Automated Early Detection of Myelodysplastic Syndrome within the General Population Using the Research Parameters of Beckman–Coulter DxH 800 Hematology Analyzer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
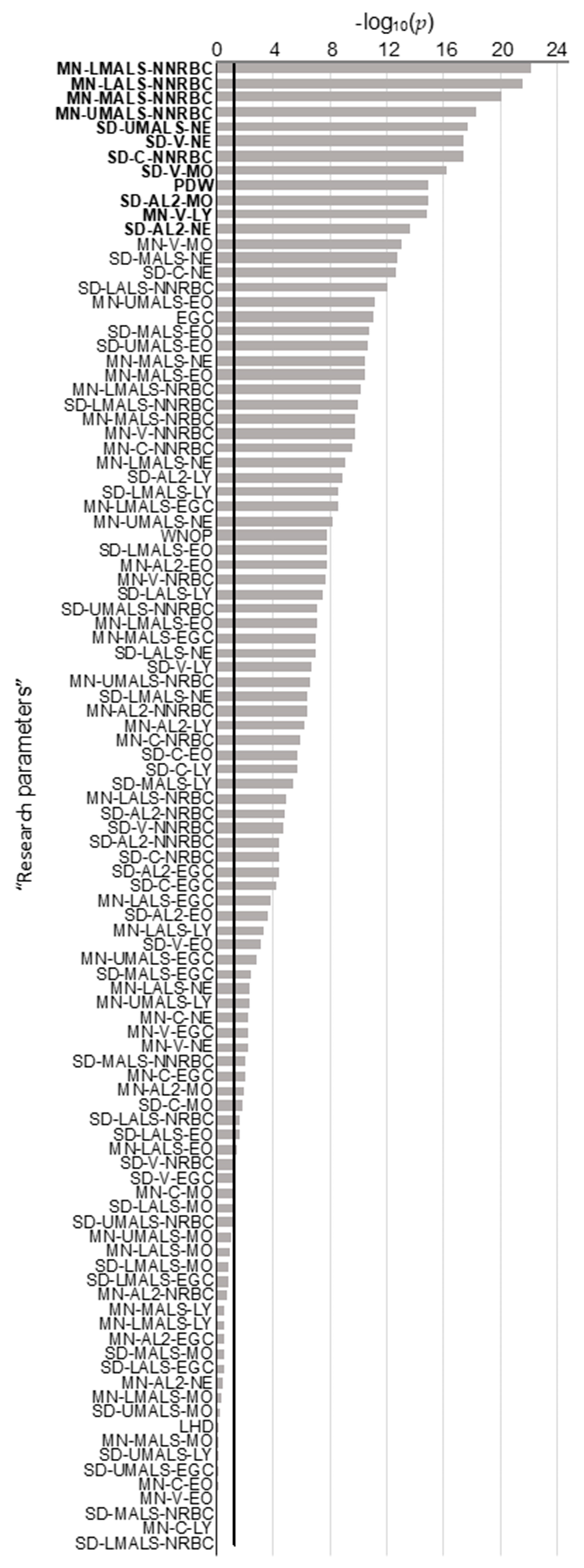
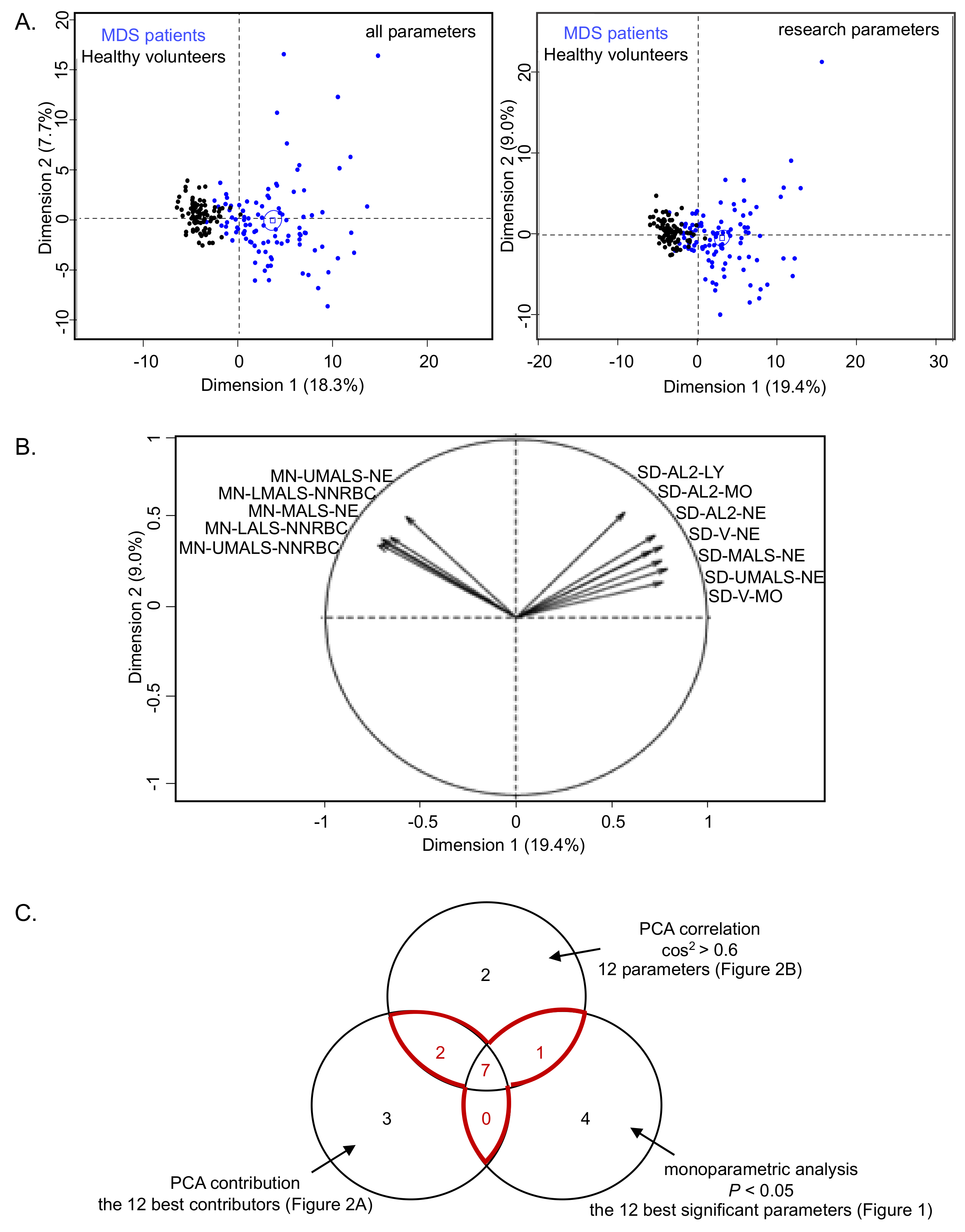
2.1. Univariate and Multivariate Analyzes
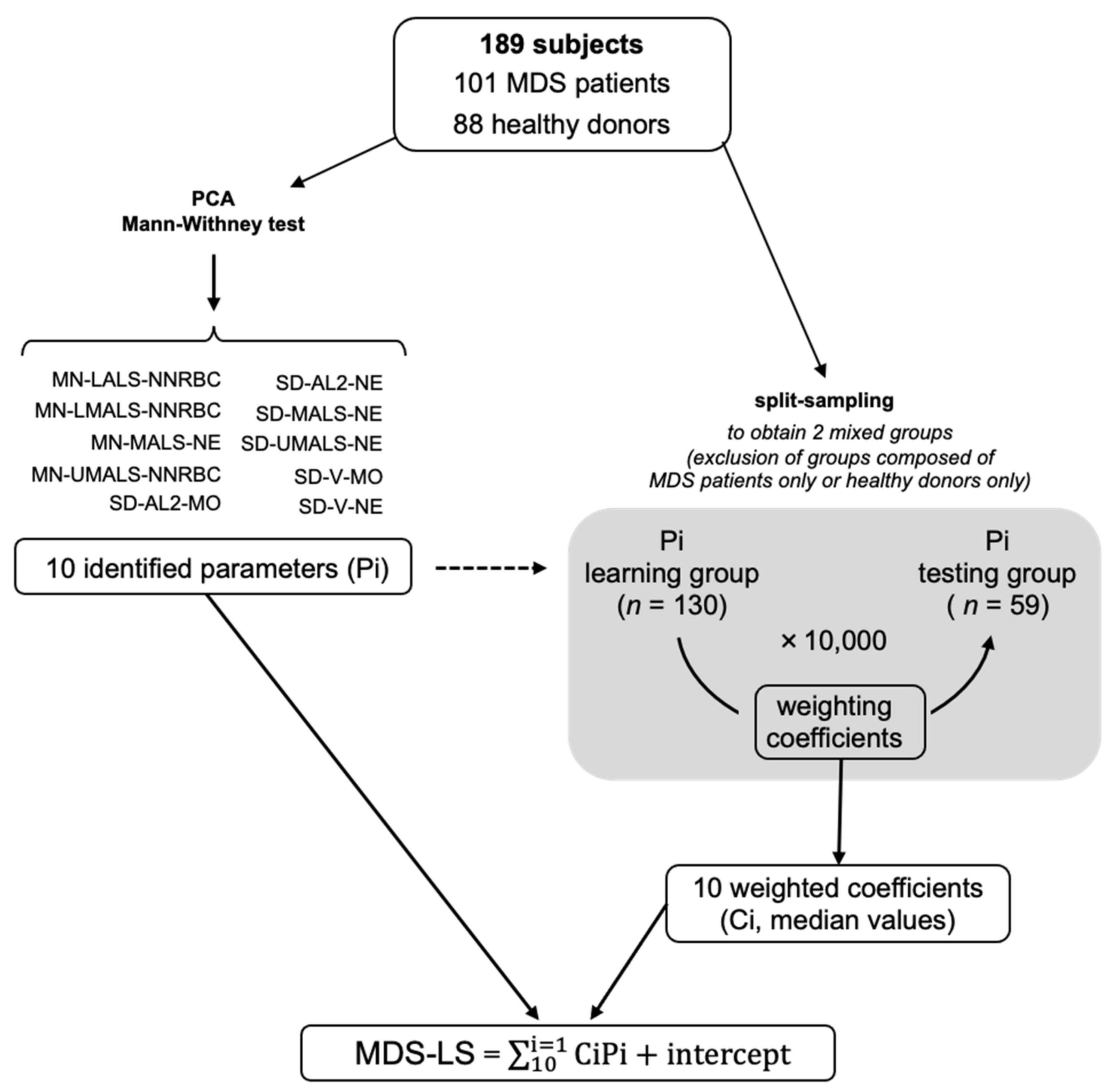
2.2. Mathematical Model Construction and Cross-Validation
2.3. Independent Testing of MDS-LS
3. Discussion
4. Materials and Methods
4.1. Cohorts Description
4.2. Data Collection
4.3. Cross-Validation Strategy to Determine the MDS-LS
4.4. Independent External Testing of the MDS-LS
4.5. Statistical Analyzes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kennedy, A.L.; Shimamura, A. Genetic Predisposition to MDS: Clinical Features and Clonal Evolution. Blood 2019, 133, 1071–1085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Platzbecker, U. Treatment of MDS. Blood 2019, 133, 1096–1107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malcovati, L.; Hellström-Lindberg, E.; Bowen, D.; Adès, L.; Cermak, J.; Canizo, C.; Della Porta, M.G.; Fenaux, P.; Gattermann, N.; Germing, U.; et al. Diagnosis and Treatment of Primary Myelodysplastic Syndromes in Adults: Recommendations from the European LeukemiaNet. Blood 2013, 122, 2943–2964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, A.M. Why Are Myelodysplastic Syndromes Unrecognized and Underdiagnosed?: A Primary Care Perspective. Am. J. Med. 2012, 125, S15–S17. [Google Scholar] [CrossRef]
- Naqvi, K.; Jabbour, E.; Bueso-ramos, C.; Pierce, S.; Borthakur, G.; Estrov, Z.; Ravandi, F.; Faderl, S.; Kantarjian, H.; Garcia-manero, G.; et al. Implications of Discrepancy in Morphologic Diagnosis of Myelodysplastic Syndrome between Referral and Tertiary Care Centers. Blood 2012, 118, 4690–4693. [Google Scholar] [CrossRef]
