RalGPS2 Interacts with Akt and PDK1 Promoting Tunneling Nanotubes Formation in Bladder Cancer and Kidney Cells Microenvironment
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmids and Constructs
2.2. Antibodies
2.3. Cell Cultures
2.4. Tunneling Nanotubes Analysis in Bladder Cancer Cell Lines
2.5. Mitochondrial Transfer via TNTs
2.6. Transfection
2.7. Time-Lapse Imaging
2.8. Western Blot Analysis
2.9. Co-Immunoprecipitation Assay
2.10. Stress Media Composition and Assay
2.11. RNA Interference
2.12. TNTs Analysis under Pharmacologically Perturbed Conditions
3. Results
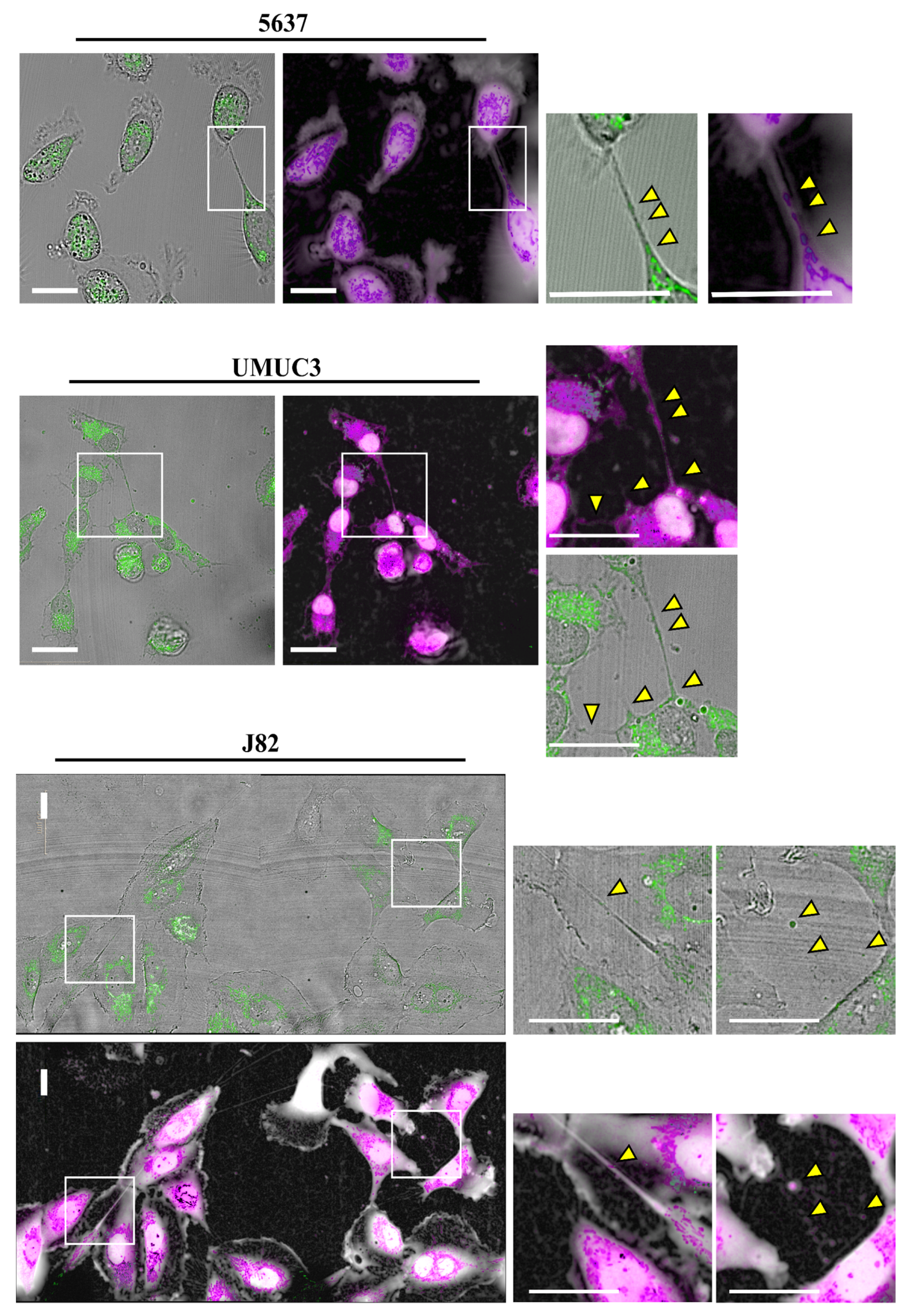
3.1. Tunneling Nanotubes Formation in Bladder Cancer Cell Lines
3.2. Mitochondrial Transfer in Bladder Cancer Cell Lines through TNTs
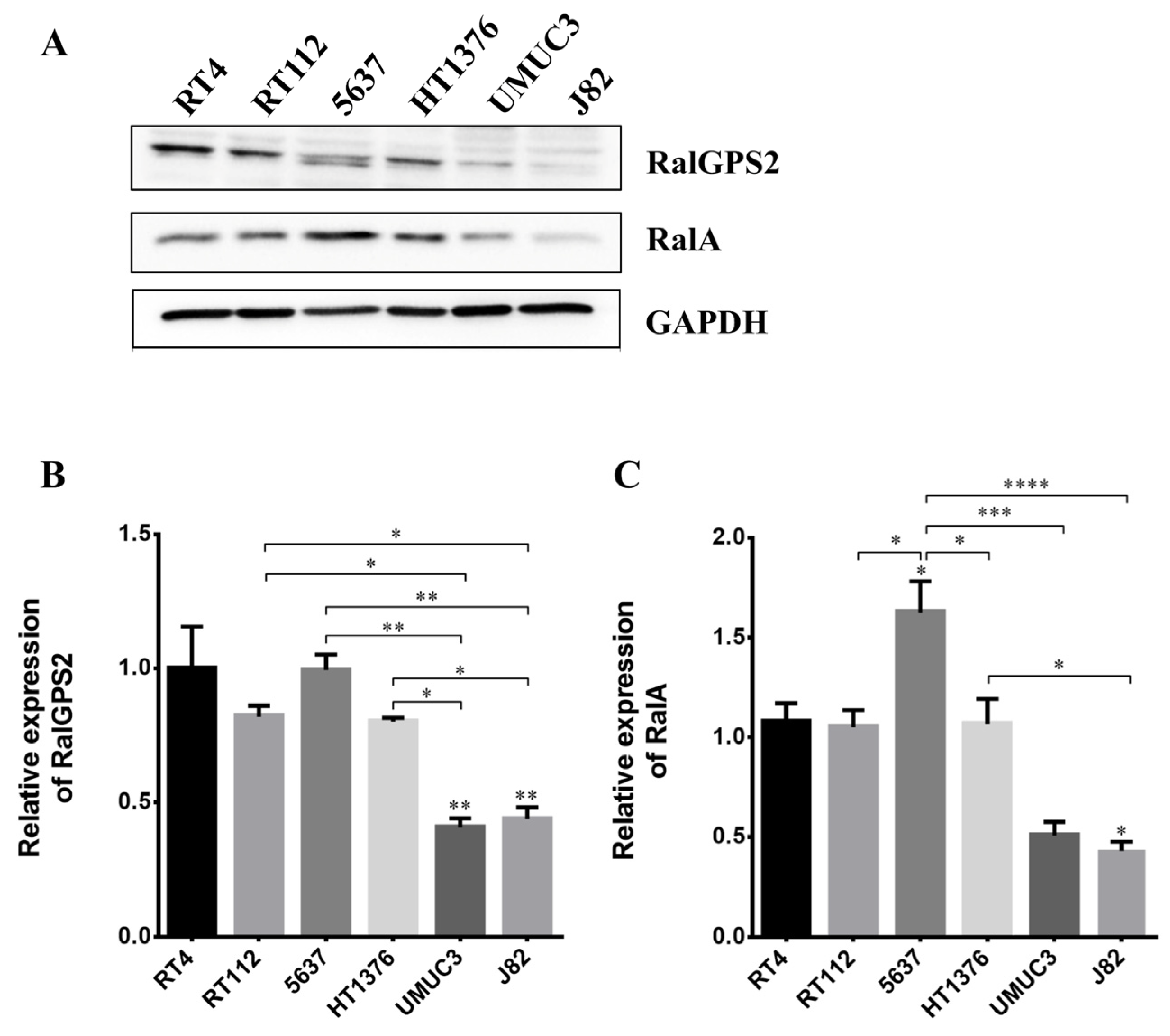
3.3. Expression of Proteins Involved in Tunneling Nanotubes Formation in Bladder Cancer Cells
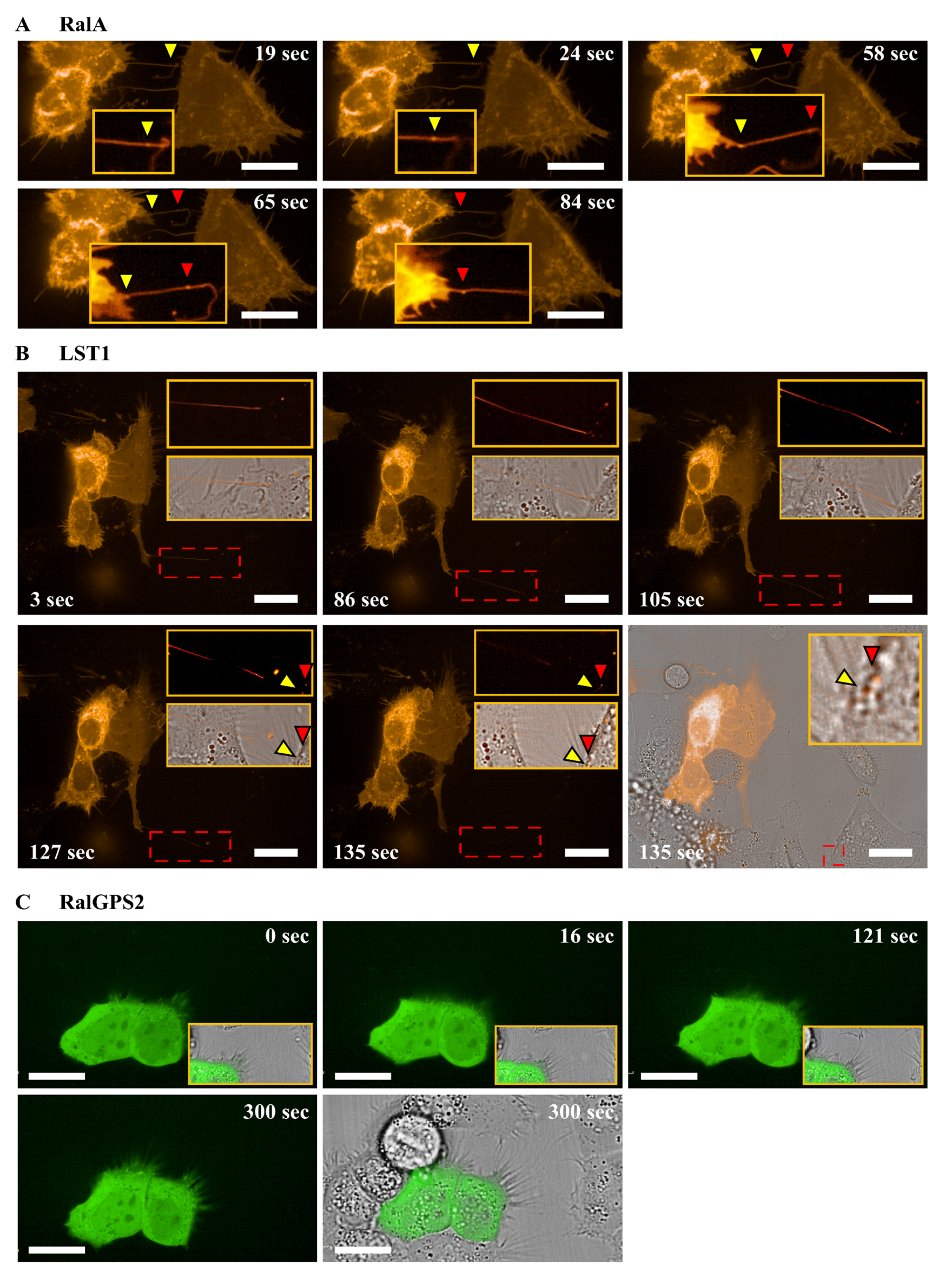
3.4. RalA and LST1 Are Transferred via TNTs in 5637 Cells
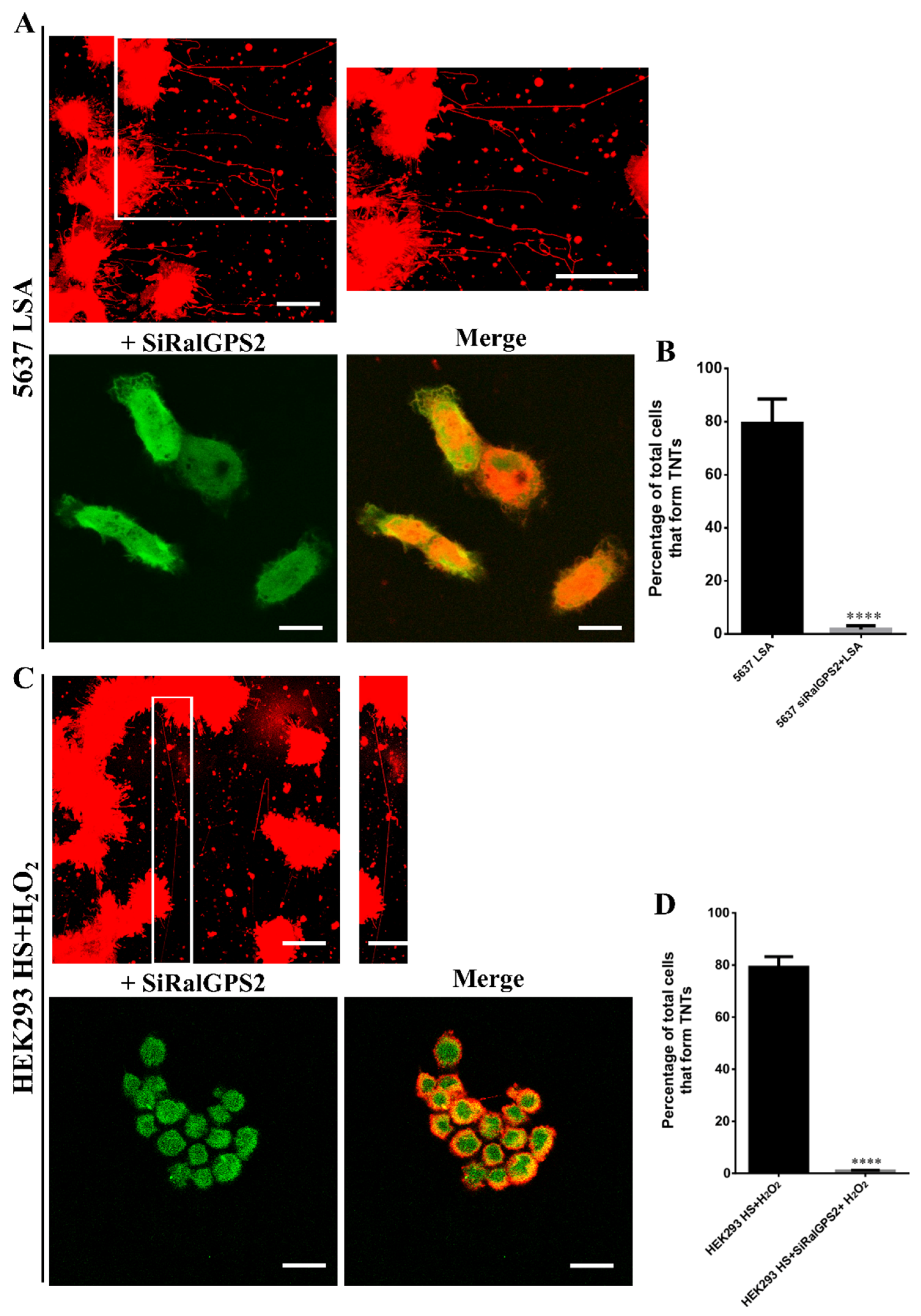
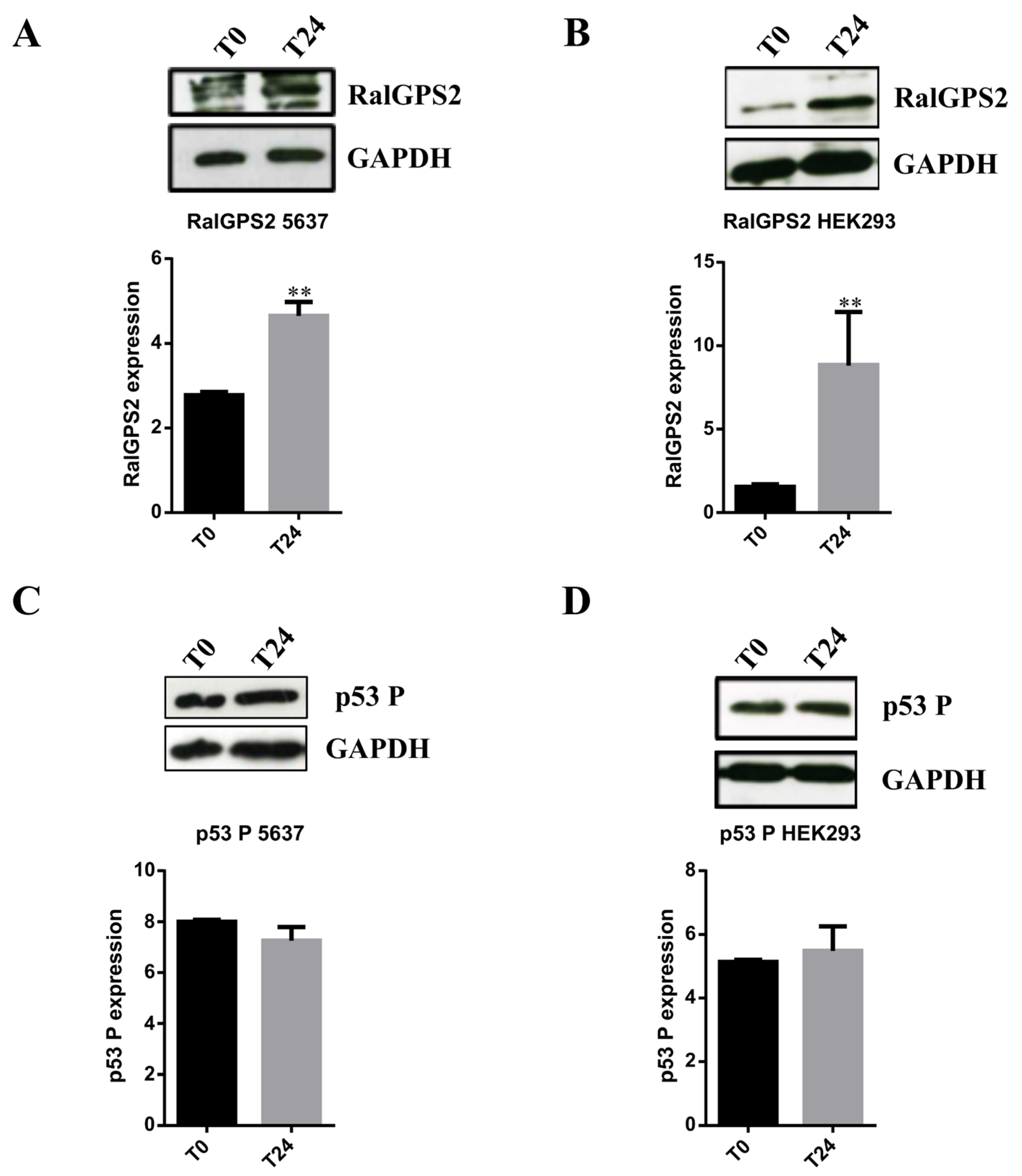
3.5. Stress Conditions Promote TNTs formation in HEK293 and 5637 Cells
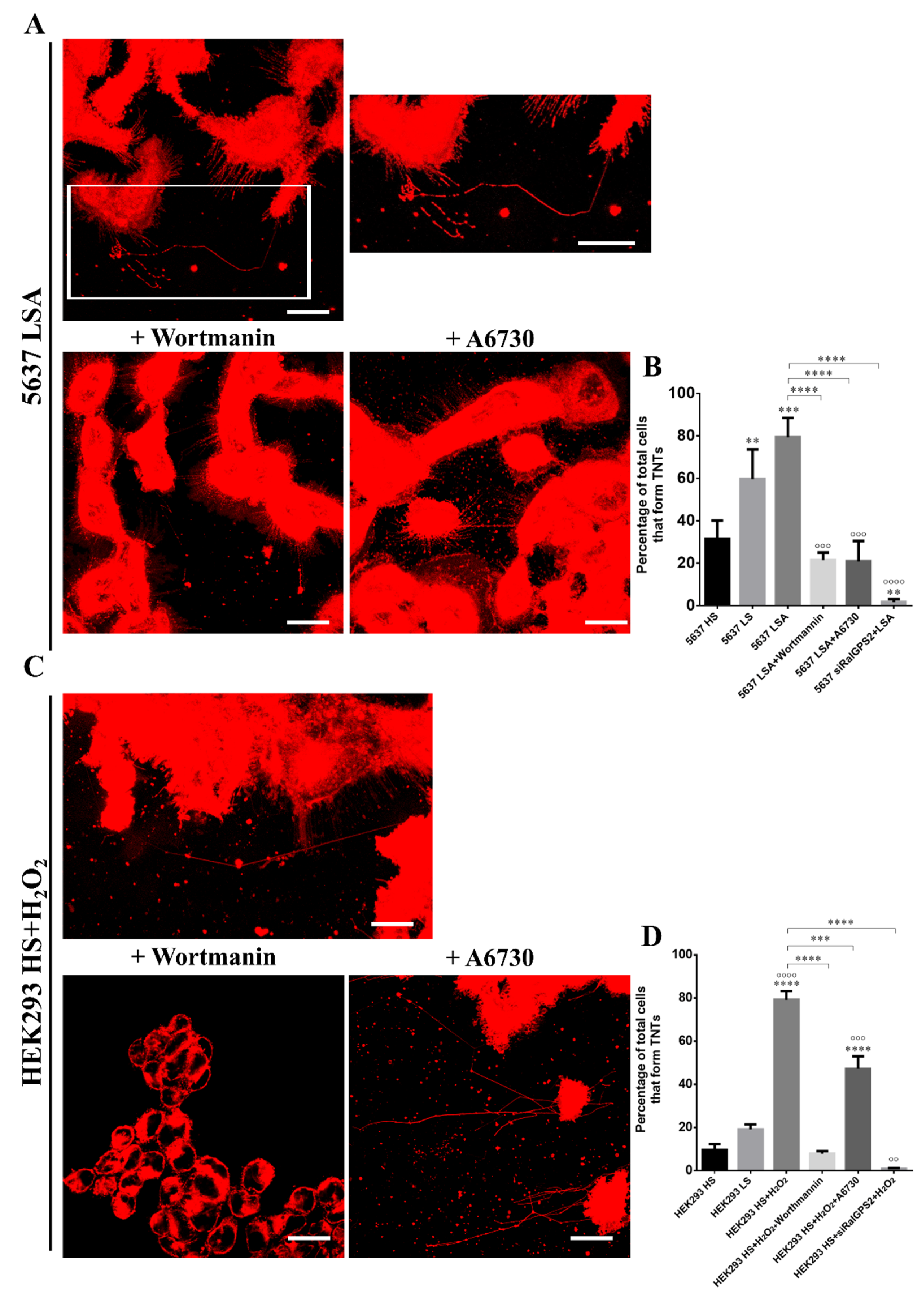
3.6. RalGPS2 Is Crucial to TNTs Formation Also in Stress Conditions in 5637 and HEK293 Cells
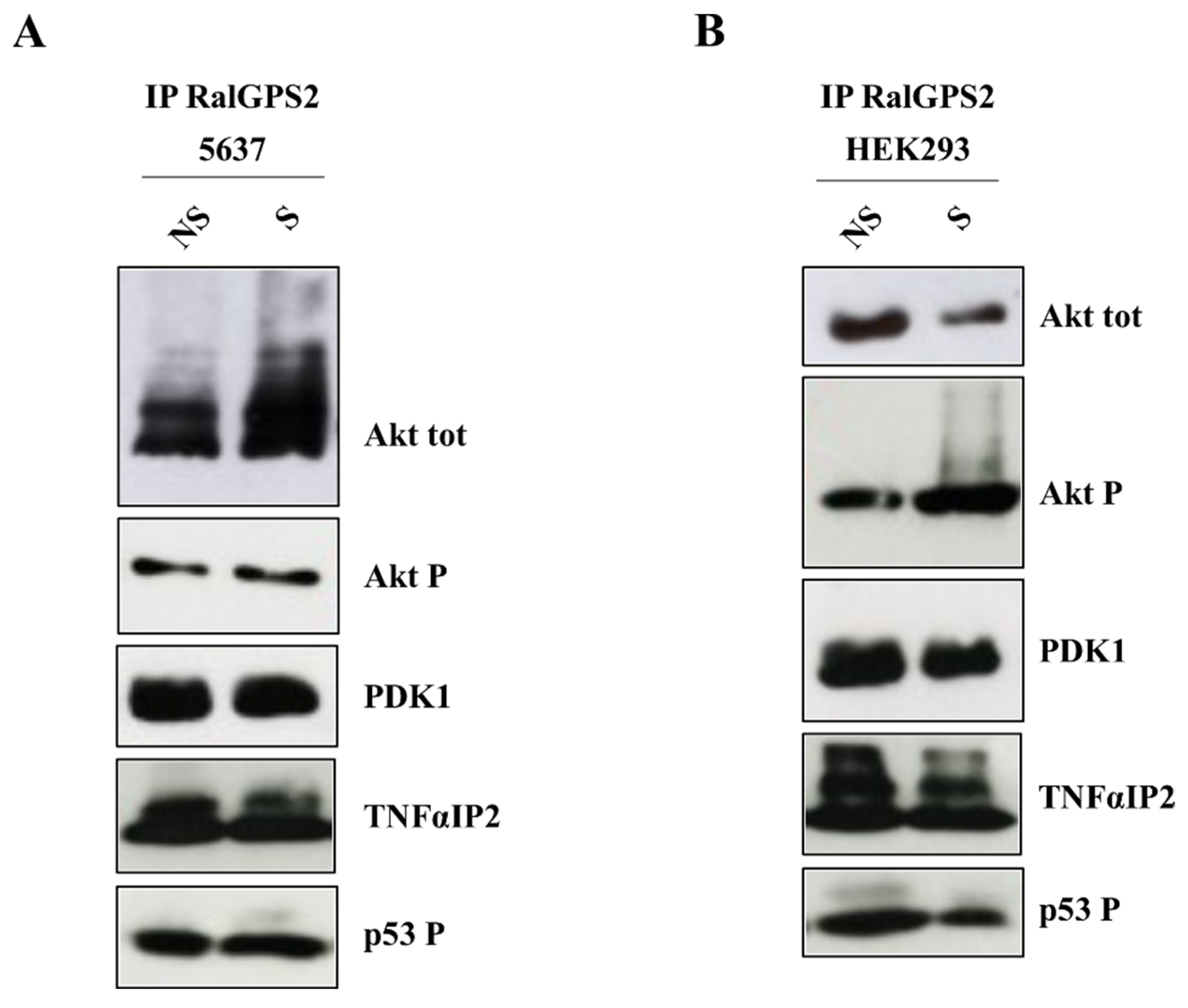
3.7. PI3K and Akt Inhibition Reduces TNTs Formation in HEK293 and 5637 Cells
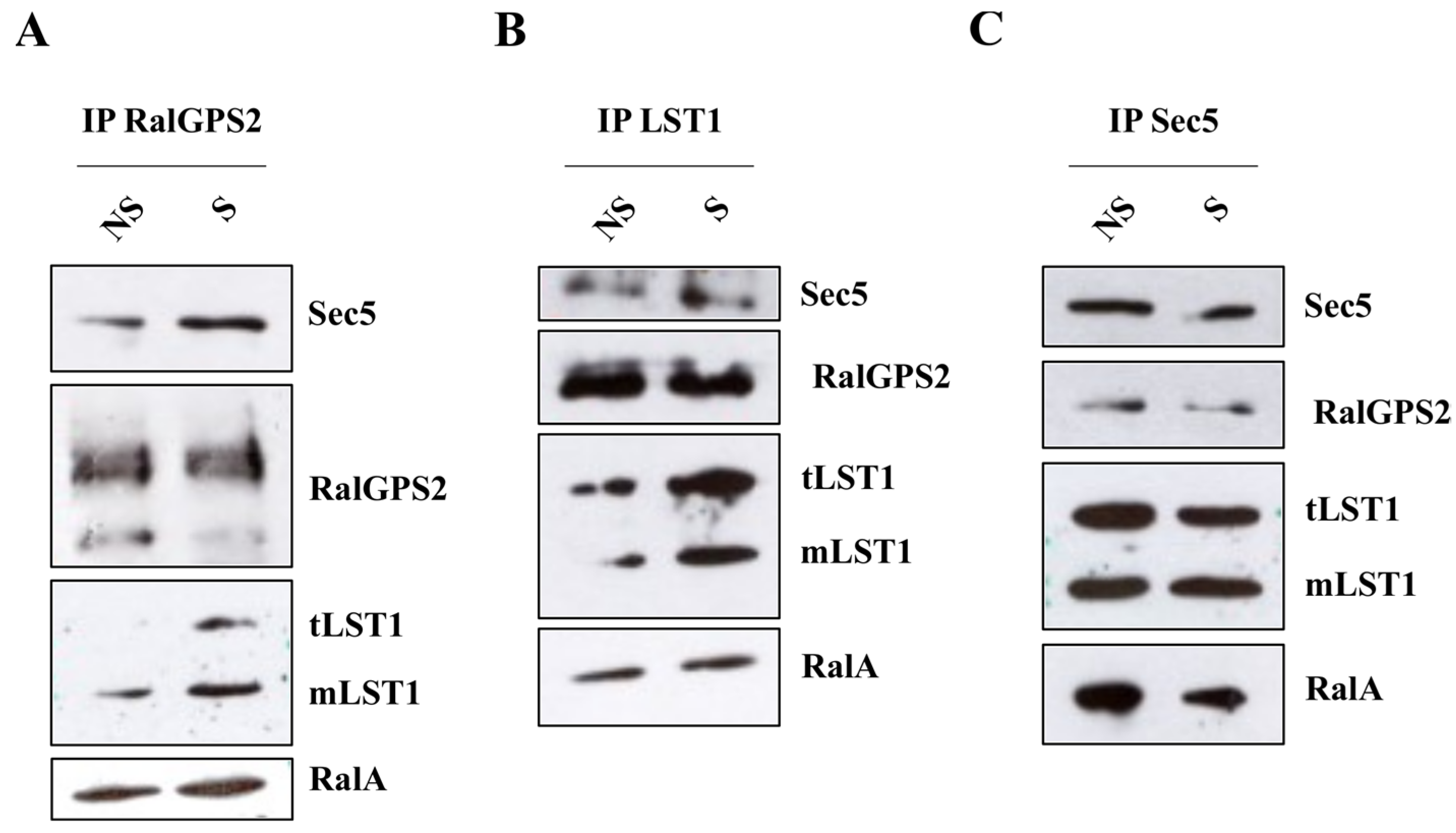
3.8. RalGPS2 Forms a Complex with LST1, RalA and Sec5 in HEK293 Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Jalanko, T.; de Jong, J.J.; Gibb, E.A.; Seiler, R.; Black, P.C. Genomic Subtyping in Bladder Cancer. Curr. Urol. Rep. 2020, 21, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Van Den Bosch, S.; Witjes, J.A. Long-Term Cancer-Specific Survival in Patients with High-Risk, Non-Muscle-Invasive Bladder Cancer and Tumour Progression: A Systematic Review. Eur. Urol. 2011, 60, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Alfred Witjes, J.; Lebret, T.; Compérat, E.M.; Cowan, N.C.; De Santis, M.; Bruins, H.M.; Hernández, V.; Espinós, E.L.; Dunn, J.; Rouanne, M.; et al. Updated 2016 EAU Guidelines on Muscle-Invasive and Metastatic Bladder Cancer. Eur. Urol. 2017, 71, 462–475. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, C.; Yang, X. Endoscopic Molecular Imaging plus Photoimmunotherapy: A New Strategy for Monitoring and Treatment of Bladder Cancer. Mol. Ther.-Oncolytics 2020, 18, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Dieleman, J.; Campbell, M.; Chapin, A.; Eldrenkamp, E.; Fan, V.Y.; Haakenstad, A.; Kates, J.; Liu, Y.; Matyasz, T.; Micah, A.; et al. Evolution and Patterns of Global Health Financing 1995-2014: Development Assistance for Health, and Government, Prepaid Private, and out-of-Pocket Health Spending in 184 Countries. Lancet 2017, 389, 1981–2004. [Google Scholar] [CrossRef] [Green Version]
- Lee, L.J.; Kwon, C.S.; Forsythe, A.; Mamolo, C.M.; Masters, E.T.; Jacobs, I.A. Humanistic and Economic Burden of Non-Muscle Invasive Bladder Cancer: Results of Two Systematic Literature Reviews. Clin. Outcomes Res. 2020, 12, 693–709. [Google Scholar] [CrossRef]
- Nishida-Aoki, N.; Gujral, T.S. Review Emerging Approaches to Study Cell–Cell Interactions in Tumor Microenvironment. Oncotarget 2019, 10, 785–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rustom, A.; Saffrich, R.; Markovic, I.; Walther, P.; Gerdes, H.H. Nanotubular Highways for Intercellular Organelle Transport. Science 2004, 303, 1007–1010. [Google Scholar] [CrossRef] [Green Version]
