Predictive Biomarkers for Outcomes of Immune Checkpoint Inhibitors (ICIs) in Melanoma: A Systematic Review
Abstract
:Simple Summary
Abstract
1. Introduction
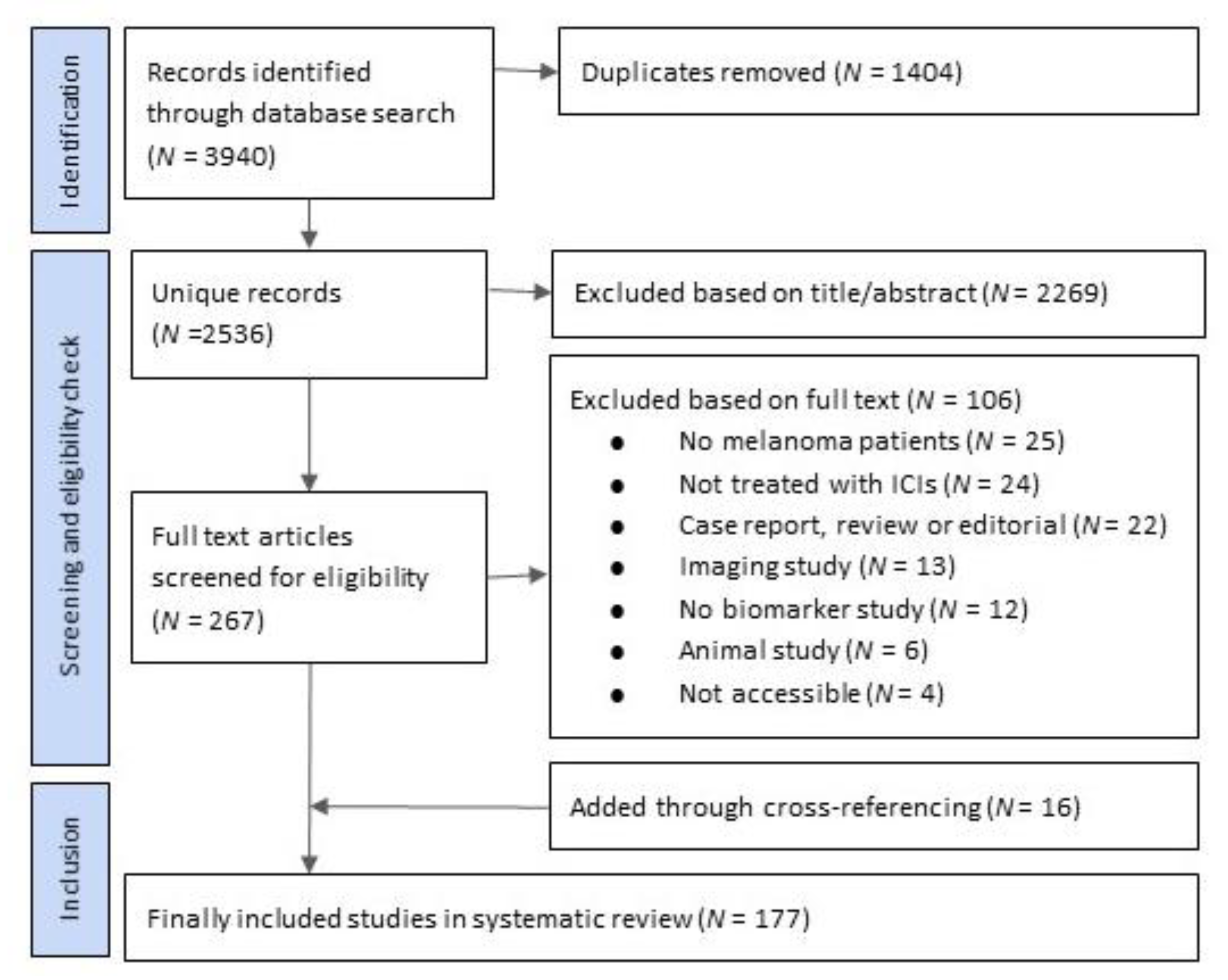
2. Materials and Methods
2.1. Search Strategy
2.2. Selection Criteria
2.3. Data Extraction and Quality Assessment
2.4. Data Presentation
3. Results
3.1. Peripheral Blood Biomarkers
3.2. Tumor Biomarkers
3.3. Gut Microbiome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Integraal Kankercentrum Nederland. Cijfers over Kanker. Available online: www.cijfersoverkanker.nl (accessed on 23 July 2021).