- Brown, M.; Wittwer, C. Flow Cytometry: Principles and Clinical Applications in Hematology. Clin. Chem. 2000, 46, 1221–1229. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.H.; Kim, Y.; Lim, J.; Kim, M.; Oh, E.J.; Lee, H.K.; Park, Y.J.; Min, W.S.; Cho, B.; Lee, K.; et al. Determination of Acute Leukemia Lineage with New Morphologic Parameters Available in the Complete Blood Cell Count. Ann. Clin. Lab. Sci. 2014, 44, 19–26. [Google Scholar]
- Shin, S.; Park, S.H.; Park, J. Incidental Identification of Plasmodium Vivax during Routine Complete Blood Count Analysis Using the UniCel DxH 800. Ann. Lab. Med. 2018, 38, 165–168. [Google Scholar] [CrossRef]
- Chaves, F.; Tierno, B.; Xu, D. Quantitative Determination of Neutrophil VCS Parameters by the Coulter Automated Hematology Analyzer New and Reliable Indicators for Acute Bacterial Infection. Am. J. Clin. Pathol. 2005, 124, 440–444. [Google Scholar] [CrossRef]
- Abiramalatha, T.; Santhanam, S.; Mammen, J.J.; Rebekah, G.; Shabeer, M.P.; Choudhury, J.; Nair, S.C. Utility of Neutrophil Volume Conductivity Scatter (VCS) Parameter Changes as Sepsis Screen in Neonates. J. Perinatol. 2016, 36, 733–738. [Google Scholar] [CrossRef]
- Crouser, E.D.; Parrillo, J.E.; Seymour, C.; Angus, D.C.; Bicking, K.; Tejidor, L.; Magari, R.; Careaga, D.; Williams, J.; Closser, D.R.; et al. Improved Early Detection of Sepsis in the ED with a Novel Monocyte Distribution Width Biomarker. Chest 2017, 152, 518–526. [Google Scholar] [CrossRef] [PubMed]
