Machine Learning for Prediction of Immunotherapy Efficacy in Non-Small Cell Lung Cancer from Simple Clinical and Biological Data
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Data
2.3. Statistical Analysis
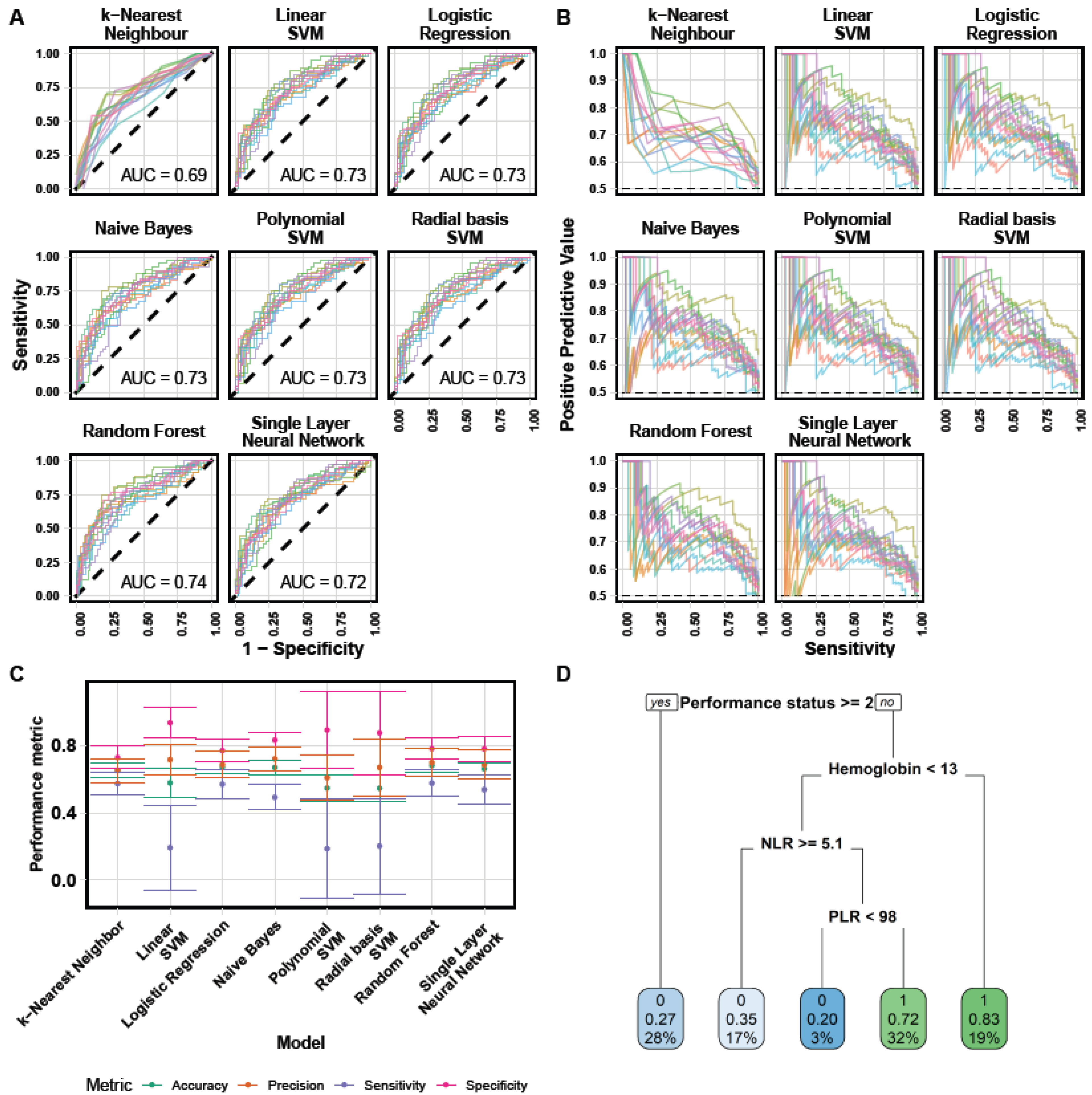
2.4. Machine Learning
3. Results
3.1. Patients and Disease Characteristics
3.2. Statistical Analysis
3.2.1. Response
3.2.2. Survival Analysis
3.3. Machine Learning
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Planchard, D.; Popat, S.; Kerr, K.; Novello, S.; Smit, E.F.; Faivre-Finn, C.; Mok, T.S.; Reck, M.; Van Schil, P.E.; Hellmann, M.D.; et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2018, 29 (Suppl. 4), iv192–iv237. [Google Scholar] [CrossRef] [PubMed]
- Reck, M.; Rodríguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csőszi, T.; Fülöp, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2016, 375, 1823–1833. [Google Scholar] [CrossRef] [Green Version]
- Gooden, M.J.M.; de Bock, G.H.; Leffers, N.; Daemen, T.; Nijman, H.W. The prognostic influence of tumour-infiltrating lymphocytes in cancer: A systematic review with meta-analysis. Br. J. Cancer 2011, 105, 93–103. [Google Scholar] [CrossRef] [Green Version]
- Balkwill, F.; Mantovani, A. Inflammation and cancer: Back to Virchow? Lancet 2001, 357, 539–545. [Google Scholar] [CrossRef]
- Hopkins, A.M.; Rowland, A.; Kichenadasse, G.; Wiese, M.D.; Gurney, H.; McKinnon, R.A.; Karapetis, C.S.; Sorich, M.J. Predicting response and toxicity to immune checkpoint inhibitors using routinely available blood and clinical markers. Br. J. Cancer 2017, 117, 913–920. [Google Scholar] [CrossRef]
- Diem, S.; Schmid, S.; Krapf, M.; Flatz, L.; Born, D.; Jochum, W.; Templeton, A.J.; Früha, M. Neutrophil-to-Lymphocyte ratio (NLR) and Platelet-to-Lymphocyte ratio (PLR) as prognostic markers in patients with non-small cell lung cancer (NSCLC) treated with nivolumab. Lung Cancer 2017, 111, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Proctor, M.J.; McMillan, D.C.; Morrison, D.S.; Fletcher, C.D.; Horgan, P.G.; Clarke, S.J. A derived neutrophil to lymphocyte ratio predicts survival in patients with cancer. Br. J. Cancer 2012, 107, 695–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrucci, P.F.; Ascierto, P.A.; Pigozzo, J.; Del Vecchio, M.; Antonini Cappellini, G.C.; Guidoboni, M.; Queirolo, P.; Savoia, P.; Mandala, M.; Simeone, E.; et al. Baseline neutrophils and derived neutrophil-to-lymphocyte ratio: Prognostic relevance in metastatic melanoma patients receiving ipilimumab. Ann. Oncol. 2016, 27, 732–738. [Google Scholar] [CrossRef] [PubMed]