- Roehlecke, C.; Schmidt, M.H.H. Tunneling Nanotubes and Tumor Microtubes in Cancer. Cancers 2020, 12, 857. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, T.; Mukherjee, S.; Pattnaik, B.; Kumar, M.; Singh, S.; Rehman, R.; Tiwari, B.K.; Jha, K.A.; Barhanpurkar, A.P.; Wani, M.R.; et al. Miro1 Regulates Intercellular Mitochondrial Transport & Enhances Mesenchymal Stem Cell Rescue Efficacy. EMBO J. 2014, 33, 994–1010. [Google Scholar] [CrossRef]
- Abounit, S.; Bousset, L.; Loria, F.; Zhu, S.; Chaumont, F.; Pieri, L.; Olivo-Marin, J.; Melki, R.; Zurzolo, C. Tunneling Nanotubes Spread Fibrillar A-synuclein by Intercellular Trafficking of Lysosomes. EMBO J. 2016, 35, 2120–2138. [Google Scholar] [CrossRef]
- Bhat, S.; Ljubojevic, N.; Zhu, S.; Fukuda, M.; Echard, A.; Zurzolo, C. Rab35 and Its Effectors Promote Formation of Tunneling Nanotubes in Neuronal Cells. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Schiller, C.; Huber, J.E.; Diakopoulos, K.N.; Weiss, E.H. Tunneling Nanotubes Enable Intercellular Transfer of MHC Class I Molecules. Hum. Immunol. 2013, 74, 412–416. [Google Scholar] [CrossRef]
- Sowinski, S.; Jolly, C.; Berninghausen, O.; Purbhoo, M.A.; Chauveau, A.; Köhler, K.; Oddos, S.; Eissmann, P.; Brodsky, F.M.; Hopkins, C.; et al. Membrane Nanotubes Physically Connect T Cells over Long Distances Presenting a Novel Route for HIV-1 Transmission. Nat. Cell Biol. 2008, 10, 211–219. [Google Scholar] [CrossRef]
- Ariazi, J.; Benowitz, A.; De Biasi, V.; Den Boer, M.L.; Cherqui, S.; Cui, H.; Douillet, N.; Eugenin, E.A.; Favre, D.; Goodman, S.; et al. Tunneling Nanotubes and Gap Junctions–Their Role in Long-Range Intercellular Communication during Development, Health, and Disease Conditions. Front. Mol. Neurosci. 2017, 10, 333. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.J.; Yang, W.M.; Li, F.; Zhu, W.; Chen, Z. Tunneling Nanotubes Mediated MicroRNA-155 Intercellular Transportation Promotes Bladder Cancer Cells’ Invasive and Proliferative Capacity. Int. J. Nanomed. 2019, 14, 9731–9743. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Gerdes, H.H. Transfer of Mitochondria via Tunneling Nanotubes Rescues Apoptotic PC12 Cells. Cell Death Differ. 2015, 22, 1181–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lou, E.; Fujisawa, S.; Morozov, A.; Barlas, A.; Romin, Y.; Dogan, Y.; Gholami, S.; Moreira, A.L.; Manova-Todorova, K.; Moore, M.A.S. Tunneling Nanotubes Provide a Unique Conduit for Intercellular Transfer of Cellular Contents in Human Malignant Pleural Mesothelioma. PLoS ONE 2012, 7, e33093. [Google Scholar] [CrossRef] [Green Version]
- Gousset, K.; Schiff, E.; Langevin, C.; Marijanovic, Z.; Caputo, A.; Browman, D.T.; Chenouard, N.; de Chaumont, F.; Martino, A.; Enninga, J.; et al. Prions Hijack Tunnelling Nanotubes for Intercellular Spread. Nat. Cell Biol. 2009, 11, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, K.; Khandare, A.; Burianovskyy, L.; Maruyama, S.; Zhang, F.; Nasjletti, A.; Goligorsky, M.S. Tunneling Nanotubes Mediate Rescue of Prematurely Senescent Endothelial Cells by Endothelial Progenitors: Exchange of Lysosomal Pool. Aging 2011, 3, 597–608. [Google Scholar] [CrossRef] [Green Version]
- Plotnikov, E.Y.; Khryapenkova, T.G.; Galkina, S.I.; Sukhikh, G.T.; Zorov, D.B. Cytoplasm and Organelle Transfer between Mesenchymal Multipotent Stromal Cells and Renal Tubular Cells in Co-Culture. Exp. Cell Res. 2010, 316, 2447–2455. [Google Scholar] [CrossRef]
- Ranzinger, J.; Rustom, A.; Abel, M.; Leyh, J.; Kihm, L.; Witkowski, M.; Scheurich, P.; Zeier, M.; Schwenger, V. Nanotube Action between Human Mesothelial Cells Reveals Novel Aspects of Inflammatory Responses. PLoS ONE 2011, 6, e29537. [Google Scholar] [CrossRef] [Green Version]
