al. CBC-ARS2 stimulates 3’-end maturation of multiple RNA families and favors cap-proximal processing. Nat. Struct. Mol. Biol. 2013, 20, 1358–1366. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, C.; Christie, J.; Pienaar, M.; Gambling, J.; Nickerson, P.E.; Alford, S.C.; Chow, R.L.; Howard, P.L. Mutagenesis of ARS2 Domains to Assess Possible Roles in Cell Cycle Progression and MicroRNA and Replication-Dependent Histone mRNA Biogenesis. Mol. Cell. Biol. 2015, 35, 3753–3767. [Google Scholar] [CrossRef] [PubMed]
- So, B.R.; Di, C.; Cai, Z.; Venters, C.C.; Guo, J.; Oh, J.M.; Arai, C.; Dreyfuss, G. A Complex of U1 snRNP with Cleavage and Polyadenylation Factors Controls Telescripting, Regulating mRNA Transcription in Human Cells. Mol. Cell 2019, 76, 590–599.e4. [Google Scholar] [CrossRef] [PubMed]
- Jarmolowski, A.; Boelens, W.C.; Izaurralde, E.; Mattaj, I.W. Nuclear export of different classes of RNA is mediated by specific factors. J. Cell Biol. 1994, 124, 627–635. [Google Scholar] [CrossRef] [PubMed]
- Fornerod, M.; Ohno, M.; Yoshida, M.; Mattaj, I.W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 1997, 90, 1051–1060. [Google Scholar] [CrossRef]
- Reed, R.; Hurt, E. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell 2002, 108, 523–531. [Google Scholar] [CrossRef]
- Katahira, J. mRNA export and the TREX complex. Biochim. Biophys. Acta 2012, 1819, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Fontoura, B.M.; Dales, S.; Blobel, G.; Zhong, H. The nucleoporin Nup98 associates with the intranuclear filamentous protein network of TPR. Proc. Natl. Acad. Sci. USA 2001, 98, 3208–3213. [Google Scholar] [CrossRef]
- Enninga, J.; Levy, D.E.; Blobel, G.; Fontoura, B.M. Role of nucleoporin induction in releasing an mRNA nuclear export block. Science 2002, 295, 1523–1525. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Seo, H.S.; Blobel, G.; Hoelz, A. Structural and functional analysis of the interaction between the nucleoporin Nup98 and the mRNA export factor Rae1. Proc. Natl. Acad. Sci. USA 2010, 107, 10406–10411. [Google Scholar] [CrossRef]
- Dufu, K.; Livingstone, M.J.; Seebacher, J.; Gygi, S.P.; Wilson, S.A.; Reed, R. ATP is required for interactions between UAP56 and two conserved mRNA export proteins, Aly and CIP29, to assemble the TREX complex. Genes Dev. 2010, 24, 2043–2053. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Zhang, H.; Wu, X.; He, Z.; Wang, L.; Yin, S.; Tian, B.; Li, G.; Cheng, H. ALYREF mainly binds to the 5’ and the 3’ regions of the mRNA in vivo. Nucleic Acids Res. 2017, 45, 9640–9653. [Google Scholar] [CrossRef] [PubMed]
- Rufener, S.C.; Muhlemann, O. eIF4E-bound mRNPs are substrates for nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol. 2013, 20, 710–717. [Google Scholar] [CrossRef]
- Calero, G.; Wilson, K.F.; Ly, T.; Rios-Steiner, J.L.; Clardy, J.C.; Cerione, R.A. Structural basis of m7GpppG binding to the nuclear cap-binding protein complex. Nat. Struct. Biol. 2002, 9, 912–917. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Dufu, K.; Lee, C.S.; Hsu, J.L.; Dias, A.; Reed, R. Human mRNA export machinery recruited to the 5’ end of mRNA. Cell 2006, 127, 1389–1400. [Google Scholar] [CrossRef]
- Dou, Y.; Barbosa, I.; Jiang, H.; Iasillo, C.; Molloy, K.R.; Schulze, W.M.; Cusack, S.; Schmid, M.; Le Hir, H.; LaCava, J.; et al. NCBP3 positively impacts mRNA biogenesis. Nucleic Acids Res. 2020, 48, 10413–10427. [Google Scholar] [CrossRef] [PubMed]
