RNA Biomarkers as a Response Measure for Survival in Patients with Metastatic Castration-Resistant Prostate Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Assessments
2.3. Biomarker Analysis and Selection
2.4. Statistical Analysis
3. Results
3.1. Patients
3.2. Biomarkers as Indicator for Survival
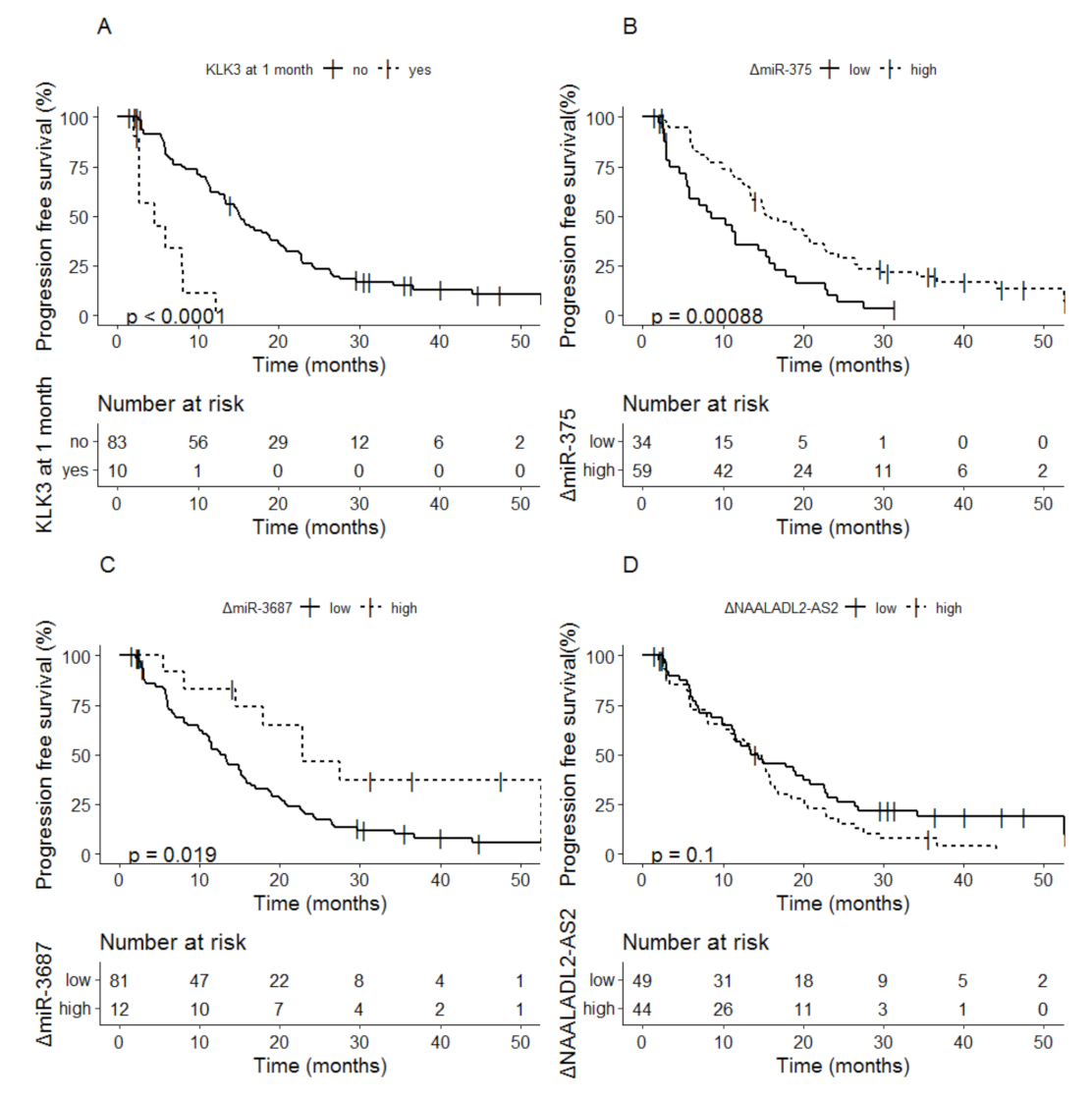
3.2.1. Baseline Biomarkers as Indicator for Progression Free Survival
3.2.2. Biomarker Dynamics at 1 Month as Indicator for Progression Free Survival
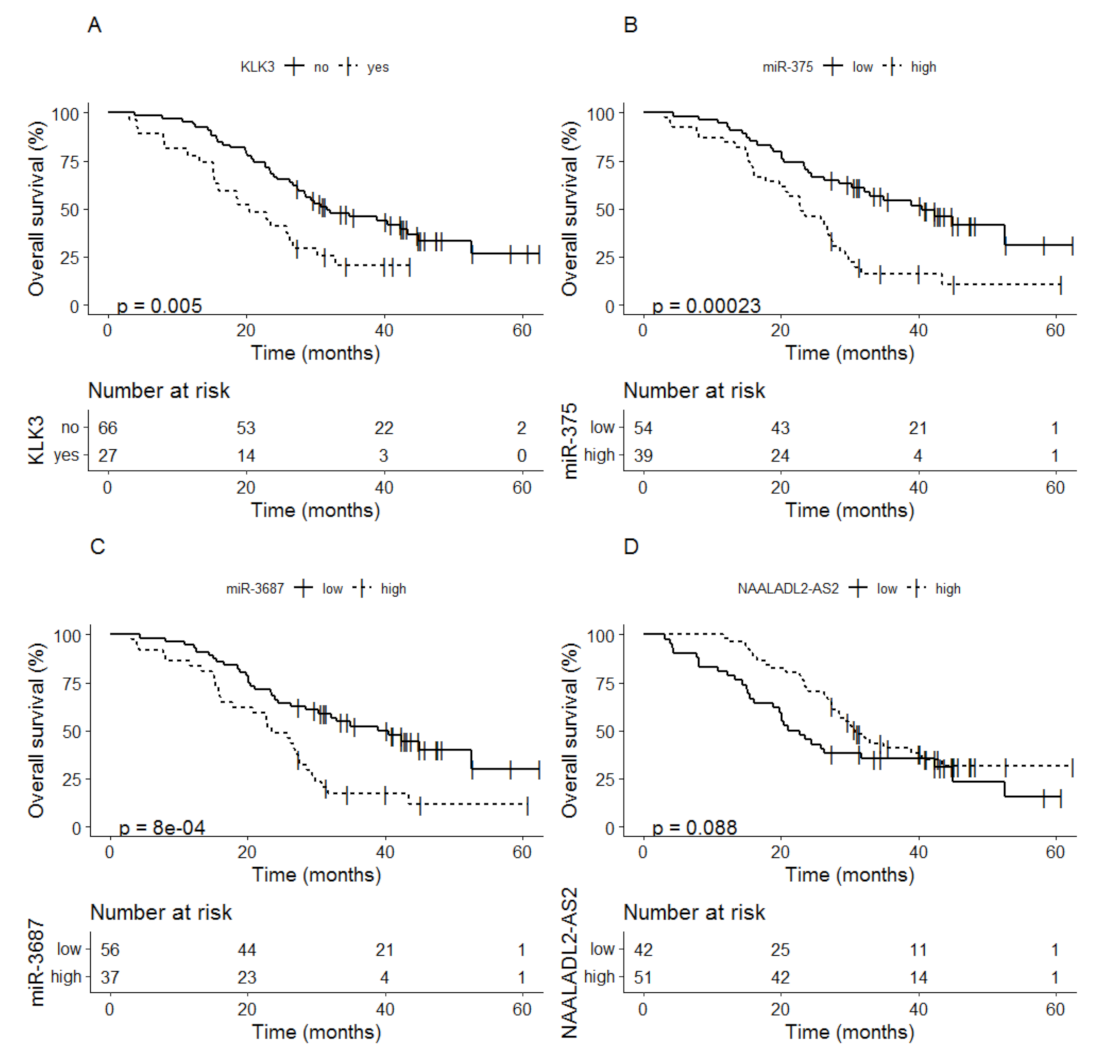
3.2.3. Baseline Biomarkers as Indicator for Overall Survival
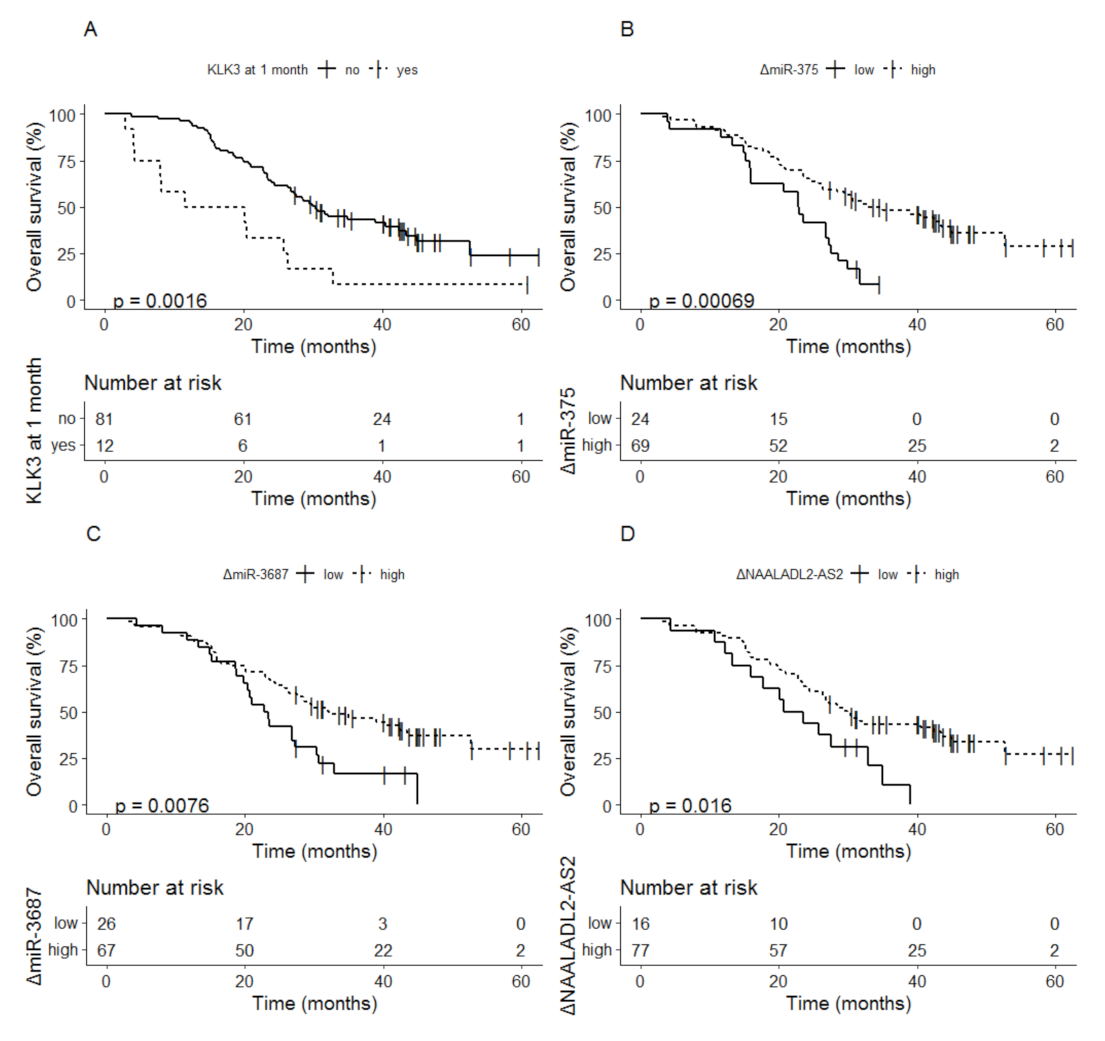
3.2.4. Biomarker Dynamics at 1 Month as Indicator for Overall Survival
3.3. Predictive Performance of Different Models
3.4. Multivariate Cox Regression of the Predictive Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed consent statement
Data Availability Statement
Conflicts of Interest
References
- Montgomery, R.B.; Mostaghel, E.A.; Vessella, R.; Hess, D.L.; Kalhorn, T.F.; Higano, C.S.; True, L.D.; Nelson, P.S. Maintenance of intratumoral androgens in metastatic prostate cancer: A mechanism for castration-resistant tumor growth. Cancer Res. 2008, 68, 4447–4454. [Google Scholar] [CrossRef] [Green Version]
- Scher, H.I.; Beer, T.M.; Higano, C.S.; Anand, A.; Taplin, M.E.; Efstathiou, E.; Rathkopf, D.; Shelkey, J.; Yu, E.Y.; Alumkal, J.; et al. Antitumour activity of MDV3100 in castration-resistant prostate cancer: A phase 1–2 study. Lancet 2010, 375, 1437–1446. [Google Scholar] [CrossRef] [Green Version]
- Attard, G.; Reid, A.H.; Yap, T.A.; Raynaud, F.; Dowsett, M.; Settatree, S.; Barrett, M.; Parker, C.; Martins, V.; Folkerd, E.; et al. Phase I clinical trial of a selective inhibitor of CYP17, abiraterone acetate, confirms that castration-resistant prostate cancer commonly remains hormone driven. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2008, 26, 4563–4571. [Google Scholar] [CrossRef]
- Beer, T.M.; Armstrong, A.J.; Rathkopf, D.E.; Loriot, Y.; Sternberg, C.N.; Higano, C.S.; Iversen, P.; Bhattacharya, S.; Carles, J.; Chowdhury, S.; et al. Enzalutamide in Metastatic Prostate Cancer before Chemotherapy. N. Engl. J. Med. 2014, 371, 424–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, C.J.; Smith, M.R.; de Bono, J.S.; Molina, A.; Logothetis, C.J.; de Souza, P.; Fizazi, K.; Mainwaring, P.; Piulats, J.M.; Ng, S.; et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N. Engl. J. Med. 2013, 368, 138–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buttigliero, C.; Tucci, M.; Bertaglia, V.; Vignani, F.; Bironzo, P.; Di Maio, M.; Scagliotti, G.V. Understanding and overcoming the mechanisms of primary and acquired resistance to abiraterone and enzalutamide in castration resistant prostate cancer. Cancer Treat. Rev. 2015, 41, 884–892. [Google Scholar] [CrossRef]