- Diakos, C.I.; Tu, D.; Gebski, V.; Yip, S.; Wilson, K.; Karapetis, C.S.; O’Callaghan, C.J.; Shapiro, J.; Tebbutt, N.; Jonker, D.J.; et al. Is the Derived Neutrophil to Lymphocyte Ratio (Dnlr) an Independent Prognostic Marker in Patients with Metastatic Colorectal Cancer (Mcrc)? Analysis of the CO.17 And CO.20 Studies. Ann. Oncol. 2016, 27, vi196. [Google Scholar] [CrossRef]
- Gu, X.; Sun, S.; Gao, X.-S.; Xiong, W.; Qin, S.; Qi, X.; Ma, M.; Li, X.; Zhou, D.; Wang, W.; et al. Prognostic Value of Platelet to Lymphocyte Ratio in Non-Small Cell Lung Cancer: Evidence from 3430 Patients. Sci. Rep. 2016, 6, 23893. [Google Scholar] [CrossRef] [PubMed]
- Mezquita, L.; Auclin, E.; Ferrara, R.; Charrier, M.; Remon, J.; Planchard, D.; Ponce, S.; Paz Ares, L.; Leroy, L.; Audigier-Valette, C.; et al. Association of the Lung Immune Prognostic Index with Immune Checkpoint Inhibitor Outcomes in Patients with Advanced Non–Small Cell Lung Cancer. JAMA Oncol. 2018, 4, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Pavan, A.; Calvetti, L.; Dal Maso, A.; Attili, I.; Del Bianco, P.; Pasello, G.; Guarneri, V.; Aprile, G.; Conte, P.F.; Bonanno, L. Peripheral Blood Markers Identify Risk of Immune-Related Toxicity in Advanced Non-Small Cell Lung Cancer Treated with Immune-Checkpoint Inhibitors. Oncologist 2019, 24, 1128–1136. [Google Scholar] [CrossRef] [Green Version]
- Barlesi, F.; Greillier, L.; Monville, F.; Foa, C.; le Treut, J.; Audigier-Valette, C.; Vély, F.; Garcia, S.; Sabatier, F.; Ciccolini, J.; et al. LBA53 Precision immuno-oncology for advanced non-small cell lung cancer (NSCLC) patients (pts) treated with PD1/L1 immune checkpoint inhibitors (ICIs): A first analysis of the PIONeeR study. Ann. Oncol. 2020, 31, S1183. [Google Scholar] [CrossRef]
- Benzekry, S. Artificial intelligence and mechanistic modeling for clinical decision making in oncology. Clin. Pharmacol. Ther. 2020, 108, 471–486. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Statistical modeling: The two cultures. Stat. Sci. 2001, 16, 199–231. [Google Scholar] [CrossRef]
- Ahn, B.-C.; So, J.-W.; Synn, C.-B.; Kim, T.H.; Kim, J.H.; Byeon, Y.; Kim, Y.S.; Heo, S.G.; Yang, S.-D.; Yun, M.R.; et al. Clinical decision support algorithm based on machine learning to assess the clinical response to anti-programmed death-1 therapy in patients with non-small-cell lung cancer. Eur. J. Cancer 2021, 153, 179–189. [Google Scholar] [CrossRef]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef] [PubMed]
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 2 October 2019).
- Cox, D.R. Regression Models and Life-Tables. J. R. Stat. Soc. Ser. B Methodol. 1972, 34, 187–202. [Google Scholar] [CrossRef]
- Therneau, T.M. A Package for Survival Analysis in R. Available online: https://CRAN.R-project.org/package=survival (accessed on 8 December 2020).