- Rambout, X.; Maquat, L.E. NCBP3: A Multifaceted Adaptive Regulator of Gene Expression. Trends Biochem. Sci. 2021, 46, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Schulze, W.M.; Cusack, S. Structural basis for mutually exclusive co-transcriptional nuclear cap-binding complexes with either NELF-E or ARS2. Nat. Commun. 2017, 8, 1302. [Google Scholar] [CrossRef] [PubMed]
- Topisirovic, I.; Siddiqui, N.; Lapointe, V.L.; Trost, M.; Thibault, P.; Bangeranye, C.; Pinol-Roma, S.; Borden, K.L. Molecular dissection of the eukaryotic initiation factor 4E (eIF4E) export-competent RNP. EMBO J. 2009, 28, 1087–1098. [Google Scholar] [CrossRef]
- Kudo, N.; Matsumori, N.; Taoka, H.; Fujiwara, D.; Schreiner, E.P.; Wolff, B.; Yoshida, M.; Horinouchi, S. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 1999, 96, 9112–9117. [Google Scholar] [CrossRef] [PubMed]
- Roux, P.P.; Topisirovic, I. Signaling Pathways Involved in the Regulation of mRNA Translation. Mol. Cell. Biol. 2018, 38, e00070-18. [Google Scholar] [CrossRef] [PubMed]
- Dever, T.E.; Dinman, J.D.; Green, R. Translation Elongation and Recoding in Eukaryotes. Cold Spring Harb. Perspect. Biol 2018, 10. [Google Scholar] [CrossRef]
- Borden, K.L. The eukaryotic translation initiation factor eIF4E wears a “cap” for many occasions. Translation 2016, 4, e1220899. [Google Scholar] [CrossRef]
- Smith, R.C.L.; Kanellos, G.; Vlahov, N.; Alexandrou, C.; Willis, A.E.; Knight, J.R.P.; Sansom, O.J. Translation initiation in cancer at a glance. J. Cell Sci. 2021, 134. [Google Scholar] [CrossRef] [PubMed]
- Kapp, L.D.; Lorsch, J.R. The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 2004, 73, 657–704. [Google Scholar] [CrossRef] [PubMed]
- Hinnebusch, A.G. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 2014, 83, 779–812. [Google Scholar] [CrossRef] [PubMed]
- Batool, A.; Aashaq, S.; Andrabi, K.I. Eukaryotic initiation factor 4E (eIF4E): A recap of the cap-binding protein. J. Cell Biochem. 2019, 120, 14201–14212. [Google Scholar] [CrossRef] [PubMed]
- Alcazar-Roman, A.R.; Wente, S.R. Inositol polyphosphates: A new frontier for regulating gene expression. Chromosoma 2008, 117, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Gross, T.; Siepmann, A.; Sturm, D.; Windgassen, M.; Scarcelli, J.J.; Seedorf, M.; Cole, C.N.; Krebber, H. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science 2007, 315, 646–649. [Google Scholar] [CrossRef] [PubMed]
- Ishigaki, Y.; Li, X.; Serin, G.; Maquat, L.E. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 2001, 106, 607–617. [Google Scholar] [CrossRef]
- Maquat, L.E.; Tarn, W.Y.; Isken, O. The pioneer round of translation: Features and functions. Cell 2010, 142, 368–374. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Park, J.; Kim, Y.K. Crosstalk between translation and the aggresome-autophagy pathway. Autophagy 2018, 14, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- Koritzinsky, M.; Magagnin, M.G.; van den Beucken, T.; Seigneuric, R.; Savelkouls, K.; Dostie, J.; Pyronnet, S.; Kaufman, R.J.; Weppler, S.A.; Voncken, J.W.; et al. Gene expression during acute and prolonged hypoxia is regulated by distinct mechanisms of translational control. EMBO J. 2006, 25, 1114–1125. [Google Scholar] [CrossRef] [PubMed]