- Pisibon, C.; Ouertani, A.; Bertolotto, C.; Ballotti, R.; Cheli, Y. Immune Checkpoints in Cancers: From Signaling to the Clinic. Cancers 2021, 13, 4573. [Google Scholar] [CrossRef] [PubMed]
- Hodi, F.S.; O’Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C.; et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010, 363, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.; Schachter, J.; Long, G.V.; Arance, A.; Grob, J.J.; Mortier, L.; Daud, A.; Carlino, M.S.; McNeil, C.; Lotem, M.; et al. Pembrolizumab versus Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2015, 372, 2521–2532. [Google Scholar] [CrossRef]
- Larkin, J.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.J.; Cowey, C.L.; Lao, C.D.; Schadendorf, D.; Dummer, R.; Smylie, M.; Rutkowski, P.; et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N. Engl. J. Med. 2015, 373, 23–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, J.; Mandala, M.; Del Vecchio, M.; Gogas, H.J.; Arance, A.M.; Cowey, C.L.; Dalle, S.; Schenker, M.; Chiarion-Sileni, V.; Marquez-Rodas, I.; et al. Adjuvant Nivolumab versus Ipilimumab in Resected Stage III or IV Melanoma. N. Engl. J. Med. 2017, 377, 1824–1835. [Google Scholar] [CrossRef] [PubMed]
- Forschner, A.; Battke, F.; Hadaschik, D.; Schulze, M.; Weißgraeber, S.; Han, C.T.; Kopp, M.; Frick, M.; Klumpp, B.; Tietze, N.; et al. Tumor mutation burden and circulating tumor DNA in combined CTLA-4 and PD-1 antibody therapy in metastatic melanoma—Results of a prospective biomarker study. J. Immunother. Cancer 2019, 7, 180. [Google Scholar] [CrossRef] [PubMed]
- Diem, S.; Kasenda, B.; Spain, L.; Martin-Liberal, J.; Marconcini, R.; Gore, M.; Larkin, J. Serum lactate dehydrogenase as an early marker for outcome in patients treated with anti-PD-1 therapy in metastatic melanoma. Br. J. Cancer 2016, 114, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Diem, S.; Hasan, A.O.; Ackermann, C.J.; Bomze, D.; Koelzer, V.H.; Jochum, W.; Speiser, D.E.; Mertz, K.D.; Flatz, L. Tumor infiltrating lymphocytes in lymph node metastases of stage III melanoma correspond to response and survival in nine patients treated with ipilimumab at the time of stage IV disease. Cancer Immunol. Immunother. 2018, 67, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Daud, A.I.; Wolchok, J.D.; Robert, C.; Hwu, W.-J.; Weber, J.S.; Ribas, A.; Hodi, F.S.; Joshua, A.; Kefford, R.; Hersey, P.; et al. Programmed Death-Ligand 1 Expression and Response to the Anti–Programmed Death 1 Antibody Pembrolizumab in Melanoma. J. Clin. Oncol. 2016, 34, 4102–4109. [Google Scholar] [CrossRef] [PubMed]
- Ribas, A.; Kefford, R.; Marshall, M.A.; Punt, C.J.; Haanen, J.B.; Marmol, M.; Garbe, C.; Gogas, H.; Schachter, J.; Linette, G.; et al. Phase III randomized clinical trial comparing tremelimumab with standard-of-care chemotherapy in patients with advanced melanoma. J. Clin. Oncol. 2013, 31, 616–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. BMJ 2009, 339, b2535. [Google Scholar] [CrossRef] [Green Version]
- Riley, R.D.; Moons, K.G.M.; Snell, K.I.E.; Ensor, J.; Hooft, L.; Altman, D.G.; Hayden, J.; Collins, G.S.; Debray, T.P.A. A guide to systematic review and meta-analysis of prognostic factor studies. BMJ 2019, 364, k4597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Page, M.J.; Moher, D.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. PRISMA 2020 explanation and elaboration: Updated guidance and exemplars for reporting systematic reviews. BMJ 2021, 372, n160. [Google Scholar] [CrossRef]
- Kelderman, S.; Heemskerk, B.; Van Tinteren, H.; Brom, R.R.H.V.D.; Hospers, G.; Eertwegh, A.J.M.V.D.; Kapiteijn, E.; De Groot, J.W.B.; Soetekouw, P.; Jansen, R.L.; et al. Lactate dehydrogenase as a selection criterion for ipilimumab treatment in metastatic melanoma. Cancer Immunol. Immunother. 2014, 63, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Martens, A.; Wistuba-Hamprecht, K.; Geukes Foppen, M.; Yuan, J.; Postow, M.A.; Wong, P.; Romano, E.; Khammari, A.; Dreno, B.; Capone, M.; et al. Baseline Peripheral Blood Biomarkers Associated with Clinical Outcome of Advanced Melanoma Patients Treated with Ipilimumab. Clin. Cancer Res. 2016, 22, 2908–2918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valpione, S.; Martinoli, C.; Fava, P.; Mocellin, S.; Campana, L.G.; Quaglino, P.; Ferrucci, P.F.; Pigozzo, J.; Astrua, C.; Testori, A.; et al. Personalised medicine: Development and external validation of a prognostic model for metastatic melanoma patients treated with ipilimumab. Eur. J. Cancer 2015, 51, 2086–2094. [Google Scholar] [CrossRef]
- Weide, B.; Martens, A.; Hassel, J.C.; Berking, C.; Postow, M.A.; Bisschop, K.; Simeone, E.; Mangana, J.; Schilling, B.; Di Giacomo, A.M.; et al. Baseline Biomarkers for Outcome of Melanoma Patients Treated with Pembrolizumab. Clin. Cancer Res. 2016, 22, 5487–5496. [Google Scholar] [CrossRef] [Green Version]