- Annala, M.; Vandekerkhove, G.; Khalaf, D.; Taavitsainen, S.; Beja, K.; Warner, E.W.; Sunderland, K.; Kollmannsberger, C.; Eigl, B.J.; Finch, D.; et al. Circulating Tumor DNA Genomics Correlate with Resistance to Abiraterone and Enzalutamide in Prostate Cancer. Cancer Discov. 2018, 8, 444–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scher, H.I.; Morris, M.J.; Stadler, W.M.; Higano, C.; Basch, E.; Fizazi, K.; Antonarakis, E.S.; Beer, T.M.; Carducci, M.A.; Chi, K.N.; et al. Trial Design and Objectives for Castration-Resistant Prostate Cancer: Updated Recommendations From the Prostate Cancer Clinical Trials Working Group 3. J. Clin. Oncol. 2016, 34, 1402–1418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belin, L.; Tan, A.; De Rycke, Y.; Dechartres, A. Progression-free survival as a surrogate for overall survival in oncology trials: A methodological systematic review. Br. J. Cancer 2020, 122, 1707–1714. [Google Scholar] [CrossRef] [PubMed]
- Chi, K.N.; Kheoh, T.; Ryan, C.J.; Molina, A.; Bellmunt, J.; Vogelzang, N.J.; Rathkopf, D.E.; Fizazi, K.; Kantoff, P.W.; Li, J.; et al. A prognostic index model for predicting overall survival in patients with metastatic castration-resistant prostate cancer treated with abiraterone acetate after docetaxel. Ann. Oncol. 2016, 27, 454–460. [Google Scholar] [CrossRef]
- Ryan, C.J.; Kheoh, T.; Li, J.; Molina, A.; De Porre, P.; Carles, J.; Efstathiou, E.; Kantoff, P.W.; Mulders, P.F.A.; Saad, F.; et al. Prognostic Index Model for Progression-Free Survival in Chemotherapy-Naïve Metastatic Castration-Resistant Prostate Cancer Treated with Abiraterone Acetate Plus Prednisone. Clin. Genitourin. Cancer 2017, 16, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Khalaf, D.J.; Avilés, C.M.; Azad, A.A.; Sunderland, K.; Todenhöfer, T.; Eigl, B.J.; Finch, D.; Le, L.; Atwell, A.; Keith, B.; et al. A prognostic model for stratifying clinical outcomes in chemotherapy-naive metastatic castration-resistant prostate cancer patients treated with abiraterone acetate. Can. Urol. Assoc. J. 2018, 12, E47–E52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armstrong, A.J.; Lin, P.; Higano, C.S.; Sternberg, C.N.; Sonpavde, G.; Tombal, B.; Templeton, A.J.; Fizazi, K.; Phung, D.; Wong, E.K.; et al. Development and validation of a prognostic model for overall survival in chemotherapy-naive men with metastatic castration-resistant prostate cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. ESMO 2018, 29, 2200–2207. [Google Scholar] [CrossRef]