- Uniacke, J.; Perera, J.K.; Lachance, G.; Francisco, C.B.; Lee, S. Cancer cells exploit eIF4E2-directed synthesis of hypoxia response proteins to drive tumor progression. Cancer Res. 2014, 74, 1379–1389. [Google Scholar] [CrossRef] [PubMed]
- Oh, N.; Kim, K.M.; Cho, H.; Choe, J.; Kim, Y.K. Pioneer round of translation occurs during serum starvation. Biochem. Biophys. Res. Commun. 2007, 362, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Weiss, B.; Allen, G.E.; Kloehn, J.; Abid, K.; Jaquier-Gubler, P.; Curran, J.A. eIF4E3 forms an active eIF4F complex during stresses (eIF4FS) targeting mTOR and re-programs the translatome. Nucleic Acids Res. 2021, 49, 5159–5176. [Google Scholar] [CrossRef] [PubMed]
- Jeong, K.; Ryu, I.; Park, J.; Hwang, H.J.; Ha, H.; Park, Y.; Oh, S.T.; Kim, Y.K. Staufen1 and UPF1 exert opposite actions on the replacement of the nuclear cap-binding complex by eIF4E at the 5’ end of mRNAs. Nucleic Acids Res. 2019, 47, 9313–9328. [Google Scholar] [CrossRef] [PubMed]
- Garre, E.; Romero-Santacreu, L.; De Clercq, N.; Blasco-Angulo, N.; Sunnerhagen, P.; Alepuz, P. Yeast mRNA cap-binding protein Cbc1/Sto1 is necessary for the rapid reprogramming of translation after hyperosmotic shock. Mol. Biol. Cell 2012, 23, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Choe, J.; Kim, K.M.; Park, S.; Lee, Y.K.; Song, O.K.; Kim, M.K.; Lee, B.G.; Song, H.K.; Kim, Y.K. Rapid degradation of replication-dependent histone mRNAs largely occurs on mRNAs bound by nuclear cap-binding proteins 80 and 20. Nucleic Acids Res. 2013, 41, 1307–1318. [Google Scholar] [CrossRef] [PubMed]
- Choe, J.; Ryu, I.; Park, O.H.; Park, J.; Cho, H.; Yoo, J.S.; Chi, S.W.; Kim, M.K.; Song, H.K.; Kim, Y.K. eIF4AIII enhances translation of nuclear cap-binding complex-bound mRNAs by promoting disruption of secondary structures in 5’UTR. Proc. Natl. Acad. Sci. USA 2014, 111, E4577–E4586. [Google Scholar] [CrossRef]
- Kim, K.M.; Cho, H.; Choi, K.; Kim, J.; Kim, B.W.; Ko, Y.G.; Jang, S.K.; Kim, Y.K. A new MIF4G domain-containing protein, CTIF, directs nuclear cap-binding protein CBP80/20-dependent translation. Genes Dev. 2009, 23, 2033–2045. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.; Su, D.; Scheliga, J.S.; Pluskal, T.; Boronat, S.; Motamedchaboki, K.; Campos, A.R.; Qi, F.; Hidalgo, E.; Yanagida, M.; et al. A Transcript-Specific eIF3 Complex Mediates Global Translational Control of Energy Metabolism. Cell Rep. 2016, 16, 1891–1902. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.S.; Kranzusch, P.J.; Doudna, J.A.; Cate, J.H. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 2016, 536, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Guo, Y. Knockdown of eIF3D inhibits breast cancer cell proliferation and invasion through suppressing the Wnt/beta-catenin signaling pathway. Int. J. Clin. Exp. Pathol. 2015, 8, 10420–10427. [Google Scholar]
- de la Parra, C.; Ernlund, A.; Alard, A.; Ruggles, K.; Ueberheide, B.; Schneider, R.J. A widespread alternate form of cap-dependent mRNA translation initiation. Nat. Commun. 2018, 9, 3068. [Google Scholar] [CrossRef] [PubMed]
- Lahr, R.M.; Fonseca, B.D.; Ciotti, G.E.; Al-Ashtal, H.A.; Jia, J.J.; Niklaus, M.R.; Blagden, S.P.; Alain, T.; Berman, A.J. La-related protein 1 (LARP1) binds the mRNA cap, blocking eIF4F assembly on TOP mRNAs. Elife 2017, 6, e24146. [Google Scholar] [CrossRef]
- Izaurralde, E.; Lewis, J.; McGuigan, C.; Jankowska, M.; Darzynkiewicz, E.; Mattaj, I.W. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell 1994, 78, 657–668. [Google Scholar] [CrossRef]