- Kuhn, M.; Wickham, H. Tidymodels: A Collection of Packages for Modeling and Machine Learning Using Tidyverse Principles. Available online: https://www.tidymodels.org (accessed on 8 December 2020).
- Becker, T.; Weberpals, J.; Jegg, A.M.; So, W.V.; Fischer, A.; Weisser, M.; Schmich, F.; Rüttinger, D.; Bauer-Mehren, A. An enhanced prognostic score for overall survival of patients with cancer derived from a large real-world cohort. Ann. Oncol. 2020, 31, 1561–1568. [Google Scholar] [CrossRef] [PubMed]
- Nicolò, C.; Périer, C.; Prague, M.; Bellera, C.; MacGrogan, G.; Saut, O.; Benzakry, S. Machine Learning and Mechanistic Modeling for Prediction of Metastatic Relapse in Early-Stage Breast Cancer. JCO Clin. Cancer Inform. 2020, 4, 259–274. [Google Scholar] [CrossRef]
- Trebeschi, S.; Drago, S.G.; Birkbak, N.J.; Kurilova, I.; Călin, A.M.; Delli Pizzi, A.; Lalezari, F.; Lambregts, D.M.J.; Rohaan, M.W.; Parmar, C.; et al. Predicting response to cancer immunotherapy using noninvasive radiomic biomarkers. Ann. Oncol. 2019, 30, 998–1004. [Google Scholar] [CrossRef] [Green Version]
- Wiesweg, M.; Mairinger, F.; Reis, H.; Goetz, M.; Walter, R.F.H.; Hager, T.; Metzenmacher, M.; Eberhardt, W.E.E.; McCutcheon, A.; Aigner, C.; et al. Machine learning-based predictors for immune checkpoint inhibitor therapy of non-small-cell lung cancer. Ann. Oncol. 2019, 30, 655–657. [Google Scholar] [CrossRef] [PubMed]
- Wiesweg, M.; Mairinger, F.; Reis, H.; Goetz, M.; Walter, R.F.H.; Hager, T.; Metzenmacher, M.; Eberhardt, W.E.E.; McCutcheonet, A.; Köster, J.; et al. Machine learning reveals a PD-L1–independent prediction of response to immunotherapy of non-small cell lung cancer by gene expression context. Eur. J. Cancer 2020, 140, 76–85. [Google Scholar] [CrossRef]
- Riley, R.D.; Ensor, J.; Snell, K.I.E.; Harrell, F.E., Jr.; Martin, G.P.; Reitsma, J.B.; Moons, K.G.M.; Collins, G.; van Smeden, M. Calculating the sample size required for developing a clinical prediction model. BMJ 2020, 368, m441. [Google Scholar] [CrossRef] [Green Version]
- Christodoulou, E.; Ma, J.; Collins, G.S.; Steyerberg, E.W.; Verbakel, J.Y.; Van Calster, B. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J. Clin. Epidemiol. 2019, 110, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, F.; Yuan, F.; Li, Y.; Ma, J.; Ou, Q.; Liu, Z.; Yang, B.; Wang, L.; Tao, H.; et al. Pretreatment hemoglobin level as a predictor to evaluate the efficacy of immune checkpoint inhibitors in patients with advanced non-small cell lung cancer. Ther. Adv. Med. Oncol. 2020, 12, 1758835920970049. [Google Scholar] [CrossRef] [PubMed]