- Ferrucci, P.F.; Ascierto, P.A.; Pigozzo, J.; Del Vecchio, M.; Maio, M.; Antonini Cappellini, G.C.; Guidoboni, M.; Queirolo, P.; Savoia, P.; Mandalà, M.; et al. Baseline neutrophils and derived neutrophil-to-lymphocyte ratio: Prognostic relevance in metastatic melanoma patients receiving ipilimumab. Ann. Oncol. 2016, 27, 732–738. [Google Scholar] [CrossRef]
- Ferrucci, P.F.; Gandini, S.; Battaglia, A.; Alfieri, S.; Di Giacomo, A.M.; Giannarelli, D.; Cappellini, G.C.A.; De Galitiis, F.; Marchetti, P.; Amato, G.; et al. Baseline neutrophil-to-lymphocyte ratio is associated with outcome of ipilimumab-treated metastatic melanoma patients. Br. J. Cancer 2015, 112, 1904–1910. [Google Scholar] [CrossRef] [Green Version]
- Iivanainen, S.; Ahvonen, J.; Knuuttila, A.; Tiainen, S.; Koivunen, J.P. Elevated CRP levels indicate poor progression-free and overall survival on cancer patients treated with PD-1 inhibitors. ESMO Open 2019, 4, e000531. [Google Scholar] [CrossRef] [Green Version]
- Bjoern, J.; Juul Nitschke, N.; Zeeberg Iversen, T.; Schmidt, H.; Fode, K.; Svane, I.M. Immunological correlates of treatment and response in stage IV malignant melanoma patients treated with Ipilimumab. OncoImmunology 2016, 5, e1100788. [Google Scholar] [CrossRef] [Green Version]
- Meyer, C.; Cagnon, L.; Costa-Nunes, C.M.; Baumgaertner, P.; Montandon, N.; Leyvraz, L.; Michielin, O.; Romano, E.; Speiser, D.E. Frequencies of circulating MDSC correlate with clinical outcome of melanoma patients treated with ipilimumab. Cancer Immunol. Immunother. 2013, 63, 247–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sade-Feldman, M.; Kanterman, J.; Klieger, Y.; Ish-Shalom, E.; Olga, M.; Saragovi, A.; Shtainberg, H.; Lotem, M.; Baniyash, M. Clinical Significance of Circulating CD33+CD11b+HLA-DR− Myeloid Cells in Patients with Stage IV Melanoma Treated with Ipilimumab. Clin. Cancer Res. 2016, 22, 5661–5672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Coaña, Y.P.; Wolodarski, M.; Poschke, I.; Yoshimoto, Y.; Yang, Y.; Nyström, M.; Edbäck, U.; Brage, S.E.; Lundqvist, A.; Masucci, G.; et al. Ipilimumab treatment decreases monocytic MDSCs and increases CD8 effector memory T cells in long-term survivors with advanced melanoma. Oncotarget 2017, 8, 21539–21553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gebhardt, C.; Sevko, A.; Jiang, H.; Lichtenberger, R.; Reith, M.; Tarnanidis, K.; Holland-Letz, T.; Umansky, L.; Beckhove, P.; Sucker, A.; et al. Myeloid Cells and Related Chronic Inflammatory Factors as Novel Predictive Markers in Melanoma Treatment with Ipilimumab. Clin. Cancer Res. 2015, 21, 5453–5459. [Google Scholar] [CrossRef] [Green Version]
- Retseck, J.; Nasr, A.; Lin, Y.; Lin, H.; Mendiratta, P.; Butterfield, L.H.; Tarhini, A.A. Long term impact of CTLA4 blockade immunotherapy on regulatory and effector immune responses in patients with melanoma. J. Transl. Med. 2018, 16, 184. [Google Scholar] [CrossRef]
- Weber, J.; Gibney, G.; Kudchadkar, R.R.; Yu, B.; Cheng, P.; Martinez, A.J.; Kroeger, J.; Richards, A.; McCormick, L.; Moberg, V.; et al. Phase I/II Study of Metastatic Melanoma Patients Treated with Nivolumab Who Had Progressed after Ipilimumab. Cancer Immunol. Res. 2016, 4, 345–353. [Google Scholar] [CrossRef] [Green Version]
- Hong, X.; Sullivan, R.J.; Kalinich, M.; Kwan, T.T.; Giobbie-Hurder, A.; Pan, S.; LiCausi, J.A.; Milner, J.D.; Nieman, L.T.; Wittner, B.S.; et al. Molecular signatures of circulating melanoma cells for monitoring early response to immune checkpoint therapy. Proc. Natl. Acad. Sci. USA 2018, 115, 2467–2472. [Google Scholar] [CrossRef] [Green Version]
- Seremet, T.; Jansen, Y.; Planken, S.; Njimi, H.; Delaunoy, M.; El Housni, H.; Awada, G.; Schwarze, J.K.; Keyaerts, M.; Everaert, H.; et al. Undetectable circulating tumor DNA (ctDNA) levels correlate with favorable outcome in metastatic melanoma patients treated with anti-PD1 therapy. J. Transl. Med. 2019, 17, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Keller, L.; Guibert, N.; Casanova, A.; Brayer, S.; Farella, M.; Delaunay, M.; Gilhodes, J.; Martin, E.; Balagué, G.; Favre, G.; et al. Early Circulating Tumour DNA Variations Predict Tumour Response in Melanoma Patients Treated with Immunotherapy. Acta Derm. Venereol. 2019, 99, 206–210. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Long, G.V.; Boyd, S.; Lo, S.; Menzies, A.M.; Tembe, V.; Guminski, A.; Jakrot, V.; Scolyer, R.A.; Mann, G.J.; et al. Circulating tumour DNA predicts response to anti-PD1 antibodies in metastatic melanoma. Ann. Oncol. 2017, 28, 1130–1136. [Google Scholar] [CrossRef] [PubMed]
- Wistuba-Hamprecht, K.; Martens, A.; Heubach, F.; Romano, E.; Foppen, M.G.; Yuan, J.; Postow, M.; Wong, P.; Mallardo, D.; Schilling, B.; et al. Peripheral CD8 effector-memory type 1 T-cells correlate with outcome in ipilimumab-treated stage IV melanoma patients. Eur. J. Cancer 2017, 73, 61–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tietze, J.K.; Wilkins, D.E.C.; Sckisel, G.D.; Bouchlaka, M.N.; Alderson, K.L.; Weiss, J.M.; Ames, E.; Bruhn, K.W.; Craft, N.; Wiltrout, R.H.; et al. Delineation of antigen-specific and antigen-nonspecific CD8+ memory T-cell responses after cytokine-based cancer immunotherapy. Blood 2012, 119, 3073–3083. [Google Scholar] [CrossRef] [PubMed]