- Antonarakis, E.S.; Lu, C.; Wang, H.; Luber, B.; Nakazawa, M.; Roeser, J.C.; Chen, Y.; Mohammad, T.A.; Chen, Y.; Fedor, H.L.; et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 2014, 371, 1028–1038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scher, H.I.; Lu, D.; Schreiber, N.A.; Louw, J.; Graf, R.P.; Vargas, H.A.; Johnson, A.; Jendrisak, A.; Bambury, R.; Danila, D.; et al. Association of AR-V7 on Circulating Tumor Cells as a Treatment-Specific Biomarker with Outcomes and Survival in Castration-Resistant Prostate Cancer. JAMA Oncol. 2016, 2, 1441–1449. [Google Scholar] [CrossRef] [PubMed]
- Bernemann, C.; Schnoeller, T.J.; Luedeke, M.; Steinestel, K.; Boegemann, M.; Schrader, A.J.; Steinestel, J. Expression of AR-V7 in Circulating Tumour Cells Does Not Preclude Response to Next Generation Androgen Deprivation Therapy in Patients with Castration Resistant Prostate Cancer. Eur. Urol. 2017, 71, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Scher, H.I.; Heller, G.; Molina, A.; Attard, G.; Danila, D.C.; Jia, X.; Peng, W.; Sandhu, S.K.; Olmos, D.; Riisnaes, R.; et al. Circulating tumor cell biomarker panel as an individual-level surrogate for survival in metastatic castration-resistant prostate cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2015, 33, 1348–1355. [Google Scholar] [CrossRef] [PubMed]
- Tolmeijer, S.H.; Boerrigter, E.; Schalken, J.A.; Geerlings, M.J.; Oort, I.M.v.; Erp, N.P.v.; Gerritsen, W.R.; Ligtenberg, M.J.L.; Mehra, N. A Systematic Review and Meta-Analysis on the Predictive Value of Cell-Free DNA–Based Androgen Receptor Copy Number Gain in Patients With Castration-Resistant Prostate Cancer. JCO Precis. Oncol. 2020, 4, 714–729. [Google Scholar] [CrossRef]
- Sumanasuriya, S.; Seed, G.; Parr, H.; Christova, R.; Pope, L.; Bertan, C.; Bianchini, D.; Rescigno, P.; Figueiredo, I.; Goodall, J.; et al. Elucidating Prostate Cancer Behaviour During Treatment via Low-pass Whole-genome Sequencing of Circulating Tumour DNA. Eur. Urol. 2021, 80, 243–253. [Google Scholar] [CrossRef]