- Salter, R.D.; Watkins, S.C. Dynamic Properties of Antigen Between Dendritic Cells. Immunol. Res. 2006, 36, 211–220. [Google Scholar] [CrossRef]
- Xu, W.; Santini, P.A.; Sullivan, J.S.; He, B.; Shan, M.; Ball, S.C.; Dyer, W.B.; Ketas, T.J.; Chadburn, A.; Cohen-Gould, L.; et al. HIV-1 Evades Virus-Specific IgG2 and IgA Responses by Targeting Systemic and Intestinal B Cells via Long-Range Intercellular Conduits. Nat. Immunol. 2009, 10, 1008–1017. [Google Scholar] [CrossRef] [Green Version]
- Gurke, S.; Barroso, J.F.V.; Hodneland, E.; Bukoreshtliev, N.V.; Schlicker, O.; Gerdes, H.H. Tunneling Nanotube (TNT)-like Structures Facilitate a Constitutive, Actomyosin-Dependent Exchange of Endocytic Organelles between Normal Rat Kidney Cells. Exp. Cell Res. 2008, 314, 3669–3683. [Google Scholar] [CrossRef]
- Thayanithy, V.; Dickson, E.L.; Steer, C.; Subramanian, S.; Lou, E. Tumor-Stromal Cross Talk: Direct Cell-to-Cell Transfer of Oncogenic MicroRNAs via Tunneling Nanotubes. Transl. Res. 2014, 164, 359–365. [Google Scholar] [CrossRef] [Green Version]
- Thayanithy, V.; Babatunde, V.; Dickson, E.L.; Wong, P.; Oh, S.; Ke, X.; Barlas, A.; Fujisawa, S.; Romin, Y.; Moreira, A.L.; et al. Tumor Exosomes Induce Tunneling Nanotubes in Lipid Raft-Enriched Regions of Human Mesothelioma Cells. Exp. Cell Res. 2014, 323, 178–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desir, S.; O’Hare, P.; Vogel, R.I.; Sperduto, W.; Sarkari, A.; Dickson, E.L.; Wong, P.; Nelson, A.C.; Fong, Y.; Steer, C.J.; et al. Chemotherapy-Induced Tunneling Nanotubes Mediate Intercellular Drug Efflux in Pancreatic Cancer. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef]
- Vidulescu, C.; Clejan, S.; O’Connor, K.C. Vesicle Traffic through Intercellular Bridges in DU 145 Human Prostate Cancer Cells. J. Cell. Mol. Med. 2004, 8, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Önfelt, B.; Nedvetzki, S.; Benninger, R.K.P.; Purbhoo, M.A.; Sowinski, S.; Hume, A.N.; Seabra, M.C.; Neil, M.A.A.; French, P.M.W.; Davis, D.M. Structurally Distinct Membrane Nanotubes between Human Macrophages Support Long-Distance Vesicular Traffic or Surfing of Bacteria. J. Immunol. 2006, 177, 8476–8483. [Google Scholar] [CrossRef] [Green Version]
- Gerdes, H.H.; Carvalho, R.N. Intercellular Transfer Mediated by Tunneling Nanotubes. Curr. Opin. Cell Biol. 2008, 20, 470–475. [Google Scholar] [CrossRef] [PubMed]
- Veranič, P.; Lokar, M.; Schütz, G.J.; Weghuber, J.; Wieser, S.; Hägerstrand, H.; Kralj-Iglič, V.; Iglič, A. Different Types of Cell-to-Cell Connections Mediated by Nanotubular Structures. Biophys. J. 2008, 95, 4416–4425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Veruki, M.L.; Bukoreshtliev, N.V.; Hartveit, E.; Gerdes, H.H. Animal Cells Connected by Nanotubes Can Be Electrically Coupled through Interposed Gap-Junction Channels. Proc. Natl. Acad. Sci. USA 2010, 107, 17194–17199. [Google Scholar] [CrossRef] [Green Version]
- Gousset, K.; Marzo, L.; Commere, P.H.; Zurzolo, C. Myo10 Is a Key Regulator of TNT Formation in Neuronal Cells. J. Cell Sci. 2013, 126, 4424–4435. [Google Scholar] [CrossRef] [Green Version]
- Schiller, C.; Diakopoulos, K.N.; Rohwedder, I.; Kremmer, E.; von Toerne, C.; Ueffing, M.; Weidle, U.H.; Ohno, H.; Weiss, E.H. LST1 Promotes the Assembly of a Molecular Machinery Responsible for Tunneling Nanotube Formation. J. Cell Sci. 2013, 126, 767–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohno, H.; Hase, K.; Kimura, S. M-Sec: Emerging Secrets of Tunneling Nanotube Formation. Commun. Integr. Biol. 2010, 3, 231–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mittal, R.; Karhu, E.; Wang, J.S.; Delgado, S.; Zukerman, R.; Mittal, J.; Jhaveri, V.M. Cell Communication by Tunneling Nanotubes: Implications in Disease and Therapeutic Applications. J. Cell. Physiol. 2019, 234, 1130–1146. [Google Scholar] [CrossRef]