- Subrahmanyam, P.B.; Dong, Z.; Gusenleitner, D.; Giobbie-Hurder, A.; Severgnini, M.; Zhou, J.; Manos, M.; Eastman, L.M.; Maecker, H.T.; Hodi, F.S. Distinct predictive biomarker candidates for response to anti-CTLA-4 and anti-PD-1 immunotherapy in melanoma patients. J. Immunother. Cancer 2018, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Gide, T.N.; Quek, C.; Menzies, A.M.; Tasker, A.T.; Shang, P.; Holst, J.; Madore, J.; Lim, S.Y.; Velickovic, R.; Wongchenko, M.; et al. Distinct Immune Cell Populations Define Response to Anti-PD-1 Monotherapy and Anti-PD-1/Anti-CTLA-4 Combined Therapy. Cancer Cell 2019, 35, 238–255.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Postow, M.A.; Manuel, M.; Wong, P.; Yuan, J.; Dong, Z.; Liu, C.; Perez, S.; Tanneau, I.; Noel, M.; Courtier, A.; et al. Peripheral T cell receptor diversity is associated with clinical outcomes following ipilimumab treatment in metastatic melanoma. J. Immunother. Cancer 2015, 3, 23. [Google Scholar] [CrossRef] [Green Version]
- Hogan, S.A.; Courtier, A.; Cheng, P.F.; Jaberg-Bentele, N.F.; Goldinger, S.M.; Manuel, M.; Perez, S.; Plantier, N.; Mouret, J.-F.; Nguyen-Kim, T.D.L.; et al. Peripheral Blood TCR Repertoire Profiling May Facilitate Patient Stratification for Immunotherapy against Melanoma. Cancer Immunol. Res. 2018, 7, 77–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capone, M.; Fratangelo, F.; Giannarelli, D.; Sorrentino, C.; Turiello, R.; Zanotta, S.; Galati, D.; Madonna, G.; Tuffanelli, M.; Scarpato, L.; et al. Frequency of circulating CD8+CD73+T cells is associated with survival in nivolumab-treated melanoma patients. J. Transl. Med. 2020, 18, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Graves, M.; CelliMarchett, G.; Van Zyl, B.; Tang, D.; Vilain, R.E.; van der Westhuizen, A.; Bowden, N.A. Monitoring Patient Response to Pembrolizumab With Peripheral Blood Exhaustion Marker Profiles. Front. Med. 2019, 6, 113. [Google Scholar] [CrossRef]
- Jacquelot, N.; Roberti, M.P.; Enot, D.P.; Rusakiewicz, S.; Ternès, N.; Jegou, S.; Woods, D.M.; Sodré, A.L.; Hansen, M.; Meirow, Y.; et al. Predictors of responses to immune checkpoint blockade in advanced melanoma. Nat. Commun. 2017, 8, 592. [Google Scholar] [CrossRef] [Green Version]
- Bochem, J.; Zelba, H.; Amaral, T.; Spreuer, J.; Soffel, D.; Eigentler, T.; Wagner, N.B.; Uslu, U.; Terheyden, P.; Meier, F.; et al. Peripheral PD-1+CD56+ T-cell frequencies correlate with outcome in stage IV melanoma under PD-1 blockade. PLoS ONE 2019, 14, e0221301. [Google Scholar] [CrossRef] [PubMed]
- Nonomura, Y.; Otsuka, A.; Nakashima, C.; Seidel, J.; Kitoh, A.; Dainichi, T.; Nakajima, S.; Sawada, Y.; Matsushita, S.; Aoki, M.; et al. Peripheral blood Th9 cells are a possible pharmacodynamic biomarker of nivolumab treatment efficacy in metastatic melanoma patients. OncoImmunology 2016, 5, e1248327. [Google Scholar] [CrossRef]
- Simeone, E.; Gentilcore, G.; Giannarelli, D.; Grimaldi, A.M.; Caracò, C.; Curvietto, M.; Esposito, A.; Paone, M.; Palla, M.; Cavalcanti, E.; et al. Immunological and biological changes during ipilimumab treatment and their potential correlation with clinical response and survival in patients with advanced melanoma. Cancer Immunol. Immunother. 2014, 63, 675–683. [Google Scholar] [CrossRef]
- Johnson, D.B.; Frampton, G.; Rioth, M.J.; Yusko, E.; Xu, Y.; Guo, X.; Ennis, R.C.; Fabrizio, D.; Chalmers, Z.R.; Greenbowe, J.; et al. Targeted Next Generation Sequencing Identifies Markers of Response to PD-1 Blockade. Cancer Immunol. Res. 2016, 4, 959–967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snyder, A.; Makarov, V.; Merghoub, T.; Yuan, J.; Zaretsky, J.M.; Desrichard, A.; Walsh, L.A.; Postow, M.A.; Wong, P.; Ho, T.S.; et al. Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma. N. Engl. J. Med. 2014, 371, 2189–2199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, M.A.; Weeder, B.; David, J.; Nellore, A.; Thompson, R.F. Burden of tumor mutations, neoepitopes, and other variants are weak predictors of cancer immunotherapy response and overall survival. Genome Med. 2020, 12, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vargas, F.A.; Furness, A.J.S.; Litchfield, K.; Joshi, K.; Rosenthal, R.; Ghorani, E.; Solomon, I.; Lesko, M.H.; Ruef, N.; Roddie, C.; et al. Fc Effector Function Contributes to the Activity of Human Anti-CTLA-4 Antibodies. Cancer Cell 2018, 33, 649–663.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Allen, E.M.; Miao, D.; Schilling, B.; Shukla, S.A.; Blank, C.; Zimmer, L.; Sucker, A.; Hillen, U.; Foppen, M.H.G.; Goldinger, S.M.; et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015, 350, 207–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riaz, N.; Havel, J.; Makarov, V.; Desrichard, A.; Urba, W.J.; Sims, J.S.; Hodi, F.S.; Martín-Algarra, S.; Mandal, R.; Sharfman, W.H.; et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell 2017, 171, 934–949.e16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, P.-L.; Roh, W.; Reuben, A.; Cooper, Z.A.; Spencer, C.N.; Prieto, P.A.; Miller, J.P.; Bassett, R.L.; Gopalakrishnan, V.; Wani, K.; et al. Analysis of Immune Signatures in Longitudinal Tumor Samples Yields Insight into Biomarkers of Response and Mechanisms of Resistance to Immune Checkpoint Blockade. Cancer Discov. 2016, 6, 827–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cristescu, R.; Mogg, R.; Ayers, M.; Albright, A.; Murphy, E.; Yearley, J.; Sher, X.; Liu, X.Q.; Lu, H.; Nebozhyn, M.; et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade–based immunotherapy. Science 2018, 362, 197. [Google Scholar] [CrossRef] [Green Version]