- Lewis, J.D.; Izaurralde, E. The role of the cap structure in RNA processing and nuclear export. Eur. J. Biochem. 1997, 247, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Dostie, J.; Lejbkowicz, F.; Sonenberg, N. Nuclear eukaryotic initiation factor 4E (eIF4E) colocalizes with splicing factors in speckles. J. Cell Biol. 2000, 148, 239–247. [Google Scholar] [CrossRef]
- Topisirovic, I.; Guzman, M.L.; McConnell, M.J.; Licht, J.D.; Culjkovic, B.; Neering, S.J.; Jordan, C.T.; Borden, K.L. Aberrant eukaryotic translation initiation factor 4E-dependent mRNA transport impedes hematopoietic differentiation and contributes to leukemogenesis. Mol. Cell. Biol. 2003, 23, 8992–9002. [Google Scholar] [CrossRef] [PubMed]
- Topisirovic, I.; Culjkovic, B.; Cohen, N.; Perez, J.M.; Skrabanek, L.; Borden, K.L. The proline-rich homeodomain protein, PRH, is a tissue-specific inhibitor of eIF4E-dependent cyclin D1 mRNA transport and growth. EMBO J. 2003, 22, 689–703. [Google Scholar] [CrossRef]
- Lejeune, F.; Ishigaki, Y.; Li, X.; Maquat, L.E. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: Dynamics of mRNP remodeling. EMBO J. 2002, 21, 3536–3545. [Google Scholar] [CrossRef]
- Matsuda, D.; Hosoda, N.; Kim, Y.K.; Maquat, L.E. Failsafe nonsense-mediated mRNA decay does not detectably target eIF4E-bound mRNA. Nat. Struct. Mol. Biol. 2007, 14, 974–979. [Google Scholar] [CrossRef]
- Bisogno, L.S.; Keene, J.D. RNA regulons in cancer and inflammation. Curr. Opin. Genet. Dev. 2018, 48, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Keene, J.D. RNA regulons: Coordination of post-transcriptional events. Nat. Rev. Genet. 2007, 8, 533–543. [Google Scholar] [CrossRef]
- Keene, J.D.; Lager, P.J. Post-transcriptional operons and regulons co-ordinating gene expression. Chromosome Res. 2005, 13, 327–337. [Google Scholar] [CrossRef]
- Coutinho de Oliveira, L.; Volpon, L.; Rahardjo, A.K.; Osborne, M.J.; Culjkovic-Kraljacic, B.; Trahan, C.; Oeffinger, M.; Kwok, B.H.; Borden, K.L.B. Structural studies of the eIF4E-VPg complex reveal a direct competition for capped RNA: Implications for translation. Proc. Natl. Acad. Sci. USA 2019, 116, 24056–24065. [Google Scholar] [CrossRef]
- Wang, J.; Alvin Chew, B.L.; Lai, Y.; Dong, H.; Xu, L.; Balamkundu, S.; Cai, W.M.; Cui, L.; Liu, C.F.; Fu, X.Y.; et al. Quantifying the RNA cap epitranscriptome reveals novel caps in cellular and viral RNA. Nucleic Acids Res. 2019, 47, e130. [Google Scholar] [CrossRef]
- Carroll, M.; Borden, K.L. The oncogene eIF4E: Using biochemical insights to target cancer. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 2013, 33, 227–238. [Google Scholar] [CrossRef]
- Culjkovic, B.; Borden, K.L. Understanding and Targeting the Eukaryotic Translation Initiation Factor eIF4E in Head and Neck Cancer. J. Oncol. 2009, 2009, 981679. [Google Scholar] [CrossRef] [PubMed]
- Zahreddine, H.A.; Culjkovic-Kraljacic, B.; Emond, A.; Pettersson, F.; Midura, R.; Lauer, M.; Del Rincon, S.; Cali, V.; Assouline, S.; Miller, W.H.; et al. The eukaryotic translation initiation factor eIF4E harnesses hyaluronan production to drive its malignant activity. Elife 2017, 6, e29830. [Google Scholar] [CrossRef]
- Truitt, M.L.; Conn, C.S.; Shi, Z.; Pang, X.; Tokuyasu, T.; Coady, A.M.; Seo, Y.; Barna, M.; Ruggero, D. Differential Requirements for eIF4E Dose in Normal Development and Cancer. Cell 2015, 162, 59–71. [Google Scholar] [CrossRef] [PubMed]