- Andresen, V.; Wang, X.; Ghimire, S.; Omsland, M.; Gjertsen, B.T.; Gerdes, H.H. Tunneling Nanotube (TNT) Formation Is Independent of P53 Expression. Cell Death Differ. 2013, 20, 1124. [Google Scholar] [CrossRef] [Green Version]
- D’Aloia, A.; Berruti, G.; Costa, B.; Schiller, C.; Ambrosini, R.; Pastori, V.; Martegani, E.; Ceriani, M. RalGPS2 Is Involved in Tunneling Nanotubes Formation in 5637 Bladder Cancer Cells. Exp. Cell Res. 2018, 362, 349–361. [Google Scholar] [CrossRef]
- Albright, C.F.; Giddings, B.W.; Liu, J.; Vito, M.; Weinberg, R.A. Characterization of a Guanine Nucleotide Dissociation Stimulator for a Ras-Related GTPase. EMBO J. 1993, 12, 339–347. [Google Scholar] [CrossRef]
- Hernandez-Muñoz, I.; Malumbres, M.; Leonardi, P.; Pellicer, A. The Rgr Oncogene (Homologous to RalGDS) Induces Transformation and Gene Expression by Activating Ras, Ral and Rho Mediated Pathways. Oncogene 2000, 19, 2745–2757. [Google Scholar] [CrossRef] [Green Version]
- Wolthuis, R.M.F.; De Ruiter, N.D.; Cool, R.H.; Bos, J.L. Stimulation of Gene Induction and Cell Growth by the Ras Effector Rlf. EMBO J. 1997, 16, 6748–6761. [Google Scholar] [CrossRef] [Green Version]
- Hase, K.; Kimura, S.; Takatsu, H.; Ohmae, M.; Kawano, S.; Kitamura, H.; Ito, M.; Watarai, H.; Hazelett, C.C.; Yeaman, C.; et al. M-Sec Promotes Membrane Nanotube Formation by Interacting with Ral and the Exocyst Complex. Nat. Cell Biol. 2009, 11, 1427–1432. [Google Scholar] [CrossRef] [PubMed]
- Ceriani, M.; Scandiuzzi, C.; Amigoni, L.; Tisi, R.; Berruti, G.; Martegani, E. Functional Analysis of RalGPS2, a Murine Guanine Nucleotide Exchange Factor for RalA GTPase. Exp. Cell Res. 2007, 313, 2293–2307. [Google Scholar] [CrossRef] [PubMed]
- Neyraud, V.; Aushev, V.N.; Hatzoglou, A.; Meunier, B.; Cascone, I.; Camonis, J. RalA and RalB Proteins Are Ubiquitinated GTPases, and Ubiquitinated RalA Increases Lipid Raft Exposure at the Plasma Membrane. J. Biol. Chem. 2012, 287, 29397–29405. [Google Scholar] [CrossRef] [Green Version]
- Schiller, C.; Nitschké, M.J.E.; Seidl, A.; Kremmer, E.; Weiss, E.H. Rat Monoclonal Antibodies Specific for LST1 Proteins. Hybridoma 2009, 28, 281–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuiverloon, T.C.M.; De Jong, F.C.; Costello, J.C.; Theodorescu, D. Systematic Review: Characteristics and Preclinical Uses of Bladder Cancer Cell Lines. Bladder Cancer 2018, 4, 169–183. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Cui, J.; Sun, X.; Zhang, Y. Tunneling-Nanotube Development in Astrocytes Depends on P53 Activation. Cell Death Differ. 2011, 18, 732–742. [Google Scholar] [CrossRef] [Green Version]
- Desir, S.; Wong, P.; Turbyville, T.; Chen, D.; Shetty, M.; Clark, C.; Zhai, E.; Romin, Y.; Manova-Todorova, K.; Starr, T.K.; et al. Intercellular Transfer of Oncogenic KRAS via Tunneling Nanotubes Introduces Intracellular Mutational Heterogeneity in Colon Cancer Cells. Cancers 2019, 11, 892. [Google Scholar] [CrossRef] [Green Version]
- Rainy, N.; Chetrit, D.; Rouger, V.; Vernitsky, H.; Rechavi, O.; Marguet, D.; Goldstein, I.; Ehrlich, M.; Kloog, Y. H-Ras Transfers from B to T Cells via Tunneling Nanotubes. Cell Death Dis. 2013, 4, e726. [Google Scholar] [CrossRef] [PubMed]
- Hekmatshoar, Y.; Nakhle, J.; Galloni, M.; Vignais, M.L. The Role of Metabolism and Tunneling Nanotube-Mediated Intercellular Mitochondria Exchange in Cancer Drug Resistance. Biochem. J. 2018, 475, 2305–2328. [Google Scholar] [CrossRef]