- Smith, M.R.; Thomas, S.; Gormley, M.; Chowdhury, S.; Olmos, D.; Oudard, S.; Feng, F.Y.; Rajpurohit, Y.; Urtishak, K.; Ricci, D.S.; et al. Blood Biomarker Landscape in Patients with High-risk Nonmetastatic Castration-Resistant Prostate Cancer Treated with Apalutamide and Androgen-Deprivation Therapy as They Progress to Metastatic Disease. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2021, 27, 4539–4548. [Google Scholar] [CrossRef]
- Annala, M.; Fu, S.; Bacon, J.V.W.; Sipola, J.; Iqbal, N.; Ferrario, C.; Ong, M.; Wadhwa, D.; Hotte, S.J.; Lo, G.; et al. Cabazitaxel versus abiraterone or enzalutamide in poor prognosis metastatic castration-resistant prostate cancer: A multicentre, randomised, open-label, phase II trial. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. ESMO 2021, 32, 896–905. [Google Scholar] [CrossRef] [PubMed]
- Benoist, G.E.; van Oort, I.M.; Boerrigter, E.; Verhaegh, G.W.; van Hooij, O.; Groen, L.; Smit, F.; de Mol, P.; Hamberg, P.; Dezentje, V.O.; et al. Prognostic Value of Novel Liquid Biomarkers in Patients with Metastatic Castration-Resistant Prostate Cancer Treated with Enzalutamide: A Prospective Observational Study. Clin. Chem. 2020, 66, 842–851. [Google Scholar] [CrossRef] [PubMed]
- Boerrigter, E.; Benoist, G.E.; van Oort, I.M.; Verhaegh, G.W.; van Hooij, O.; Groen, L.; Smit, F.; Oving, I.M.; de Mol, P.; Smilde, T.J.; et al. Liquid biopsy reveals KLK3 mRNA as a prognostic marker for progression free survival in patients with metastatic castration-resistant prostate cancer undergoing first-line abiraterone acetate and prednisone treatment. Mol. Oncol. 2021, 15, 2453–2465. [Google Scholar] [CrossRef] [PubMed]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef]
- Lausen, B.; Schumacher, M. Maximally selected rank statistics. Biometrics 1992, 48, 73–85. [Google Scholar] [CrossRef]
- Rescigno, P.; Lorente, D.; Bianchini, D.; Ferraldeschi, R.; Kolinsky, M.P.; Sideris, S.; Zafeiriou, Z.; Sumanasuriya, S.; Smith, A.D.; Mehra, N.; et al. Prostate-specific Antigen Decline After 4 Weeks of Treatment with Abiraterone Acetate and Overall Survival in Patients with Metastatic Castration-resistant Prostate Cancer. Eur. Urol. 2016, 70, 724–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tibshirani, R. The lasso method for variable selection in the Cox model. Stat. Med. 1997, 16, 385–395. [Google Scholar] [CrossRef] [Green Version]
- Uno, H.; Cai, T.; Pencina, M.J.; D’Agostino, R.B.; Wei, L.J. On the C-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat. Med. 2011, 30, 1105–1117. [Google Scholar] [CrossRef] [Green Version]