- Hamid, O.; Robert, C.; Daud, A.; Hodi, F.S.; Hwu, W.J.; Kefford, R.; Wolchok, J.D.; Hersey, P.; Joseph, R.; Weber, J.S.; et al. Five-year survival outcomes for patients with advanced melanoma treated with pembrolizumab in KEYNOTE-001. Ann. Oncol. 2019, 30, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Karachaliou, N.; Gonzalez-Cao, M.; Crespo, G.; Drozdowskyj, A.; Aldeguer, E.; Gimenez-Capitan, A.; Teixido, C.; Molina-Vila, M.A.; Viteri, S.; De Los Llanos Gil, M.; et al. Interferon gamma, an important marker of response to immune checkpoint blockade in non-small cell lung cancer and melanoma patients. Ther. Adv. Med. Oncol. 2018, 10, 1758834017749748. [Google Scholar] [CrossRef] [PubMed]
- Roh, W.; Chen, P.-L.; Reuben, A.; Spencer, C.N.; Prieto, P.A.; Miller, J.P.; Gopalakrishnan, V.; Wang, F.; Cooper, Z.A.; Reddy, S.M.; et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci. Transl. Med. 2017, 9, eaaah3560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, R.-R.; Chasalow, S.D.; Wang, L.; Hamid, O.; Schmidt, H.; Cogswell, J.; Alaparthy, S.; Berman, D.; Jure-Kunkel, M.; Siemers, N.O.; et al. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol. Immunother. 2011, 61, 1019–1031. [Google Scholar] [CrossRef] [PubMed]
- Hugo, W.; Zaretsky, J.M.; Sun, L.; Song, C.; Moreno, B.H.; Hu-Lieskovan, S.; Berent-Maoz, B.; Pang, J.; Chmielowski, B.; Cherry, G.; et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell 2016, 165, 35–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodig, S.J.; Gusenleitner, D.; Jackson, D.G.; Gjini, E.; Giobbie-Hurder, A.; Jin, C.; Chang, H.; Lovitch, S.B.; Horak, C.; Weber, J.S.; et al. MHC proteins confer differential sensitivity to CTLA-4 and PD-1 blockade in untreated metastatic melanoma. Sci. Transl. Med. 2018, 10, eaar3342. [Google Scholar] [CrossRef] [Green Version]
- Tarhini, A.A.; Lin, Y.; Lin, H.-M.; Vallabhaneni, P.; Sander, C.; LaFramboise, W.; Hamieh, L. Expression profiles of immune-related genes are associated with neoadjuvant ipilimumab clinical benefit. OncoImmunology 2017, 6, e1231291. [Google Scholar] [CrossRef] [Green Version]
- Varn, F.S.; Wang, Y.; Cheng, C. A B cell-derived gene expression signature associates with an immunologically active tumor microenvironment and response to immune checkpoint blockade therapy. OncoImmunology 2018, 8, e1513440. [Google Scholar] [CrossRef] [Green Version]
- Ayers, M.; Lunceford, J.; Nebozhyn, M.; Murphy, E.; Loboda, A.; Kaufman, D.R.; Albright, A.; Cheng, J.D.; Kang, S.P.; Shankaran, V.; et al. IFN-γ–related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Investig. 2017, 127, 2930–2940. [Google Scholar] [CrossRef] [PubMed]
- Ribas, A.; Shin, D.; Zaretsky, J.; Frederiksen, J.; Cornish, A.; Avramis, E.; Seja, E.; Kivork, C.; Siebert, J.; Kaplan-Lefko, P.; et al. PD-1 Blockade Expands Intratumoral Memory T Cells. Cancer Immunol. Res. 2016, 4, 194–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, P.F.; Wei, W.; Smithy, J.W.; Acs, B.; Toki, M.; Blenman, K.R.; Zelterman, D.; Kluger, H.M.; Rimm, D.L. Multiplex Quantitative Analysis of Tumor-Infiltrating Lymphocytes and Immunotherapy Outcome in Metastatic Melanoma. Clin. Cancer Res. 2019, 25, 2442–2449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamid, O.; Schmidt, H.; Nissan, A.; Ridolfi, L.; Aamdal, S.; Hansson, J.; Guida, M.; Hyams, D.M.; Gómez, H.; Bastholt, L.; et al. A prospective phase II trial exploring the association between tumor microenvironment biomarkers and clinical activity of ipilimumab in advanced melanoma. J. Transl. Med. 2011, 9, 204. [Google Scholar] [CrossRef] [Green Version]
- Mastracci, L.; Fontana, V.; Queirolo, P.; Carosio, R.; Grillo, F.; Morabito, A.; Banelli, B.; Tanda, E.; Boutros, A.; Dozin, B.; et al. Response to ipilimumab therapy in metastatic melanoma patients: Potential relevance of CTLA-4+ tumor infiltrating lymphocytes and their in situ localization. Cancer Immunol. Immunother. 2020, 69, 653–662. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [Green Version]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.-L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti–PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef] [Green Version]