- Crouser, E.D.; Parrillo, J.E.; Seymour, C.W.; Angus, D.C.; Bicking, K.; Esguerra, V.G.; Peck-Palmer, O.M.; Magari, R.T.; Julian, M.W.; Kleven, J.M.; et al. Monocyte Distribution Width: A Novel Indicator of Sepsis-2 and Sepsis-3 in High-Risk Emergency Department Patients. Crit. Care Med. 2019, 47, 1018–1025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, Y.J.; Kim, J.H.; Park, Y.J.; Kahng, J.; Lee, H.; Lee, K.Y.; Kim, M.Y.; Han, K.; Lee, W. Evaluation of Cell Population Data on the UniCel DxH 800 Coulter Cellular Analysis System as a Screening for Viral Infection in Children. Int. J. Lab. Hematol. 2012, 34, 283–289. [Google Scholar] [CrossRef]
- Czader, M.; Orazi, A. World Health Organization Classification of Myelodysplastic Syndromes. Curr. Pharm. Des. 2012, 18, 3149–3162. [Google Scholar] [CrossRef] [PubMed]
- Le Roux, G.; Vlad, A.; Eclache, V.; Malanquin, C.; Collon, J.F.; Gantier, M.; Schillinger, F.; Peltier, J.Y.; Savin, B.; Letestu, R.; et al. Routine Diagnostic Procedures of Myelodysplastic Syndromes: Value of a Structural Blood Cell Parameter (NEUT-X) Determined by the Sysmex XE-2100TM. Int. J. Lab. Hematol. 2010, 32, e237–e243. [Google Scholar] [CrossRef]
- Miguel, A.; Orero, M.; Simon, R.; Collado, R.; Perez, P.L.; Pacios, A.; Iglesias, R.; Martinez, A.; Carbonell, F. Automated Neutrophil Morphology and Its Utility in the Assessment of Neutrophil Dysplasia. Lab. Hematol. 2007, 13, 98–102. [Google Scholar] [CrossRef]
- Haschke-Becher, E.; Vockenhuber, M.; Niedetzky, P.; Totzke, U.; Gabriel, C. A New High-Throughput Screening Method for the Detection of Chronic Lymphatic Leukemia and Myelodysplastic Syndrome. Clin. Chem. Lab. Med. 2008, 46, 85–88. [Google Scholar] [CrossRef]