- De Benedetti, A.; Harris, A.L. eIF4E expression in tumors: Its possible role in progression and malignacies. Int. J. Biochem. Cell Biol. 1999, 31, 59–72. [Google Scholar] [CrossRef]
- Graff, J.R.; Boghaert, E.R.; De Benedetti, A.; Tudor, D.L.; Zimmer, C.C.; Chan, S.K.; Zimmer, S.G. Reduction of translation initiation factor 4E decreases the malignancy of ras-transformed cloned rat embryo fibroblasts. Int. J. Cancer 1995, 60, 255–263. [Google Scholar] [CrossRef]
- Liu, T.; Li, R.; Zhao, H.; Deng, J.; Long, Y.; Shuai, M.T.; Li, Q.; Gu, H.; Chen, Y.Q.; Leng, A.M. eIF4E promotes tumorigenesis and modulates chemosensitivity to cisplatin in esophageal squamous cell carcinoma. Oncotarget 2016, 7, 66851–66864. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Fu, J.; Lu, C.; Mapara, M.Y.; Raza, S.; Hengst, U.; Lentzsch, S. Elevated Translation Initiation Factor eIF4E Is an Attractive Therapeutic Target in Multiple Myeloma. Mol. Cancer Ther. 2016, 15, 711–719. [Google Scholar] [CrossRef]
- Ruggero, D.; Montanaro, L.; Ma, L.; Xu, W.; Londei, P.; Cordon-Cardo, C.; Pandolfi, P.P. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat. Med. 2004, 10, 484–486. [Google Scholar] [CrossRef] [PubMed]
- Wendel, H.G.; Silva, R.L.; Malina, A.; Mills, J.R.; Zhu, H.; Ueda, T.; Watanabe-Fukunaga, R.; Fukunaga, R.; Teruya-Feldstein, J.; Pelletier, J.; et al. Dissecting eIF4E action in tumorigenesis. Genes Dev. 2007, 21, 3232–3237. [Google Scholar] [CrossRef]
- Sonenberg, N.; Gingras, A.C. The mRNA 5’ cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell Biol. 1998, 10, 268–275. [Google Scholar] [CrossRef]
- Kentsis, A.; Dwyer, E.C.; Perez, J.M.; Sharma, M.; Chen, A.; Pan, Z.Q.; Borden, K.L. The RING domains of the promyelocytic leukemia protein PML and the arenaviral protein Z repress translation by directly inhibiting translation initiation factor eIF4E. J. Mol. Biol. 2001, 312, 609–623. [Google Scholar] [CrossRef]
- Rousseau, D.; Kaspar, R.; Rosenwald, I.; Gehrke, L.; Sonenberg, N. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc. Natl. Acad. Sci. USA 1996, 93, 1065–1070. [Google Scholar]
- Culjkovic-Kraljacic, B.; Fernando, T.M.; Marullo, R.; Calvo-Vidal, N.; Verma, A.; Yang, S.; Tabbo, F.; Gaudiano, M.; Zahreddine, H.; Goldstein, R.L.; et al. Combinatorial targeting of nuclear export and translation of RNA inhibits aggressive B-cell lymphomas. Blood 2016, 127, 858–868. [Google Scholar] [CrossRef] [PubMed]
- Assouline, S.; Culjkovic, B.; Cocolakis, E.; Rousseau, C.; Beslu, N.; Amri, A.; Caplan, S.; Leber, B.; Roy, D.C.; Miller, W.H., Jr.; et al. Molecular targeting of the oncogene eIF4E in acute myeloid leukemia (AML): A proof-of-principle clinical trial with ribavirin. Blood 2009, 114, 257–260. [Google Scholar] [CrossRef] [PubMed]
- Graff, J.R.; Konicek, B.W.; Vincent, T.M.; Lynch, R.L.; Monteith, D.; Weir, S.N.; Schwier, P.; Capen, A.; Goode, R.L.; Dowless, M.S.; et al. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J. Clin. Investig. 2007, 117, 2638–2648. [Google Scholar] [CrossRef]
- Pettersson, F.; Yau, C.; Dobocan, M.C.; Culjkovic-Kraljacic, B.; Retrouvey, H.; Puckett, R.; Flores, L.M.; Krop, I.E.; Rousseau, C.; Cocolakis, E.; et al. Ribavirin treatment effects on breast cancers overexpressing eIF4E, a biomarker with prognostic specificity for luminal B-type breast cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2011, 17, 2874–2884. [Google Scholar] [CrossRef]