- Frankel, A.E.; Coughlin, L.A.; Kim, J.; Froehlich, T.W.; Xie, Y.; Frenkel, E.P.; Koh, A. Metagenomic Shotgun Sequencing and Unbiased Metabolomic Profiling Identify Specific Human Gut Microbiota and Metabolites Associated with Immune Checkpoint Therapy Efficacy in Melanoma Patients. Neoplasia 2017, 19, 848–855. [Google Scholar] [CrossRef]
- Simpson, T.R.; Li, F.; Montalvo-Ortiz, W.; Sepulveda, M.A.; Bergerhoff, K.; Arce, F.; Roddie, C.; Henry, J.Y.; Yagita, H.; Wolchok, J.D.; et al. Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti–CTLA-4 therapy against melanoma. J. Exp. Med. 2013, 210, 1695–1710. [Google Scholar] [CrossRef]
- Selby, M.J.; Engelhardt, J.J.; Quigley, M.; Henning, K.A.; Chen, T.; Srinivasan, M.; Korman, A.J. Anti-CTLA-4 Antibodies of IgG2a Isotype Enhance Antitumor Activity through Reduction of Intratumoral Regulatory T Cells. Cancer Immunol. Res. 2013, 1, 32–42. [Google Scholar] [CrossRef] [Green Version]
- Robert, L.; Tsoi, J.; Wang, X.; Emerson, R.; Homet, B.; Chodon, T.; Mok, S.; Huang, R.R.; Cochran, A.J.; Comin-Anduix, B.; et al. CTLA4 Blockade Broadens the Peripheral T-Cell Receptor Repertoire. Clin. Cancer Res. 2014, 20, 2424–2432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, H.; Park, J.-H.; Kiyotani, K.; Zewde, M.G.; Miyashita, A.; Jinnin, M.; Kiniwa, Y.; Okuyama, R.; Tanaka, R.; Fujisawa, Y.; et al. Intratumoral expression levels of PD-L1, GZMA, and HLA-A along with oligoclonal T cell expansion associate with response to nivolumab in metastatic melanoma. OncoImmunology 2016, 5, e1204507. [Google Scholar] [CrossRef] [Green Version]
- Jessurun, C.A.C.; Vos, J.A.M.; Limpens, J.; Luiten, R.M. Biomarkers for Response of Melanoma Patients to Immune Checkpoint Inhibitors: A Systematic Review. Front. Oncol. 2017, 7, 233. [Google Scholar] [CrossRef] [PubMed]
- Woods, D.M.; Laino, A.S.; Winters, A.F.; Alexandre, J.M.; Freeman, D.; Rao, V.; Adavani, S.S.; Weber, J.S.; Chattopadhyay, P.K. Nivolumab and ipilimumab are associated with distinct immune landscape changes and response-associated immunophenotypes. JCI Insight 2020, 5, e137066. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.C.; Postow, M.A.; Orlowski, R.J.; Mick, R.; Bengsch, B.; Manne, S.; Xu, W.; Harmon, S.; Giles, J.R.; Wenz, B.; et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature 2017, 545, 60–65. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Yang, M.; Wang, Q.; Song, F.; Li, X.; Chen, K. The new identified biomarkers determine sensitivity to immune check-point blockade therapies in melanoma. OncoImmunology 2019, 8, 1608132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krieg, C.; Nowicka, M.; Guglietta, S.; Schindler, S.; Hartmann, F.J.; Weber, L.M.; Dummer, R.; Robinson, M.D.; Levesque, M.P.; Becher, B. High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat. Med. 2018, 24, 144–153. [Google Scholar] [CrossRef]
- Deshpande, R.P.; Sharma, S.; Watabe, K. The Confounders of Cancer Immunotherapy: Roles of Lifestyle, Metabolic Disorders and Sociological Factors. Cancers 2020, 12, 2983. [Google Scholar] [CrossRef]



| Biomarker | N of Studies | N of Patients | Median Patients per Article (IQR) | Response | PFS | OS | Quality Assessment |
|---|---|---|---|---|---|---|---|
| LDH | 20 | 2539 | 86 (53–86) | LDH was associated with response in 4/10 studies, not associated in 6/10 studies. | LDH was associated with PFS in 3/3 studies. | LDH was associated with OS in 12/16 studies, not associated in 4/16 studies. | 2/20 high risk, 14/20, moderate, 4/20 low risk of bias |
| NLR | 11 | 1632 | 78 (43–184) | NLR was associated with response in 3/5 studies, not associated in 2/5 studies. | NLR was associated with PFS in 4/5 studies, not associated in 1/5 studies. | NLR was associated with OS in 7/10 studies, not associated in 3/10 studies | 1/11 high risk, 6/11 moderate risk, 4/11 low risk of bias |
| TMB | 7 | 724 | 64 (56–174) | TMB was associated with response in 3/6 studies, not associated in 3/6 studies. | TMB was associated with OS in 2/4 studies, not associated in 2/4 studies. | 3/7 high risk, 4/7 moderate, 0/7 low risk of bias | |
| Neoantigen load (NAL) | 5 | 385 | 64 (54–107) | NAL was associated with response in 2/3 studies, not associated in 1/3 studies. | NAL was associated with OS in 2/2 studies. | 2/5 high risk 3/5 moderate, 0/5 low risk of bias | |
| PD-L1 expression on tumor cells | 5 | 637 | 111 (48–214) | PD-L1 was not associated with response in 5/5 studies. | PD-L1 was not associated with PFS in 1/1 studies. | PDL-1 was not associated with OS in 3/3 studies. | 2/5 high risk, 3/5 moderate, 0/5 low risk of bias |
| MDSCs | 4 | 726 | 48 (22–475) | MDSCs were associated with response in 3/3 studies. | MDSCs were associated with OS in 2/2 studies. | 1/4 high risk, 3/4 moderate risk, 0/3 low risk of bias | |
| T-cell inflamed GEP | 4 | 304 | 58 (33–192) | GEP was associated with response in 4/4 studies. | 2/4 high risk, 2/4 moderate, 0/4 low risk of bias | ||
| Tregs in tumor tissue | 4 | 169 | 38 (31–58) | Tregs were associated with response in 2/4 studies, not associated in 2/4 studies. | Tregs were associated with OS in 2/2 studies. | 0/4 high risk, 4/4 moderate, 0/4 low risk of bias | |