- Raess, P.W.; van de Geijn, G.J.M.; Njo, T.L.; Klop, B.; Sukhachev, D.; Wertheim, G.; Mcaleer, T.; Master, S.R.; Bagg, A. Automated Screening for Myelodysplastic Syndromes through Analysis of Complete Blood Count and Cell Population Data Parameters. Am. J. Hematol. 2014, 89, 369–374. [Google Scholar] [CrossRef]
- Kim, S.Y.; Park, Y.; Kim, H.; Kim, J.; Kwon, G.C.; Koo, S.H. Discriminating Myelodysplastic Syndrome and Other Myeloid Malignancies from Non-Clonal Disorders by Multiparametric Analysis of Automated Cell Data. Clin. Chim. Acta 2018, 480, 56–64. [Google Scholar] [CrossRef]
- Shestakova, A.; Nael, A.; Nora, V.; Rezk, S.; Zhao, X. Automated Leukocyte Parameters Are Useful in the Assessment of Myelodysplastic Syndromes. Cytom. Part B Clin. Cytom. 2020, 1–13. [Google Scholar] [CrossRef]
- Rocco, V.; Maconi, M.; Gioia, M.; Silvestri, M.G.; Tanca, D.; Catalano, T.; Avino, D.; Di Palma, A.; Rovetti, A.; Danise, P. Possibility of Myelodysplastic Syndromes Screening Using a Complete Blood Automated Cell Count. Leuk. Res. 2011, 35, 1623–1627. [Google Scholar] [CrossRef] [PubMed]
- Boutault, R.; Peterlin, P.; Boubaya, M.; Sockel, K.; Chevallier, P.; Garnier, A.; Guillaume, T.; Le Bourgeois, A.; Debord, C.; Godon, C.; et al. A Novel Complete Blood Count-Based Score to Screen for Myelodysplastic Syndrome in Cytopenic Patients. Br. J. Haematol. 2018, 183, 736–746. [Google Scholar] [CrossRef] [PubMed]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 Revision to the World Health Organization Classification of Myeloid Neoplasms and Acute Leukemia. Blood 2016, 127, 2391–2406. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Tang, H.; Chen, K.; Chen, Y.; Xu, D. Biological Variations of Hematologic Parameters Determined by UniCel DxH 800 Hematology Analyzer. Arch. Pathol. Lab. Med. 2013, 137, 1106–1110. [Google Scholar] [CrossRef] [Green Version]