- Dall’Olio, F.G.; Maggio, I.; Massucci, M.; Mollica, V.; Fragomeno, B.; Ardizzoni, A. ECOG performance status ≥2 as a prognostic factor in patients with advanced non small cell lung cancer treated with immune checkpoint inhibitors—A systematic review and meta-analysis of real world data. Lung Cancer 2020, 145, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, M.; Kasinath, P.; High, R.; Yu, F.; Teply, B.A. Impact of Performance Status on Response and Survival Among Patients Receiving Checkpoint Inhibitors for Advanced Solid Tumors. JCO Oncol. Pract. 2021. [Google Scholar] [CrossRef]
- Melosky, B.; Chu, Q.; Juergens, R.A.; Leighl, N.; Ionescu, D.; Tsao, M.-S.; McLeod, D.; Hirsh, V. Breaking the biomarker code: PD-L1 expression and checkpoint inhibition in advanced NSCLC. Cancer Treat. Rev. 2018, 65, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Lium, Y.; Dong, Z.; Jiang, T.; Hou, L.; Wu, F.; Gao, G.; He, Y.; Zhao, J.; Li, X.; Zhao, C.; et al. Heterogeneity of PD-L1 Expression Among the Different Histological Components and Metastatic Lymph Nodes in Patients with Resected Lung Adenosquamous Carcinoma. Clin. Lung Cancer 2018, 19, e421–e430. [Google Scholar]
- Boffa, D.J.; Graf, R.P.; Salazar, M.C.; Hoag, J.; Lu, D.; Krupa, R.; Louw, J.; Dugan, L.; Wang, Y.; Landers, M.; et al. Cellular Expression of PD-L1 in the Peripheral Blood of Lung Cancer Patients is Associated with Worse Survival. Cancer Epidemiol. Prev. Biomark. 2017, 26, 1139–1145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.J.R.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Børresen-Dale, A.-L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berland, L.; Heeke, S.; Humbert, O.; Macocco, A.; Long-Mira, E.; Lassalle, S.; Lespinet-Fabre, V.; Lalvée, S.; Bordone, O.; Cohenet, C.; et al. Current views on tumor mutational burden in patients with nonsmall cell lung cancer treated by immune checkpoint inhibitors. J. Thorac. Dis. 2018, 11, S71–S80. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.S.; Stojanov, P.; Polak, P.; Kryukov, G.V.; Cibulskis, K.; Sivachenko, A.; Carter, S.L.; Stewart, C.; Mermel, C.H.; Roberts, S.A.; et al. Mutational heterogeneity in cancer and the search for new cancer genes. Nature 2013, 499, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, H.; Sanchez-Vega, F.; La, K.; Chatila, W.; Jonsson, P.; Halpenny, D.; Plodkowski, A.; Long, N.; Sauter, J.L.; Rekhtman, N.; et al. Molecular Determinants of Response to Anti–Programmed Cell Death (PD)-1 and Anti–Programmed Death-Ligand 1 (PD-L1) Blockade in Patients with Non–Small-Cell Lung Cancer Profiled with Targeted Next-Generation Sequencing. J. Clin. Oncol. 2018, 36, 633–641. [Google Scholar] [CrossRef]
- Jeanson, A.; Tomasini, P.; Souquet-Bressand, M.; Brandone, N.; Boucekine, M.; Grangeon, M.; Chaleat, S.; Khobta, N.; Milia, J.; Mhanna, L.; et al. Efficacy of Immune Checkpoint Inhibitors in KRAS-Mutant Non-Small Cell Lung Cancer (NSCLC). J. Thorac. Oncol. 2019, 14, 1095–1101. [Google Scholar] [CrossRef]