| monocytic MDSCs | 4 | 168 | 39 (32–55) | moMDSCs were associated with response in 2/2 studies. | moMDSCs were associated with PFS in 2/2 studies | moMDSCs were associated with OS in 1/1 studies. | 3/4 high risk 1/4 moderate risk, 0/4 low risk of bias |
| Tregs in blood | 3 | 741 | 95 (31–615) | Tregs were associated with response in 1/1 studies. | Tregs were associated with RFS in 1/1 studies | Tregs were associated with OS in 2/2 studies. | 1/3 high risk, 2/3 moderate risk, 0/3 as low risk of bias |
| CD8 memory T-cells in blood | 3 | 90 | 30 (17–43) | CD8 memory T-cells were associated with response in 2/2 studies. | CD8 memory T-cells were associated with OS in 3/3 studies. | 2/3 high risk, 1/3 moderate risk 0/1 low risk of bias | |
| TILs | 3 | 90 | 17 (9–64) | TILs were associated with response in 3/3 studies. | TILs were not associated with PFS in 1/1 studies. | TILs were not associated with OS in 1/1 studies. | 1/3 high risk, 2/3 moderate, 0/3 low risk of bias |
| TCR diversity in blood | 2 | 54 | 27 (N/A) | TCR diversity was associated with response in 2/2 studies. | TCR diversity was not associated with OS in 1/1 studies. | 1/2 high risk, 1/2 moderate risk, 0/2 low risk of bias | |
| NK cells in blood | 2 | 63 | 32 (N/A) | NK cells were associated with response in 1/2 studies, not associated in 1/2 studies. | 1/2 high risk, 1/2 moderate, risk 0/2 low risk of bias |
| Biomarker | N of Studies | N of Patients | Median Patients per Study (IQR) | Response | PFS | OS | Quality Assessment |
|---|---|---|---|---|---|---|---|
| LDH | 20 | 2274 | 78 (39–152) | LDH was associated with response in 4/10 studies, not associated in 6/10 studies. | LDH was associated with PFS in 10/11 studies, not associated in 1/11 studies. | LDH was associated with OS in 13/13 studies. | 5/20 high, 12/20 moderate, 3/20 low risk of bias |
| PD-L1 expression on tumor cells | 12 | 1481 | 52 (30–68) | PD-L1 was associated with response in 7/12 studies, not associated in 5/12 studies. | PD-L1 was associated with PFS in 2/5 studies, not associated in 3/5 studies. | PD-L1 was associated with OS in 3/4 studies, not associated in 1/4 studies. | 8/12 high, 4/12 moderate, 0/12 low risk of bias |
| T-cell inflamed GEP | 9 | 1237 | 58 (33–192) | GEP was associated with response in 7/9 studies, not associated in 2/9 studies. | GEP was associated with PFS in 1/2 studies, not associated in 1/2 studies | GEP was not associated with OS in 2/2 studies. | 4/9 high, 5/9 moderate, 0/9 low risk of bias |
| NLR | 8 | 732 | 77 (41–138) | NLR was associated with response 1/3 studies, not associated in 2/3 studies. | NLR was associated with PFS in 5/5 studies. | NLR was associated with OS in 6/6 studies. | 1/8 high, 6/8 moderate, 1/8 low risk of bias |
| TMB | 8 | 68 | 52 (41–67) | TMB was associated with response in 3/6 studies, not associated in 3/6 studies. | TMB was associated with PFS in 2/2 studies. | TMB was associated with OS in 3/4 studies, not associated in 1/4 studies. | 4/8 high, 4/8 moderate, 0/8 low risk of bias |
| NK cells in blood | 5 | 128 | 20 (13–41) | NK cells were associated with response in 3/4 studies, not associated in 1/4 studies. | NK cells were not associated with OS in 2/2 studies. | 4/5 high, 1/5 moderate, 0/5 low risk of bias | |
| TCR diversity in tumor | 4 | 184 | 52 (22–57) | TCR diversity was associated with response in 3/4 studies, not associated in 1/4 studies. | 2/4 high, 2/4 moderate, 0/4 low risk of bias | ||
| Gut microbiomes | 2 | 104 | 52 (N/A) | Gut microbiomes were associated with response in 2/2 studies | Gut microbiomes were associated with PFS in 1/1 studies | 0/2 high, 2/2 moderate, 0/2 low risk | |
| CD8 memory T-cells in blood | 2 | 29 | 15 (N/A) | CD8 memory cells were associated with response in 1/2 studies, not associated in 1/2 studies. | CD8 memory cells were not associated with OS in 1/1 studies. | 2/2 high, 0/2 moderate, 0/2 low risk of bias | |
| TILs | 2 | 121 | 60 (N/A) | TILs were associated with response in 2/2 studies | 2/2 high, 0/2 moderate, 0/2 low risk of bias | ||
| ctDNA | 1 | 85 | N/A | ctDNA was associated with PFS in 1/1 studies. | ctDNA was associated with OS in 1/1 studies. | 0/1 high, 1/1 moderate, 0/1 low risk of bias | |
| MDSCs | 1 | 92 | N/A | MDSCs were associated with response in 1/1 studies. | MDSCs were associated with PFS in 1/1 studies. | MDSCs were associated with OS in 1/1 studies. | 0/1 high, 1/1 moderate, 0/1 low risk of bias |
| Tregs in blood | 1 | 46 | N/A | Tregs were not associated with response in 1/1 studies. | 1/1 high, 0/1 moderate, 0/1 low risk of bias | ||
| TCR diversity in blood | 1 | 38 | N/A | TCR diversity was associated with response in 1/1 studies. | 0/1 high, 1/1 moderate, 0/1 low risk of bias |
| Biomarker | N of Studies | N of Patients | Median Patients per Study (IQR) | Response | PFS | OS | Quality Assessment |
|---|---|---|---|---|---|---|---|