- Urrechaga, E. The New Mature Red Cell Parameter, Low Haemoglobin Density of the Beckman-Coulter LH750: Clinical Utility in the Diagnosis of Iron Deficiency. Int. J. Lab. Hematol. 2010, 32, e144–e150. [Google Scholar] [CrossRef]
- Dopsaj, V.; Martinovic, J.; Dopsaj, M. Early Detection of Iron Deficiency in Elite Athletes: Could Microcytic Anemia Factor (Maf) Be Useful? Int. J. Lab. Hematol. 2014, 36, 37–44. [Google Scholar] [CrossRef]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Steyerberg, E.W.; Harrell, F.E.; Borsboom, G.J.J.; Eijkemans, M.J.; Vergouwe, Y.; Habbema, J.D.F. Internal Validation of Predictive Models. J. Clin. Epidemiol. 2001, 54, 774–781. [Google Scholar] [CrossRef]

| Characteristics | #7 | #17 | #30 | #35 | #197 | #199 | #244 | #245 | #249 | #253 | #256 | Flag |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age | 86 | 71 | 82 | 90 | 90 | 68 | 79 | 83 | 89 | 86 | 77 | |
| Gender | F | F | M | F | M | M | F | M | M | F | F | |
| MDS groups | MDS-SLD | MDS-EB-1 | MDS-MLD | MDS-RS-SLD | MDS-SLD | MDS-SLD | MDS-RS-MLD | MDS-MLD | MDS-SLD | MDS-MLD | Del(5q) | |
| Delay from diagnosis (months) | 0 | 1 | 5 | 60 | 1 | 0 | 6 | NA | 0 | 0 | 0 | |
| Cytogenetics | normal | normal | normal | normal | tri15 | normal | del(11q), tri8 | NA | normal | normal | del(5q) | |
| IPPS-R | 1 | 3 | 2 | 2 | 4 | 2 | 4.5 | NA | 2.5 | 1 | 2 | |
| RCC (<120 days) | N | N | N | Y | N | N | Y | NA | Y | N | Y | |
| Growth factors | N | N | N | N | N | N | N | NA | N | N | N | |
| Other treatment | N | N | N | N | N | N | N | NA | N | N | N | |
| RBC (T/L) | 3.6 | 3.2 | 2.8 | 2.6 | 2.3 | 2.7 | 3.3 | 4.4 | 2.9 | 3.8 | 3.5 | |
| Hb (g/L) | 104 | 109 | 82 | 81 | 82 | 87 | 100 | 130 | 96 | 114 | 113 | <80 |