- Donders, A.R.; van der Heijden, G.J.; Stijnen, T.; Moons, K.G. Review: A gentle introduction to imputation of missing values. J. Clin. Epidemiol. 2006, 59, 1087–1091. [Google Scholar] [CrossRef]
- White, I.R.; Royston, P. Imputing missing covariate values for the Cox model. Stat. Med. 2009, 28, 1982–1998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moons, K.G.; Donders, R.A.; Stijnen, T.; Harrell, F.E., Jr. Using the outcome for imputation of missing predictor values was preferred. J. Clin. Epidemiol. 2006, 59, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Sharp, A.; Welti, J.C.; Lambros, M.B.K.; Dolling, D.; Rodrigues, D.N.; Pope, L.; Aversa, C.; Figueiredo, I.; Fraser, J.; Ahmad, Z.; et al. Clinical Utility of Circulating Tumour Cell Androgen Receptor Splice Variant-7 Status in Metastatic Castration-resistant Prostate Cancer. Eur. Urol. 2019, 76, 676–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heller, G.; McCormack, R.; Kheoh, T.; Molina, A.; Smith, M.R.; Dreicer, R.; Saad, F.; de Wit, R.; Aftab, D.T.; Hirmand, M.; et al. Circulating Tumor Cell Number as a Response Measure of Prolonged Survival for Metastatic Castration-Resistant Prostate Cancer: A Comparison with Prostate-Specific Antigen Across Five Randomized Phase III Clinical Trials. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2018, 36, 572–580. [Google Scholar] [CrossRef]
- Lorente, D.; Olmos, D.; Mateo, J.; Bianchini, D.; Seed, G.; Fleisher, M.; Danila, D.C.; Flohr, P.; Crespo, M.; Figueiredo, I.; et al. Decline in Circulating Tumor Cell Count and Treatment Outcome in Advanced Prostate Cancer. Eur. Urol. 2016, 70, 985–992. [Google Scholar] [CrossRef] [Green Version]
- Beer, T.M.; Armstrong, A.J.; Rathkopf, D.; Loriot, Y.; Sternberg, C.N.; Higano, C.S.; Iversen, P.; Evans, C.P.; Kim, C.S.; Kimura, G.; et al. Enzalutamide in Men with Chemotherapy-naive Metastatic Castration-resistant Prostate Cancer: Extended Analysis of the Phase 3 PREVAIL Study. Eur. Urol. 2017, 71, 151–154. [Google Scholar] [CrossRef] [Green Version]
- Ryan, C.J.; Smith, M.R.; Fizazi, K.; Saad, F.; Mulders, P.F.; Sternberg, C.N.; Miller, K.; Logothetis, C.J.; Shore, N.D.; Small, E.J.; et al. Abiraterone acetate plus prednisone versus placebo plus prednisone in chemotherapy-naive men with metastatic castration-resistant prostate cancer (COU-AA-302): Final overall survival analysis of a randomised, double-blind, placebo-controlled phase 3 study. Lancet Oncol. 2015, 16, 152–160. [Google Scholar] [CrossRef]