| LDH | 2 | 295 | 148 | LDH was associated with response in 1/2 studies, not associated in 1/2 studies. | LDH was associated with PFS in 1/1 studies. | LDH was associated with OS in 2/2 studies. | 0/2 high risk, 1/2 moderate, 1/2 low risk of bias |
| NLR | 1 | 209 | N/A | NLR was not associated with response in 1/1 studies. | NLR was associated with OS in 1/1 studies. | 0/1 high risk, 1/1 moderate 0/1 low risk of bias | |
| TCR diversity | 1 | 80 | N/A | TCR diversity was associated with PFS in 1/1 studies. | 0/1 high risk 1/1 moderate, 0/1 low risk of bias | ||
| Memory T-cells in tumor tissue | 1 | 57 | N/A | Memory T-cells were associated with PFS in 1/1 studies. | 0/1 high risk 1/1 moderate, 0/1 low risk of bias | ||
| T-cell inflamed GEP | 1 | 57 | N/A | GEP was associated with response in 1/1 studies. | 0/1 high risk 1/1 moderate, 0/1 low risk of bias | ||
| ctDNA | 1 | 35 | N/A | ctDNA was associated with response in 1/1 studies. | ctDNA was associated with PFS in 1/1 studies. | ctDNA was associated with OS in 1/1 studies. | 1/1 high risk, 0/1 moderate 0/1 low risk |
| TMB | 1 | 35 | N/A | TMB was associated with response in 1/1 studies. | TMB was not associated with OS in 1/1 studies. | 1/1 high risk, 0/1 moderate, 0/1 low risk of bias |
| Biomarker | N of Studies | N of Patients | Median Patients per Study (IQR) | Response | PFS | OS | Quality Assessment |
|---|---|---|---|---|---|---|---|
| TMB | 5 | 861 | 91 (68–317) | TMB was associated with response in 4/5 studies, not associated in 1/5 studies. | TMB was associated with PFS in 1/2 studies, not associated in 1/2 studies. | TMB was associated with OS in 1/4 studies, not associated in 3/4 studies. | 2/5 high risk, 3/5 moderate, 0/5 low risk of bias |
| PD-L1 expression on tumor cells | 5 | 298 | 51 (1–84) | PD-L1 expression was associated with response in 2/3 studies, not associated in 1/3 studies. | PD-L1 expression was not associated with PFS in 1/1 studies. | PD-L1 was associated with OS in 1/4 studies, not associated with OS in 3/4 studies. | 2/5 high risk, 3/5 moderate, 0/5 low risk of bias |
| Circulating tumor cells | 3 | 190 | 82 (22–86) | CTCs were associated with response in 3/3 studies. | CTCs were associated with PFS in 3/3 studies. | CTCs were associated with OS in 2/2 studies. | 1/3 high risk, 2/3 moderate 0/3 low risk |
| LDH | 2 | 141 | 71 (N/A) | LDH was not associated with response in 1/1 studies. | LDH was associated with PFS in 1/2 studies. | LDH was associated with OS in 1-2 studies, not associated with OS in 1/2 studies. | 0/2 high risk, 2/2 moderate, 0/2 low risk of bias |
| Neoantigen load (NAL) | 2 | 423 | 212 (N/A) | NAL was associated with response in 1/2 studies, not associated in 1/2 studies. | NAL was associated with OS in 1/2 studies, not associated with OS in 1/2 studies. | 2/2 high risk, 0/2 moderate, 0/2 low risk of bias | |
| TILs in tumor tissue | 2 | 123 | 62 (N/A) | TILs were associated with response in 1/1 studies. | TILs were not associated with PFS in 1/1 studies. | TILs were associated with OS in 2/2 studies. | 1/2 high risk 1/2 moderate, 0/1 low risk of bias |
| Gut microbiomes | 2 | 66 | 33 (N/A) | Gut microbiomes were associated with response in 2/2 studies. | 0/2 high risk, 2/2 moderate, 0/2 low risk | ||
| NLR | 1 | 32 | N/A | NLR was not associated with response in 1/1 studies. | 1/1 high risk, 0/1 moderate 0/1 low risk of bias | ||
| Tregs in tumor tissue | 1 | 32 | N/A | Tregs were not associated with OS in 1/1 studies. | 1/1 high risk, 0/1 moderate 0/1 low risk of bias |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baltussen, J.C.; Welters, M.J.P.; Verdegaal, E.M.E.; Kapiteijn, E.; Schrader, A.M.R.; Slingerland, M.; Liefers, G.-J.; van der Burg, S.H.; Portielje, J.E.A.; de Glas, N.A. Predictive Biomarkers for Outcomes of Immune Checkpoint Inhibitors (ICIs) in Melanoma: A Systematic Review. Cancers 2021, 13, 6366. https://doi.org/10.3390/cancers13246366
Baltussen JC, Welters MJP, Verdegaal EME, Kapiteijn E, Schrader AMR, Slingerland M, Liefers G-J, van der Burg SH, Portielje JEA, de Glas NA. Predictive Biomarkers for Outcomes of Immune Checkpoint Inhibitors (ICIs) in Melanoma: A Systematic Review. Cancers. 2021; 13(24):6366. https://doi.org/10.3390/cancers13246366
Chicago/Turabian StyleBaltussen, Joosje C., Marij J. P. Welters, Elizabeth M. E. Verdegaal, Ellen Kapiteijn, Anne M. R. Schrader, Marije Slingerland, Gerrit-Jan Liefers, Sjoerd H. van der Burg, Johanneke E. A. Portielje, and Nienke A. de Glas. 2021. "Predictive Biomarkers for Outcomes of Immune Checkpoint Inhibitors (ICIs) in Melanoma: A Systematic Review" Cancers 13, no. 24: 6366. https://doi.org/10.3390/cancers13246366
APA StyleBaltussen, J. C., Welters, M. J. P., Verdegaal, E. M. E., Kapiteijn, E., Schrader, A. M. R., Slingerland, M., Liefers, G.-J., van der Burg, S. H., Portielje, J. E. A., & de Glas, N. A. (2021). Predictive Biomarkers for Outcomes of Immune Checkpoint Inhibitors (ICIs) in Melanoma: A Systematic Review. Cancers, 13(24), 6366. https://doi.org/10.3390/cancers13246366