| Hct (%) | 31.2 | 32.0 | 25.1 | 24.3 | 23.9 | 27.3 | 30.9 | 38.7 | 28.4 | 34.9 | 34.5 | |
| MCV (fL) | 85.8 | 100.6 | 90.9 | 93.1 | 102.2 | 101.2 | 93.0 | 87.4 | 97.9 | 92.6 | 99.0 | >105 |
| MCH (pg/cell) | 28.6 | 34.3 | 29.9 | 31.0 | 35.0 | 32.3 | 30.1 | 29.2 | 33.0 | 30.3 | 32.4 | |
| MCHC (g/dL) | 33.3 | 34.1 | 32.8 | 33.3 | 34.3 | 31.9 | 32.4 | 33.5 | 33.7 | 32.7 | 32.7 | >36.0 |
| RDW (%) | 14.6 | 15.0 | 17.8 | 16.6 | 15.8 | 16.9 | 18.9 | 15.4 | 22.0 | 14.3 | 20.1 | >22.0 |
| Plt (G/L) | 227 | 146 | 394 | 373 | 111 | 383 | 182 | 104 | 188 | 257 | 447 | <100 |
| MPV (fL) | 9.3 | 11.3 | 8.8 | 9.4 | 8.5 | 7.8 | 10.7 | 10.8 | 8.6 | 8.6 | 10.7 | <7.0 |
| WBC(G/L) | 5.7 | 7.4 | 8.4 | 8.4 | 3.9 | 8.2 | 5.5 | 5.3 | 9.6 | 4.5 | 2.8 | |
| % NE | 67.5 | 83.4 | 79.0 | 74.8 | 78.7 | 79.0 | 61.4 | 76.5 | 79.8 | 46.0 | 52.6 | |
| % LY | 20.9 | 7.4 | 15.1 | 10.0 | 11.6 | 10.7 | 22.7 | 8.5 | 6.0 | 39.9 | 30.9 | |
| % MO | 8.2 | 8.3 | 3.4 | 10.4 | 6.9 | 7.1 | 10.7 | 12.8 | 11.2 | 10.2 | 9.4 | >20.0 |
| % EO | 2.4 | 0.4 | 1.3 | 2.5 | 2.3 | 2.6 | 4.2 | 1.6 | 1.9 | 3.5 | 4.1 | |
| % BA | 1.0 | 0.5 | 1.2 | 2.3 | 0.5 | 0.6 | 1.0 | 0.6 | 1.1 | 0.4 | 3.0 | |
| Abs NE (G/L) | 3.8 | 6.2 | 6.6 | 6.3 | 3.1 | 6.5 | 3.4 | 4.0 | 7.7 | 2.1 | 1.5 | <1.5 |
| Abs LY (G/L) | 1.2 | 0.5 | 1.3 | 0.8 | 0.5 | 0.9 | 1.3 | 0.5 | 0.6 | 1.8 | 0.9 | >4.0 |
| Abs MO (G/L) | 0.5 | 0.6 | 0.3 | 0.9 | 0.3 | 0.6 | 0.6 | 0.7 | 1.1 | 0.5 | 0.3 | >1.5 |
| Abs EO (G/L) | 0.1 | 0.0 | 0.1 | 0.2 | 0.1 | 0.2 | 0.2 | 0.1 | 0.2 | 0.2 | 0.1 | >1.5 |
| Abs BA (G/L) | 0.1 | 0.0 | 0.1 | 0.2 | 0.0 | 0.0 | 0.1 | 0.0 | 0.1 | 0.0 | 0.1 | >0.3 |
| % NRBC | 0.1 | 0.0 | 0.0 | 0.3 | 0.1 | 0.2 | 0.1 | 0.1 | 0.5 | 0.1 | 0.1 | >2.0 |