- Mazieres, J.; Drilon, A.; Lusque, A.; Mhanna, L.; Cortot, A.B.; Mezquita, L.; Thai, A.A.; Mascaux, C.; Couraud, S.; Veillon, R.; et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: Results from the IMMUNOTARGET registry. Ann. Oncol. 2019, 30, 1321–1328. [Google Scholar] [CrossRef] [PubMed]
- Skoulidis, F.; Goldberg, M.E.; Greenawalt, D.M.; Hellmann, M.D.; Awad, M.M.; Gainor, J.F.; Schrock, A.B.; Hartmaier, R.J.; Trabucco, S.E.; Gay, L.; et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov. 2018, 8, 822–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Bullock, K.; Gurjao, C.; David Braun, D.; Shukla, S.A.; Bossé, D.; Lalani, A.-K.A.; Gopal, S.; Jin, C.; Horak, C.; et al. Metabolomic adaptations and correlates of survival to immune checkpoint blockade. Nat. Commun. 2019, 10, 4346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatae, R.; Chamoto, K.; Kim, Y.H.; Sonomura, K.; Taneishi, K.; Kawaguchi, S.; Yoshida, H.; Ozasa, H.; Sakamori, Y.; Akrami, M.; et al. Combination of host immune metabolic biomarkers for the PD-1 blockade cancer immunotherapy. JCI Insight 2020, 5, e133501. [Google Scholar] [CrossRef] [Green Version]
- Ghini, V.; Laera, L.; Fantechi, B.; del Monte, F.; Benelli, M.; McCartney, A.; Tenori, L.; Luchinat, C.; Pozzessere, D. Metabolomics to Assess Response to Immune Checkpoint Inhibitors in Patients with Non-Small-Cell Lung Cancer. Cancers 2020, 12, 3574. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Limkin, E.J.; Vakalopoulou, M.; Dercle, L.; Champiat, S.; Han, S.R.; Verlingue, L.; Brandao, D.; Lancia, A.; Ammari, S.; et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: An imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018, 19, 1180–1191. [Google Scholar] [CrossRef]
- Welch, M.L.; McIntosh, C.; Haibe-Kains, B.; Milosevic, M.F.; Dekker, L.W.A.; Huan, S.H.; Purdie, T.G.; O’Sullivan, B.; Aerts, H.J.W.L.; Jaffray, D.A. Vulnerabilities of radiomic signature development: The need for safeguards. Radiother. Oncol. J. Eur. Soc. Ther. Radiol. Oncol. 2019, 130, 2–9. [Google Scholar] [CrossRef] [Green Version]
- Herbst, R.S.; Baas, P.; Kim, D.-W.; Felip, E.; Pérez-Gracia, J.L.; Han, J.-Y.; Molina, J.; Kim, L.-H.; Dubos Arvis, C.; Ahn, M.-J.; et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): A randomised controlled trial. Lancet 2016, 387, 1540–1550. [Google Scholar] [CrossRef]
- Mezquita, L.; Preeshagul, I.; Auclin, E.; Saravia, D.; Hendriks, L.; Rizvi, H.; Park, H.R.W.; Nadal, E.; Martin-Romano, P.; Ruffinelli, J.C.; et al. Predicting immunotherapy outcomes under therapy in patients with advanced NSCLC using dNLR and its early dynamics. Eur. J. Cancer 2021, 151, 211–220. [Google Scholar] [CrossRef]