- Urtishak, K.A.; Wang, L.S.; Culjkovic-Kraljacic, B.; Davenport, J.W.; Porazzi, P.; Vincent, T.L.; Teachey, D.T.; Tasian, S.K.; Moore, J.S.; Seif, A.E.; et al. Targeting EIF4E signaling with ribavirin in infant acute lymphoblastic leukemia. Oncogene 2019, 38, 2241–2262. [Google Scholar] [CrossRef]
- Kraljacic, B.C.; Arguello, M.; Amri, A.; Cormack, G.; Borden, K. Inhibition of eIF4E with ribavirin cooperates with common chemotherapies in primary acute myeloid leukemia specimens. Leukemia 2011, 25, 1197–1200. [Google Scholar] [CrossRef] [PubMed]
- Assouline, S.; Culjkovic-Kraljacic, B.; Bergeron, J.; Caplan, S.; Cocolakis, E.; Lambert, C.; Lau, C.J.; Zahreddine, H.A.; Miller, W.H., Jr.; Borden, K.L. A phase I trial of ribavirin and low-dose cytarabine for the treatment of relapsed and refractory acute myeloid leukemia with elevated eIF4E. Haematologica 2015, 100, e7–e9. [Google Scholar] [CrossRef] [PubMed]
- Assouline, S.; Cocolakis, E.; Borden, K.L. The Development of Novel Therapies for the Treatment of Acute Myeloid Leukemia (AML). Cancers 2012, 4, 1161–1179. [Google Scholar] [CrossRef] [PubMed]
- Dunn, L.A.; Fury, M.G.; Sherman, E.J.; Ho, A.A.; Katabi, N.; Haque, S.S.; Pfister, D.G. Phase I study of induction chemotherapy with afatinib, ribavirin, and weekly carboplatin and paclitaxel for stage IVA/IVB human papillomavirus-associated oropharyngeal squamous cell cancer. Head Neck 2017, 40, 233–241. [Google Scholar] [CrossRef]
- Zismanov, V.; Attar-Schneider, O.; Lishner, M.; Heffez Aizenfeld, R.; Tartakover Matalon, S.; Drucker, L. Multiple myeloma proteostasis can be targeted via translation initiation factor eIF4E. Int. J. Oncol. 2015, 46, 860–870. [Google Scholar] [CrossRef]
- Attar-Schneider, O.; Pasmanik-Chor, M.; Tartakover-Matalon, S.; Drucker, L.; Lishner, M. eIF4E and eIF4GI have distinct and differential imprints on multiple myeloma’s proteome and signaling. Oncotarget 2015, 6, 4315–4329. [Google Scholar] [CrossRef]
- Attar-Schneider, O.; Drucker, L.; Gottfried, M. Migration and epithelial-to-mesenchymal transition of lung cancer can be targeted via translation initiation factors eIF4E and eIF4GI. Lab. Investig. 2016, 96, 1004–1015. [Google Scholar] [CrossRef][Green Version]
- Pettersson, F.; del Rincon, S.V.; Emond, A.; Huor, B.; Ngan, E.; Ng, J.; Dobocan, M.C.; Siegel, P.M.; Miller, W.H. Genetic and pharmacolgic inhibition of eIF4E reduces breast cancer cell migration, invasion and metastasis. Cancer Res. 2015, 75, 1102–1112. [Google Scholar] [CrossRef]
- Kentsis, A.; Topisirovic, I.; Culjkovic, B.; Shao, L.; Borden, K.L. Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc. Natl. Acad. Sci. USA 2004, 101, 18105–18110. [Google Scholar] [CrossRef]
- Yi, T.; Kabha, E.; Papadopoulos, E.; Wagner, G. 4EGI-1 targets breast cancer stem cells by selective inhibition of translation that persists in CSC maintenance, proliferation and metastasis. Oncotarget 2014, 5, 6028–6037. [Google Scholar] [CrossRef] [PubMed]
- De, A.; Jacobson, B.A.; Peterson, M.S.; Jay-Dixon, J.; Kratzke, M.G.; Sadiq, A.A.; Patel, M.R.; Kratzke, R.A. 4EGI-1 represses cap-dependent translation and regulates genome-wide translation in malignant pleural mesothelioma. Investig. New Drugs 2018, 36, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Volpon, L.; Osborne, M.J.; Zahreddine, H.; Romeo, A.A.; Borden, K.L. Conformational changes induced in the eukaryotic translation initiation factor eIF4E by a clinically relevant inhibitor, ribavirin triphosphate. Biochem. Biophys. Res. Commun. 2013, 434, 614–619. [Google Scholar] [CrossRef] [PubMed]