- Tagawa, S.T.; Ramaswamy, K.; Huang, A.; Mardekian, J.; Schultz, N.M.; Wang, L.; Sandin, R.; Lechpammer, S.; George, D.J. Survival outcomes in patients with chemotherapy-naive metastatic castration-resistant prostate cancer treated with enzalutamide or abiraterone acetate. Prostate Cancer Prostatic Dis. 2021, 24, 1032–1040. [Google Scholar] [CrossRef] [PubMed]
- Heller, G.; Fizazi, K.; McCormack, R.; Molina, A.; MacLean, D.; Webb, I.J.; Saad, F.; de Bono, J.S.; Scher, H.I. The Added Value of Circulating Tumor Cell Enumeration to Standard Markers in Assessing Prognosis in a Metastatic Castration-Resistant Prostate Cancer Population. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 1967–1973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zedan, A.H.; Osther, P.J.S.; Assenholt, J.; Madsen, J.S.; Hansen, T.F. Circulating miR-141 and miR-375 are associated with treatment outcome in metastatic castration resistant prostate cancer. Sci. Rep. 2020, 10, 227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrandah, A.M.; Mora, R.A.; Chan, E.K.L. Emerging microRNAs in cancer diagnosis, progression, and immune surveillance. Cancer Lett. 2018, 438, 126–132. [Google Scholar] [CrossRef]
- Ciszkowicz, E.; Porzycki, P.; Semik, M.; Kaznowska, E.; Tyrka, M. MiR-93/miR-375: Diagnostic Potential, Aggressiveness Correlation and Common Target Genes in Prostate Cancer. Int. J. Mol. Sci. 2020, 21, 5667. [Google Scholar] [CrossRef] [PubMed]
- Porzycki, P.; Ciszkowicz, E.; Semik, M.; Tyrka, M. Combination of three miRNA (miR-141, miR-21, and miR-375) as potential diagnostic tool for prostate cancer recognition. Int. Urol. Nephrol. 2018, 50, 1619–1626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bidarra, D.; Constâncio, V.; Barros-Silva, D.; Ramalho-Carvalho, J.; Moreira-Barbosa, C.; Antunes, L.; Maurício, J.; Oliveira, J.; Henrique, R.; Jerónimo, C. Circulating MicroRNAs as Biomarkers for Prostate Cancer Detection and Metastasis Development Prediction. Front. Oncol. 2019, 9, 900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brase, J.C.; Johannes, M.; Schlomm, T.; Falth, M.; Haese, A.; Steuber, T.; Beissbarth, T.; Kuner, R.; Sultmann, H. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int. J. Cancer 2011, 128, 608–616. [Google Scholar] [CrossRef] [PubMed]

| Patient Characteristics at Baseline | Total (n = 93) | Iluminate Enzalutamide (n = 40) | Optimum Abiraterone Acetate (n = 53) |
|---|---|---|---|
| Age at baseline (years) | 73 (67–78) | 74 (69–78) | 71 (65–78) |
| Weight at baseline (kg) | 85.7 (79.0–92.5) | 85 (78–91) | 86 (80–93) |