- Parker, I.; Evans, K.T.; Ellefsen, K.; Lawson, D.A.; Smith, I.F. Lattice Light Sheet Imaging of Membrane Nanotubes between Human Breast Cancer Cells in Culture and in Brain Metastases. Sci. Rep. 2017, 7, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Bénard, M.; Schapman, D.; Lebon, A.; Monterroso, B.; Bellenger, M.; Le Foll, F.; Pasquier, J.; Vaudry, H.; Vaudry, D.; Galas, L. Structural and Functional Analysis of Tunneling Nanotubes (TnTs) Using GCW STED and Gconfocal Approaches. Biol. Cell 2015, 107, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Pinto, G.; Saenz-De-Santa-Maria, I.; Chastagner, P.; Perthame, E.; Delmas, C.; Toulas, C.; Moyal-Jonathan-Cohen, E.; Brou, C.; Zurzolo, C. Patient-Derived Glioblastoma Stem Cells Transfer Mitochondria through Tunneling Nanotubes in Tumor Organoids. Biochem. J. 2021, 478, 21–39. [Google Scholar] [CrossRef] [PubMed]
- Vignais, M.L.; Caicedo, A.; Brondello, J.M.; Jorgensen, C. Cell Connections by Tunneling Nanotubes: Effects of Mitochondrial Trafficking on Target Cell Metabolism, Homeostasis, and Response to Therapy. Stem. Cells Int. 2017, 2017. [Google Scholar] [CrossRef] [Green Version]
- Torab, P.; Yan, Y.; Yamashita, H.; Warrick, J.I.; Raman, J.D.; Degraff, D.J.; Wong, P.K. Three-Dimensional Microtumors for Probing Heterogeneity of Invasive Bladder Cancer. Anal. Chem. 2020, 92, 8768–8775. [Google Scholar] [CrossRef] [PubMed]
- Joneson, T.; White, M.A.; Wigler, M.H.; Bar-Sagi, D. Stimulation of Membrane Ruffling and MAP Kinase Activation by Distinct Effectors of RAS. Science 1996, 271, 810–812. [Google Scholar] [CrossRef] [PubMed]
- Desir, S.; Dickson, E.L.; Vogel, R.I.; Thayanithy, V.; Wong, P.; Teoh, D.; Geller, M.A.; Steer, C.J.; Subramanian, S.; Lou, E. Tunneling Nanotube Formation Is Stimulated by Hypoxia in Ovarian Cancer Cells. Oncotarget 2016, 7, 43150–43161. [Google Scholar] [CrossRef] [Green Version]
- Sisakhtnezhad, S.; Khosravi, L. Emerging Physiological and Pathological Implications of Tunneling Nanotubes Formation between Cells. Eur. J. Cell Biol. 2015, 94, 429–443. [Google Scholar] [CrossRef]
- Ceriani, M.; Amigoni, L.; Scandiuzzi, C.; Berruti, G.; Martegani, E. The PH-PxxP Domain of RalGPS2 Promotes PC12 Cells Differentiation Acting as a Dominant Negative for RalA GTPase Activation. Neurosci. Res. 2010, 66, 290–298. [Google Scholar] [CrossRef]
- Nguyen, D.; Liao, W.; Zeng, S.X.; Lu, H. Reviving the Guardian of the Genome: Small Molecule Activators of P53. Pharmacol. Ther. 2017, 178, 92–108. [Google Scholar] [CrossRef]
- Yuan, Y.; Jiang, Y.C.; Sun, C.K.; Chen, Q.M. Role of the Tumor Microenvironment in Tumor Progression and the Clinical Applications (Review). Oncol. Rep. 2016, 35, 2499–2515. [Google Scholar] [CrossRef] [Green Version]
- Cirri, P.; Chiarugi, P. Cancer Associated Fibroblasts: The Dark Side of the Coin. Am. J. Cancer Res. 2011, 1, 482–497. [Google Scholar] [PubMed]
- Cirri, P.; Chiarugi, P. Cancer-Associated-Fibroblasts and Tumour Cells: A Diabolic Liaison Driving Cancer Progression. Cancer Metastasis Rev. 2012, 31, 195–208. [Google Scholar] [CrossRef]
- Lou, E. A Ticket to Ride: The Implications of Direct Intercellular Communication via Tunneling Nanotubes in Peritoneal and Other Invasive Malignancies. Front. Oncol. 2020, 10, 2478. [Google Scholar] [CrossRef] [PubMed]
- Pinto, G.; Brou, C.; Zurzolo, C. Tunneling Nanotubes: The Fuel of Tumor Progression? Trends Cancer 2020, 6, 874–888. [Google Scholar] [CrossRef] [PubMed]
- Osswald, M.; Jung, E.; Sahm, F.; Solecki, G.; Venkataramani, V.; Blaes, J.; Weil, S.; Horstmann, H.; Wiestler, B.; Syed, M.; et al. Brain Tumour Cells Interconnect to a Functional and Resistant Network. Nature 2015, 528, 93–98. [Google Scholar] [CrossRef]