- Kurtz, D.M.; Esfahani, M.S.; Scherer, F.; Soo, J.; Jin, M.C.; Liu, C.L.; Newman, A.M.; Dührsen, U.; Hüttmann, A.; Casasnovas, O.; et al. Dynamic Risk Profiling Using Serial Tumor Biomarkers for Personalized Outcome Prediction. Cell 2019, 178, 699–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Claret, L.; Jin, J.Y.; Ferté, C.; Winter, H.; Girish, S.; Stroh, M.; He, P.; Ballinger, M.; Sandler, A.; Joshi, A.; et al. A Model of Overall Survival Predicts Treatment Outcomes with Atezolizumab versus Chemotherapy in Non-Small Cell Lung Cancer Based on Early Tumor Kinetics. Clin. Cancer Res. 2018, 24, 3292–3298. [Google Scholar] [CrossRef] [Green Version]
- Serre, R.; Benzekry, S.; Padovani, L.; Christophe Meille, C.; André, N.; Ciccolini, J.; Barlesi, F.; Muracciole, X.; Barbolosi, D. Mathematical Modeling of Cancer Immunotherapy and Its Synergy with Radiotherapy. Cancer Res. 2016, 76, 4931–4940. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Variable | N = 298 1 |
|---|---|
| Age | 62 (55, 69) |
| Sex | |
| Female | 99 (33%) |
| Male | 199 (67%) |
| Tobacco status | |
| Former Smoker | 140 (47%) |
| Non smoker | 36 (12%) |
| Smoker | 122 (41%) |
| Brain metastases | 72 (29%) |
| Performance status | |
| ≥2 | 26 (8.9%) |
| 0–1 | 265 (91%) |
| Mutation profile | |
| ALK | 1 (0.8%) |
| BRAF | 9 (7.4%) |
| EGFR | 14 (11%) |
| KRAS | 87 (71%) |
| Other mutation | 5 (4.1%) |
| ROS1 | 2 (1.6%) |
| Wild type | 4 (3.3%) |
| Immunotherapy type | |
| anti-CTLA-4 | 3 (1.0%) |
| anti-PD-1 | 266 (89%) |
| anti-PD-L1 | 26 (8.7%) |
| Combination | 3 (1.0%) |
| Previous treatment | |
| Chemotherapy | 281 (95%) |
| Chemotherapy + immunotherapy | 5 (1.7%) |
| Targeted therapy | 11 (3.7%) |
| Response | |
| Complete response | 2 (0.7%) |
| Partial response | 44 (15%) |
| Progressive disease | 131 (45%) |
| Stable disease | 113 (39%) |
| Univariable Logistic Regression | Multivariable Logistic Regression | |||||
|---|---|---|---|---|---|---|
| Variable | Odds Ratio [95% CI] | p | Signif | Odds Ratio [95% CI] | p | Signif |
| Lymphocytes | 1.1 [0.83, 1.4] | 0.678 | 0.98 [0.15, 5.2] | 0.984 | ||
| NLR | 0.49 [0.31, 0.73] | 0.000879 | *** | 0.68 [0.098, 1.9] | 0.651 | |
| Platelets | 1 [0.82, 1.3] | 0.762 | 1.3 [0.72, 2.4] | 0.404 | ||
| PLR | 0.84 [0.64, 1.1] | 0.156 | 1.1 [0.5, 2.4] | 0.788 | ||
| Leukocytes | 0.68 [0.5, 0.89] | 0.00791 | ** | 0.6 [0.0022, 3 × 102] | 0.847 | |
| Hemoglobin | 1.9 [1.5, 2.5] | 9.26 × 10−7 | *** | 1.8 [1.3, 2.4] | 0.000122 | *** |
| dNLR | 0.63 [0.47, 0.83] | 0.00155 | ** | 0.8 [0.33, 2.7] | 0.689 | |
| Neutrophils | 0.62 [0.45, 0.83] | 0.00232 | ** | 1.5 [0.0047, 2.7 × 102] | 0.863 | |
| Monocytes | 0.87 [0.69, 1.1] | 0.226 | 0.86 [0.5, 1.4] | 0.545 | ||