| Abs NRBC (G/L) | 0.01 | 0.00 | 0.00 | 0.02 | 0.00 | 0.02 | 0.01 | 0.00 | 0.05 | 0.01 | 0.00 | |
| MDS-LS | −6.5 | −8.7 | −54.4 | −10.4 | −16.5 | −41.9 | −23.1 | −16.3 | −4.0 | −2.6 | −21.5 | <0.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ravalet, N.; Foucault, A.; Picou, F.; Gombert, M.; Renoult, E.; Lejeune, J.; Vallet, N.; Lachot, S.; Rault, E.; Gyan, E.; et al. Automated Early Detection of Myelodysplastic Syndrome within the General Population Using the Research Parameters of Beckman–Coulter DxH 800 Hematology Analyzer. Cancers 2021, 13, 389. https://doi.org/10.3390/cancers13030389
Ravalet N, Foucault A, Picou F, Gombert M, Renoult E, Lejeune J, Vallet N, Lachot S, Rault E, Gyan E, et al. Automated Early Detection of Myelodysplastic Syndrome within the General Population Using the Research Parameters of Beckman–Coulter DxH 800 Hematology Analyzer. Cancers. 2021; 13(3):389. https://doi.org/10.3390/cancers13030389
Chicago/Turabian StyleRavalet, Noémie, Amélie Foucault, Frédéric Picou, Martin Gombert, Emmanuel Renoult, Julien Lejeune, Nicolas Vallet, Sébastien Lachot, Emmanuelle Rault, Emmanuel Gyan, and et al. 2021. "Automated Early Detection of Myelodysplastic Syndrome within the General Population Using the Research Parameters of Beckman–Coulter DxH 800 Hematology Analyzer" Cancers 13, no. 3: 389. https://doi.org/10.3390/cancers13030389
APA StyleRavalet, N., Foucault, A., Picou, F., Gombert, M., Renoult, E., Lejeune, J., Vallet, N., Lachot, S., Rault, E., Gyan, E., Bene, M. C., & Herault, O. (2021). Automated Early Detection of Myelodysplastic Syndrome within the General Population Using the Research Parameters of Beckman–Coulter DxH 800 Hematology Analyzer. Cancers, 13(3), 389. https://doi.org/10.3390/cancers13030389