| Hb (mmol/L) | 8.0 (7.5–8.5) | 8.1 (7.6–8.5) | 7.9 (7.4–8.4) |
| LDH (U/L) | 230.0 (201–264) | 230 (193–261) | 230 (206–264) |
| ALP (U/L) | 91.0 (73–125) | 94 (76–125) | 85 (71–125) |
| Albumin (g/L) | 41 (37–43) | 41 (37–43) | 41 (37–44) |
| PSA (ng/mL) | 45.0 (23.0–110) | 49 (23–95) | 39 (23–130) |
| PSA doubling time (months) | 3.3 (2.2–5.8) | 4.1 (2.3–6.4) | 3.3 (2.3–6.1) |
| DHEAS (µmol/L) | 1.7 (1.0–3.1) | 1.9 (1.1–3.6) | 1.6 (0.9–2.5) |
| Gleason score at diagnosis | |||
| ≤7 | 31 (33.3) | 19 (47.5) | 13 (24.5) |
| ≥8 | 58 (62.4) | 20 (50) | 37 (69.8) |
| Missing | 4 (4.3) | 1 (2.5) | 3 (5.7) |
| Ethnicity/Race | |||
| White | 89 (95.7) | 36 (90.0) | 53 (100) |
| Asian | 2 (2.2) | 2 (5.0) | |
| Missing | 2 (2.2) | 2 (5.0) | |
| ECOG performance status | |||
| 0 | 62 (67.4) | 28 (70.0) | 34 (64.2) |
| 1 | 27 (29.3) | 10 (25.0) | 17 (32.1) |
| 2 | 3 (3.3) | 1 (2.5) | 2 (3.8) |
| Missing | 1 (1.1) | 1 (2.5) | |
| Pre-treatment docetaxel a | 19 (20.4) | 3 (7.5) | 16 (30.2) |
| Previous treatments | |||
| Prostatectomy | 41 (44.1) | 21 (52.5) | 20 (37.7) |
| Radiation | 38 (40.9) | 17 (42.5) | 21 (39.6) |
| Anti-androgenpre-treatment | 49 (52.7) | 11 (27.5) | 38 (71.7) |
| Other b | 4 (4.3) | 3 (7.5) | 1 (1.9) |
| Missing | 34 (36.6) | 19 (47.5) | 15 (28.3) |
| Spread of disease | |||
| Lymph only | 14 (15.1) | 9 (22.5) | 5 (9.4) |
| Bone only | 25 (26.9) | 11 (27.5) | 14 (26.4) |
| Both bone and lymph | 36 (38.7) | 12 (30.0) | 24 (45.3) |
| Visceral + lymph node/bone | 16 (17.2) | 7 (17.5) | 9 (17.0) |
| Median time to progressionMonths (95% CI) | 14.5 (11.8–17.2) | 15.2 (6.1–24.3) | 13.4 (10.0–16.8) |
| Median overall survivalMonths (95% CI) | 28.4 (24.6–32.2) | 26.8 (18.3–35.2) | 28.4 (23.9–33.0) |
| Median follow-up timeMonths (range) | 27.4 (3.0–62.4) | 27.7 (7.8–62.4) | 27.4 (3.0–60.8) |
| Model Variables | Progression Free Survival | Overall Survival | ||
|---|---|---|---|---|
| Harell’s C-Index # | Standard Error | Harell’s C-Index | Standard Error | |
| Model 1: clinical parameters * only | 0.71 | 0.030 | 0.70 | 0.036 |
| Model 2: model 1 + KLK3 | 0.73 | 0.029 | 0.72 | 0.034 |
| Model 3: model 1 + miR-375 | 0.74 | 0.029 | 0.72 | 0.035 |
| Model 4: model 1 + miR-3687 | 0.73 | 0.031 | 0.70 | 0.035 |
| Model 5: model 1 + NAALADL2-AS2 | 0.71 | 0.030 | 0.71 | 0.036 |
| Model 6: model 1 + miR-375 + KLK3 | 0.74 | 0.028 | 0.74 | 0.034 |
| Model 7: model 1 + miR-375 + NAALADL2-AS2 | 0.73 | 0.030 | 0.73 | 0.035 |
| Model 8: model 1 + KLK3 + NAALADL2-AS2 | 0.72 | 0.030 | 0.72 | 0.035 |