- Kabaso, D.; Lokar, M.; Kralj-Iglič, V.; Veranič, P.; Iglič, A. Temperature and Cholera Toxin B Are Factors That Influence Formation of Membrane Nanotubes in RT4 and T24 Urothelial Cancer Cell Lines. Int. J. Nanomed. 2011, 6, 495–509. [Google Scholar] [CrossRef] [Green Version]
- Gerdes, H.H.; Rustom, A.; Wang, X. Tunneling Nanotubes, an Emerging Intercellular Communication Route in Development. Mech. Dev. 2013, 130, 381–387. [Google Scholar] [CrossRef]
- Ng, W.L.; Bassler, B.L. Bacterial Quorum-Sensing Network Architectures. Annu. Rev. Genet. 2009, 43, 197–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, R.; Shirakawa, R.; Nishiyama, H.; Kobayashi, T.; Kawato, M.; Kanno, T.; Nishizawa, K.; Matsui, Y.; Ohbayashi, T.; Horiguchi, M.; et al. Downregulation of Ral GTPase-Activating Protein Promotes Tumor Invasion and Metastasis of Bladder Cancer. Oncogene 2013, 32, 894–902. [Google Scholar] [CrossRef] [Green Version]
- Pasquier, J.; Guerrouahen, B.S.; Al Thawadi, H.; Ghiabi, P.; Maleki, M.; Abu-Kaoud, N.; Jacob, A.; Mirshahi, M.; Galas, L.; Rafii, S.; et al. Preferential Transfer of Mitochondria from Endothelial to Cancer Cells through Tunneling Nanotubes Modulates Chemoresistance. J. Transl. Med. 2013, 11, 94. [Google Scholar] [CrossRef] [Green Version]
- Oliva, C.R.; Moellering, D.R.; Gillespie, G.Y.; Griguer, C.E. Acquisition of Chemoresistance in Gliomas Is Associated with Increased Mitochondrial Coupling and Decreased ROS Production. PLoS ONE 2011, 6, 9–13. [Google Scholar] [CrossRef] [Green Version]
- Omsland, M.; Bruserud, Ø.; Gjertsen, B.T.; Andresen, V. Tunneling Nanotube (TNT) Formation Is Downregulated by Cytarabine and NF-ΚB Inhibition in Acute Myeloid Leukemia (AML). Oncotarget 2017, 8, 7946–7963. [Google Scholar] [CrossRef] [Green Version]
- Spees, J.L.; Olson, S.D.; Whitney, M.J.; Prockop, D.J. Mitochondrial Transfer between Cells Can Rescue Aerobic Respiration. Proc. Natl. Acad. Sci. USA 2006, 103, 1283–1288. [Google Scholar] [CrossRef] [Green Version]
- Denise, C.; Paoli, P.; Calvani, M.; Taddei, M.L.; Giannoni, E.; Kopetz, S.; Kazmi, S.M.A.; Pia, M.M.; Pettazzoni, P.; Sacco, E.; et al. 5-Fluorouracil Resistant Colon Cancer Cells Are Addicted to OXPHOS to Survive and Enhance Stem-like Traits. Oncotarget 2015, 6, 41706–41721. [Google Scholar] [CrossRef] [Green Version]
- Moskalenko, S.; Henry, D.O.; Rosse, C.; Mirey, G.; Camonis, J.H.; White, M.A. The Exocyst Is a Ral Effector Complex. Nat. Cell Biol. 2002, 4, 66–72. [Google Scholar] [CrossRef]
- Nauta, T.D.; van den Broek, M.; Gibbs, S.; van der Pouw-Kraan, T.C.T.M.; Oudejans, C.B.; van Hinsbergh, V.W.M.; Koolwijk, P. Identification of HIF-2α-Regulated Genes That Play a Role in Human Microvascular Endothelial Sprouting during Prolonged Hypoxia in Vitro. Angiogenesis 2017, 20, 39–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santha, S.; Ling, X.; Aljahdali, I.A.M.; Rasam, S.S.; Wang, X.; Liao, J.; Wang, J.; Fountzilas, C.; Li, Q.; Qu, J.; et al. Mutant Kras as a Biomarker Plays a Favorable Role in FL118-Induced Apoptosis, Reactive Oxygen Species (ROS) Production and Modulation of Survivin, Mcl-1 and XIAP in Human Bladder Cancer. Cancers 2020, 12, 3413. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Teng, R.H.; Tsai, Y.C.; Ke, H.S.; Huang, J.Y.; Chen, C.C.; Kao, Y.L.; Kuo, C.C.; Bell, W.R.; Shieh, B. H-Ras Oncogene Counteracts the Growth-Inhibitory Effect of Genistein in T24 Bladder Carcinoma Cells. Br. J. Cancer 2005, 92, 80–88. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Oh, S.; Monia, B.P.; Stacey, D.W. Antisense Oligonucleotides Demonstrate a Dominant Role of C-Ki-RAS Proteins in Regulating the Proliferation of Diploid Human Fibroblasts. J. Biol. Chem. 1996, 271, 28259–28265. [Google Scholar] [CrossRef] [Green Version]