- Kentsis, A.; Volpon, L.; Topisirovic, I.; Soll, C.E.; Culjkovic, B.; Shao, L.; Borden, K.L. Further evidence that ribavirin interacts with eIF4E. RNA 2005, 11, 1762–1766. [Google Scholar] [CrossRef]
- Zahreddine, H.A.; Culjkovic-Kraljacic, B.; Assouline, S.; Gendron, P.; Romeo, A.A.; Morris, S.J.; Cormack, G.; Jaquith, J.B.; Cerchietti, L.; Cocolakis, E.; et al. The sonic hedgehog factor GLI1 imparts drug resistance through inducible glucuronidation. Nature 2014, 511, 90–93. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, T.; Maeda, T.; Shinojima, T.; Nagata, H.; Ryuchi, R.; Oya, M. A clinical study to evaluate the efficacy and safety of docetaxal with ribavirin in patients with progressive castration resistant prostate cancer who have previously received docetaxol alone. J. Clin. Oncol. 2017. [Google Scholar] [CrossRef]
- Hong, D.S.; Kurzrock, R.; Oh, Y.; Wheler, J.; Naing, A.; Brail, L.; Callies, S.; Andre, V.; Kadam, S.K.; Nasir, A.; et al. A phase 1 dose escalation, pharmacokinetic, and pharmacodynamic evaluation of eIF-4E antisense oligonucleotide LY2275796 in patients with advanced cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2011, 17, 6582–6591. [Google Scholar] [CrossRef]
- Topisirovic, I.; Ruiz-Gutierrez, M.; Borden, K.L. Phosphorylation of the eukaryotic translation initiation factor eIF4E contributes to its transformation and mRNA transport activities. Cancer Res. 2004, 64, 8639–8642. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.; Blaydes, J.P. MNK1 and EIF4E are downstream effectors of MEKs in the regulation of the nuclear export of HDM2 mRNA. Oncogene 2008, 27, 1645–1649. [Google Scholar] [CrossRef]
- Dinner, S.; Platanias, L.C. Targeting the mTOR Pathway in Leukemia. J. Cell. Biochem. 2016, 117, 1745–1752. [Google Scholar] [CrossRef]
- Joshi, S.; Platanias, L.C. Mnk kinases in cytokine signaling and regulation of cytokine responses. Biomol. Concepts 2015, 6, 85. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Joshi, S.; Platanias, L.C. Mnk kinase pathway: Cellular functions and biological outcomes. World J. Biol. Chem. 2014, 5, 321–333. [Google Scholar] [CrossRef]
- Ning, Y.; Liu, W.; Guan, X.; Xie, X.; Zhang, Y. CPSF3 is a promising prognostic biomarker and predicts recurrence of non-small cell lung cancer. Oncol. Lett. 2019, 18, 2835–2844. [Google Scholar] [CrossRef] [PubMed]
- Ross, N.T.; Lohmann, F.; Carbonneau, S.; Fazal, A.; Weihofen, W.A.; Gleim, S.; Salcius, M.; Sigoillot, F.; Henault, M.; Carl, S.H.; et al. CPSF3-dependent pre-mRNA processing as a druggable node in AML and Ewing’s sarcoma. Nat. Chem. Biol. 2020, 16, 50–59. [Google Scholar] [CrossRef]
- Kakegawa, J.; Sakane, N.; Suzuki, K.; Yoshida, T. JTE-607, a multiple cytokine production inhibitor, targets CPSF3 and inhibits pre-mRNA processing. Biochem. Biophys. Res. Commun. 2019, 518, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, A.; Tan, Y.; Wang, S.; Ma, Q.; Chen, X.; He, Z. NCBP1 promotes the development of lung adenocarcinoma through up-regulation of CUL4B. J. Cell. Mol. Med. 2019, 23, 6965–6977. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).