| Eosinophils | 1.3 [0.97, 1.9] | 0.139 | 1.1 [0.75, 1.8] | 0.582 | ||
| Basophils | 1.2 [0.95, 1.8] | 0.177 | 1.2 [0.89, 1.8] | 0.321 | ||
| BMI | 1.2 [0.95, 1.5] | 0.123 | 1 [0.76, 1.3] | 0.997 | ||
| Performance status | 0.5 [0.39, 0.64] | 6.21 × 10−8 | *** | 0.58 [0.44, 0.75] | 7.79 × 10−5 | *** |
| Model | Accuracy | ROC AUC | PPV | NPV | Sensitivity | Specificity |
|---|---|---|---|---|---|---|
| Random Forest | 0.68 ± 0.04 | 0.74 ± 0.03 | 0.70 ± 0.08 | 0.68 ± 0.06 | 0.58 ± 0.08 | 0.78 ± 0.06 |
| Logistic Regression | 0.67 ± 0.04 | 0.73 ± 0.03 | 0.69 ± 0.08 | 0.67 ± 0.06 | 0.57 ± 0.09 | 0.77 ± 0.07 |
| Naive Bayes | 0.67 ± 0.04 | 0.73 ± 0.03 | 0.72 ± 0.07 | 0.65 ± 0.06 | 0.49 ± 0.07 | 0.83 ± 0.05 |
| Single Layer Neural Network | 0.66 ± 0.03 | 0.72 ± 0.03 | 0.69 ± 0.09 | 0.66 ± 0.06 | 0.54 ± 0.09 | 0.78 ± 0.07 |
| k-Nearest Neighbour | 0.66 ± 0.04 | 0.69 ± 0.04 | 0.65 ± 0.07 | 0.66 ± 0.06 | 0.58 ± 0.07 | 0.73 ± 0.07 |
| Linear SVM | 0.58 ± 0.09 | 0.73 ± 0.03 | 0.72 ± 0.09 | 0.58 ± 0.10 | 0.19 ± 0.25 | 0.94 ± 0.09 |
| Polynomial SVM | 0.55 ± 0.08 | 0.73 ± 0.03 | 0.61 ± 0.13 | 0.58 ± 0.13 | 0.19 ± 0.29 | 0.89 ± 0.23 |
| Radial basis SVM | 0.55 ± 0.08 | 0.73 ± 0.03 | 0.67 ± 0.17 | 0.56 ± 0.06 | 0.20 ± 0.28 | 0.88 ± 0.25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benzekry, S.; Grangeon, M.; Karlsen, M.; Alexa, M.; Bicalho-Frazeto, I.; Chaleat, S.; Tomasini, P.; Barbolosi, D.; Barlesi, F.; Greillier, L. Machine Learning for Prediction of Immunotherapy Efficacy in Non-Small Cell Lung Cancer from Simple Clinical and Biological Data. Cancers 2021, 13, 6210. https://doi.org/10.3390/cancers13246210
Benzekry S, Grangeon M, Karlsen M, Alexa M, Bicalho-Frazeto I, Chaleat S, Tomasini P, Barbolosi D, Barlesi F, Greillier L. Machine Learning for Prediction of Immunotherapy Efficacy in Non-Small Cell Lung Cancer from Simple Clinical and Biological Data. Cancers. 2021; 13(24):6210. https://doi.org/10.3390/cancers13246210
Chicago/Turabian StyleBenzekry, Sébastien, Mathieu Grangeon, Mélanie Karlsen, Maria Alexa, Isabella Bicalho-Frazeto, Solène Chaleat, Pascale Tomasini, Dominique Barbolosi, Fabrice Barlesi, and Laurent Greillier. 2021. "Machine Learning for Prediction of Immunotherapy Efficacy in Non-Small Cell Lung Cancer from Simple Clinical and Biological Data" Cancers 13, no. 24: 6210. https://doi.org/10.3390/cancers13246210
APA StyleBenzekry, S., Grangeon, M., Karlsen, M., Alexa, M., Bicalho-Frazeto, I., Chaleat, S., Tomasini, P., Barbolosi, D., Barlesi, F., & Greillier, L. (2021). Machine Learning for Prediction of Immunotherapy Efficacy in Non-Small Cell Lung Cancer from Simple Clinical and Biological Data. Cancers, 13(24), 6210. https://doi.org/10.3390/cancers13246210