- Lim, K.H.; Baines, A.T.; Fiordalisi, J.J.; Shipitsin, M.; Feig, L.A.; Cox, A.D.; Der, C.J.; Counter, C.M. Activation of RalA Is Critical for Ras-Induced Tumorigenesis of Human Cells. Cancer Cell 2005, 7, 533–545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oxford, G.; Owens, C.R.; Titus, B.J.; Foreman, T.L.; Herlevsen, M.C.; Smith, S.C.; Theodorescu, D. RalA and RalB: Antagonistic Relatives in Cancer Cell Migration. Cancer Res. 2005, 65, 7111–7120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, S.C.; Oxford, G.; Baras, A.S.; Owens, C.; Havaleshko, D.; Brautigan, D.L.; Safo, M.K.; Theodorescu, D. Expression of Ral GTPases, Their Effectors, and Activators in Human Bladder Cancer. Clin. Cancer Res. 2007, 13, 3803–3813. [Google Scholar] [CrossRef] [Green Version]
- Smith, S.C.; Theodorescu, D. The Ral GTPase Pathway in Metastatic Bladder Cancer: Key Mediator and Therapeutic Target. Urol. Oncol. Semin. Orig. Investig. 2009, 27, 42–47. [Google Scholar] [CrossRef] [Green Version]
- Martegani, E.; Ceriani, M.; Tisi, R.; Berruti, G. Cloning and Characterization of a New Ral-GEF Expressed in Mouse Testis. Ann. N. Y. Acad. Sci. 2002, 973, 135–137. [Google Scholar] [CrossRef] [PubMed]
- Chinnery, H.R.; Pearlman, E.; McMenamin, P.G. Cutting Edge: Membrane Nanotubes In Vivo: A Feature of MHC Class II + Cells in the Mouse Cornea. J. Immunol. 2008, 180, 5779–5783. [Google Scholar] [CrossRef] [Green Version]
- Arkwright, P.D.; Luchetti, F.; Tour, J.; Roberts, C.; Ayub, R.; Morales, A.P.; Rodríguez, J.J.; Gilmore, A.; Canonico, B.; Papa, S.; et al. Fas Stimulation of T Lymphocytes Promotes Rapid Intercellular Exchange of Death Signals via Membrane Nanotubes. Cell Res. 2010, 20, 72–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Islam, M.N.; Das, S.R.; Emin, M.T.; Wei, M.; Sun, L.; Westphalen, K.; Rowlands, D.J.; Quadri, S.K.; Bhattacharya, S.; Bhattacharya, J. Mitochondrial Transfer from Bone-Marrow–Derived Stromal Cells to Pulmonary Alveoli Protects against Acute Lung Injury. Nat. Med. 2012, 18, 759–765. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Zaguilán, R.; Seftor, E.A.; Seftor, R.E.B.; Chu, Y.W.; Gillies, R.J.; Hendrix, M.J.C. Acidic PH Enhances the Invasive Behavior of Human Melanoma Cells. Clin. Exp. Metastasis 1996, 14, 176–186. [Google Scholar] [CrossRef]
- Butterfield, D.A.; Lauderback, C.M. Lipid Peroxidation and Protein Oxidation in Alzheimer’s Disease Brain: Potential Causes and Consequences Involving Amyloid β-Peptide-Associated Free Radical Oxidative Stress. Free Radic. Biol. Med. 2002, 32, 1050–1060. [Google Scholar] [CrossRef]
- Howe, C.J.; LaHair, M.M.; McCubrey, J.A.; Franklin, R.A. Redox Regulation of the Calcium/Calmodulin-Dependent Protein Kinases. J. Biol. Chem. 2004, 279, 44573–44581. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Moir, R.D.; Tanzi, R.E.; Bush, A.I.; Rogers, J.T. Redox-Active Metals, Oxidative Stress, and Alzheimer’s Disease Pathology. Ann. N. Y. Acad. Sci. 2004, 1012, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.J.; Wang, X.; Shaffer, K.M.; Sasaki, C.Y.; Ma, W. Characterization of H2O2-Induced Acute Apoptosis in Cultured Neural Stem/Progenitor Cells. FEBS Lett. 2004, 570, 102–106. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.Y.; Kim, C.G.; Jho, E.-H.; Rho, M.-S.; Kim, Y.S.; Kim, Y.-H.; Lee, Y.H. Hydrogen Peroxide Negatively Modulates Wnt Signaling through Downregulation of β-Catenin. Cancer Lett. 2004, 212, 225–231. [Google Scholar] [CrossRef]
- Van Rossum, G.S.A.T.; Drummen, G.P.C.; Verkleij, A.J.; Post, J.A.; Boonstra, J. Activation of Cytosolic Phospholipase A2 in Her14 Fibroblasts by Hydrogen Peroxide: A P42/44MAPK-Dependent and Phosphorylation- Independent Mechanism. Biochim. Et Biophys. Acta-Mol. Cell Biol. Lipids 2004, 1636, 183–195. [Google Scholar] [CrossRef]