| Model 9: model 1 + miR-375 + KLK3 + NAALADL2-AS2 | 0.74 | 0.029 | 0.74 | 0.034 |
| Variables | Progression Free Survival | Overall Survival | ||
|---|---|---|---|---|
| HR (95% CI) | p-Value | HR (95% CI) | p-Value | |
| Baseline log PSA | 1.29 (0.83–2.00) | 0.26 | 1.32 (0.79–2.20) | 0.28 |
| Baseline log Hb | 9.06 × 10−5 (7.84 × 10−7–0.01) | <0.001 | 5.81 × 10−6 (3.37 × 10−8–1.00 × 10−3) | <0.001 |
| Baseline log ALP | 1.71 (0.55–5.29) | 0.35 | 2.46 (0.71–8.47) | 0.15 |
| Baseline log LDH | 0.23 (1.12 × 10−2–4.56) | 0.32 | 0.02 (9.17 × 10−4–0.34) | 0.009 |
| Delta PSA at 1 month | 2.97 (1.65–5.34) | <0.001 | 1.65 (0.93–2.91) | 0.08 |
| KLK3 detectable at baseline | 0.89 (0.42–1.86) | 0.75 | 1.53 (0.75–3.14) | 0.24 |
| KLK3 detectable at 1 month | 4.00 (1.47–10.85) | 0.007 | 1.50 (0.58–3.90) | 0.40 |
| Baseline log miR-375 | 1.93 (1.26–2.95) | 0.003 | 1.27 (0.78–2.08) | 0.33 |
| Delta miR-375 at 1 month | 0.97 (0.65–1.46) | 0.89 | 0.75 (0.44–1.28) | 0.29 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boerrigter, E.; Benoist, G.E.; van Oort, I.M.; Verhaegh, G.W.; de Haan, A.F.J.; van Hooij, O.; Groen, L.; Smit, F.; Oving, I.M.; de Mol, P.; et al. RNA Biomarkers as a Response Measure for Survival in Patients with Metastatic Castration-Resistant Prostate Cancer. Cancers 2021, 13, 6279. https://doi.org/10.3390/cancers13246279
Boerrigter E, Benoist GE, van Oort IM, Verhaegh GW, de Haan AFJ, van Hooij O, Groen L, Smit F, Oving IM, de Mol P, et al. RNA Biomarkers as a Response Measure for Survival in Patients with Metastatic Castration-Resistant Prostate Cancer. Cancers. 2021; 13(24):6279. https://doi.org/10.3390/cancers13246279
Chicago/Turabian StyleBoerrigter, Emmy, Guillemette E. Benoist, Inge M. van Oort, Gerald W. Verhaegh, Anton F. J. de Haan, Onno van Hooij, Levi Groen, Frank Smit, Irma M. Oving, Pieter de Mol, and et al. 2021. "RNA Biomarkers as a Response Measure for Survival in Patients with Metastatic Castration-Resistant Prostate Cancer" Cancers 13, no. 24: 6279. https://doi.org/10.3390/cancers13246279
APA StyleBoerrigter, E., Benoist, G. E., van Oort, I. M., Verhaegh, G. W., de Haan, A. F. J., van Hooij, O., Groen, L., Smit, F., Oving, I. M., de Mol, P., Smilde, T. J., Somford, D. M., Hamberg, P., Dezentjé, V. O., Mehra, N., van Erp, N. P., & Schalken, J. A. (2021). RNA Biomarkers as a Response Measure for Survival in Patients with Metastatic Castration-Resistant Prostate Cancer. Cancers, 13(24), 6279. https://doi.org/10.3390/cancers13246279