- Zhu, D.; Tan, K.S.; Zhang, X.; Sun, A.Y.; Sun, G.Y.; Lee, J.C.M. Hydrogen Peroxide Alters Membrane and Cytoskeleton Properties and Increases Intercellular Connections in Astrocytes. J. Cell Sci. 2005, 118, 3695–3703. [Google Scholar] [CrossRef] [Green Version]
- Abounit, S.; Zurzolo, C. Wiring through Tunneling Nanotubes—From Electrical Signals to Organelle Transfer. J. Cell Sci. 2012, 125, 1089–1098. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Wang, Y.; Zhang, J.; Tu, J.; Wang, X.J.; Su, X.D.; Wang, L.; Zhang, Y. Tunneling-Nanotube Direction Determination in Neurons and Astrocytes. Cell Death Dis. 2012, 3, e438. [Google Scholar] [CrossRef] [PubMed]
- Rosich, L.; Montraveta, A.; Xargay-Torrent, S.; Lo´pez-Guerra, M.; Rolda´n, J.; Aymerich, M.; Salaverria, I.; Bea`, S.; Campo, E.; Pe´rez-Gala´n, P.; et al. Dual PI3K/MTOR Inhibition Is Required to Effectively Impair Microenvironment Survival Signals in Mantle Cell Lymphoma. Oncotarget 2014, 5, 6788–6800. [Google Scholar] [CrossRef]
- Wang, H.; Duan, L.; Zou, Z.; Li, H.; Yuan, S.; Chen, X.; Zhang, Y.; Li, X.; Sun, H.; Zha, H.; et al. Activation of the PI3K/Akt/MTOR/P70S6K Pathway Is Involved in S100A4-Induced Viability and Migration in Colorectal Cancer Cells. Int. J. Med Sci. 2014, 11, 841–849. [Google Scholar] [CrossRef] [Green Version]
- Xue, G.; Hemmings, B.A. PKB/Akt-Dependent Regulation of Cell Motility. J. Natl. Cancer Inst. 2013, 105, 393–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chauveau, A.; Aucher, A.; Eissmann, P.; Vivier, E.; Davis, D.M. Membrane Nanotubes Facilitate Long-Distance Interactions between Natural Killer Cells and Target Cells. Proc. Natl. Acad. Sci. USA 2010, 107, 5545–5550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellacosa, A.; Kumar, C.C.; Cristofano, A.D.; Testa, J.R. Activation of AKT Kinases in Cancer: Implications for Therapeutic Targeting. Adv. Cancer Res. 2005, 94, 29–86. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Wong, R.; Feig, L.A. RalGDS Couples Growth Factor Signaling to Akt Activation. Mol. Cell. Biol. 2008, 28, 2851–2859. [Google Scholar] [CrossRef] [Green Version]
- Fruman, D.A.; Chiu, H.; Hopkins, B.D.; Bagrodia, S.; Cantley, L.C.; Abraham, R.T. The PI3K Pathway in Human Disease. Cell 2017, 170, 605–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasquale, V.; Ducci, G.; Campioni, G.; Ventrici, A.; Assalini, C.; Busti, S.; Vanoni, M.; Vago, R.; Sacco, E. Profiling and Targeting of Energy and Redox Metabolism in Grade 2 Bladder Cancer Cells with Different Invasiveness Properties. Cells 2020, 9, 2669. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Aloia, A.; Arrigoni, E.; Costa, B.; Berruti, G.; Martegani, E.; Sacco, E.; Ceriani, M. RalGPS2 Interacts with Akt and PDK1 Promoting Tunneling Nanotubes Formation in Bladder Cancer and Kidney Cells Microenvironment. Cancers 2021, 13, 6330. https://doi.org/10.3390/cancers13246330
D’Aloia A, Arrigoni E, Costa B, Berruti G, Martegani E, Sacco E, Ceriani M. RalGPS2 Interacts with Akt and PDK1 Promoting Tunneling Nanotubes Formation in Bladder Cancer and Kidney Cells Microenvironment. Cancers. 2021; 13(24):6330. https://doi.org/10.3390/cancers13246330
Chicago/Turabian StyleD’Aloia, Alessia, Edoardo Arrigoni, Barbara Costa, Giovanna Berruti, Enzo Martegani, Elena Sacco, and Michela Ceriani. 2021. "RalGPS2 Interacts with Akt and PDK1 Promoting Tunneling Nanotubes Formation in Bladder Cancer and Kidney Cells Microenvironment" Cancers 13, no. 24: 6330. https://doi.org/10.3390/cancers13246330





