Artificial Intelligence for Histology-Based Detection of Microsatellite Instability and Prediction of Response to Immunotherapy in Colorectal Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Histological and Clinical Predictors of Microsatellite Instability
3. What Is Deep Learning and How Does It Apply to Digital Pathology?
4. Application of Deep Learning to Digital Pathology in Oncology
5. Predicting MSI Status with Deep Learning
6. Predicting Response to Immunotherapy with Deep Learning
7. Future Directions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Araghi, M.; Soerjomataram, I.; Jenkins, M.; Brierley, J.; Morris, E.; Bray, F.; Arnold, M. Global trends in colorectal cancer mortality: Projections to the year 2035. Int. J. Cancer 2019, 144, 2992–3000. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Sauer, A.G.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef] [PubMed]
- Lal, N.; White, B.S.; Goussous, G.; Pickles, O.; Mason, M.J.; Beggs, A.D.; Taniere, P.; Willcox, B.E.; Guinney, J.; Middleton, G. KRAS Mutation and Consensus Molecular Subtypes 2 and 3 Are Independently Associated with Reduced Immune Infiltration and Reactivity in Colorectal Cancer. Clin. Cancer Res. 2018, 24, 224–233. [Google Scholar] [CrossRef] [PubMed]
- Becht, E.; De Reyniès, A.; Giraldo, N.A.; Pilati, C.; Buttard, B.; Lacroix, L.; Selves, J.; Sautès-Fridman, C.; Laurent-Puig, P.; Fridman, W.H. Immune and Stromal Classification of Colorectal Cancer Is Associated with Molecular Subtypes and Relevant for Precision Immunotherapy. Clin. Cancer Res. 2016, 22, 4057–4066. [Google Scholar] [CrossRef] [PubMed]
- Guinney, J.; Dienstmann, R.; Wang, X.; De Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Seppälä, T.T.; Bohm, J.; Friman, M.; Lahtinen, L.; Väyrynen, V.M.J.; Liipo, T.K.E.; Ristimäki, A.P.; Kairaluoma, M.V.J.; Kellokumpu, I.H.; Kuopio, T.H.I.; et al. Combination of microsatellite instability and BRAF mutation status for subtyping colorectal cancer. Br. J. Cancer 2015, 112, 1966–1975. [Google Scholar] [CrossRef]
- Sjoquist, K.M.; Renfro, L.A.; Simes, R.J.; Tebbutt, N.C.; Clarke, S.; Seymour, M.T.; Adams, R.; Maughan, T.S.; Saltz, L.; Goldberg, R.M.; et al. Personalizing Survival Predictions in Advanced Colorectal Cancer: The ARCAD Nomogram Project. J. Natl. Cancer Inst. 2018, 110, 638–648. [Google Scholar] [CrossRef]
- Hynes, S.O.; Coleman, H.G.; Kelly, P.J.; Irwin, S.; O’Neill, R.F.; Gray, R.T.; Mcgready, C.; Dunne, P.D.; McQuaid, S.; James, J.A.; et al. Back to the future: Routine morphological assessment of the tumour microenvironment is prognostic in stage II/III colon cancer in a large population-based study. Histopathology 2017, 71, 12–26. [Google Scholar] [CrossRef]
- Mlecnik, B.; Bindea, G.; Angell, H.K.; Maby, P.; Angelova, M.; Tougeron, D.; Church, S.E.; Lafontaine, L.; Fischer, M.; Fredriksen, T.; et al. Integrative Analyses of Colorectal Cancer Show Immunoscore Is a Stronger Predictor of Patient Survival Than Microsatellite Instability. Immunity 2016, 44, 698–711. [Google Scholar] [CrossRef]
- Rozek, L.S.; Schmit, S.L.; Greenson, J.K.; Tomsho, L.P.; Rennert, H.S.; Rennert, G.; Gruber, S.B. Tumor-Infiltrating Lymphocytes, Crohn’s-Like Lymphoid Reaction, and Survival from Colorectal Cancer. J. Natl. Cancer Inst. 2016, 108. [Google Scholar] [CrossRef] [PubMed]
- Li, G.-M. Mechanisms and functions of DNA mismatch repair. Cell Res. 2007, 18, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Vilar, E.; Gruber, S.B. Microsatellite instability in colorectal cancer—The stable evidence. Nat. Rev. Clin. Oncol. 2010, 7, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Cerretelli, G.; Ager, A.; Arends, M.J.; Frayling, I.M. Molecular pathology of Lynch syndrome. J. Pathol. 2020, 250, 518–531. [Google Scholar] [CrossRef]
- Boland, P.M.; Yurgelun, M.B.; Boland, C.R. Recent progress in Lynch syndrome and other familial colorectal cancer syndromes. CA Cancer J. Clin. 2018, 68, 217–231. [Google Scholar] [CrossRef] [PubMed]
- Sinicrope, F.A.; Rego, R.L.; Halling, K.C.; Foster, N.; Sargent, D.J.; La Plant, B.; French, A.J.; Laurie, J.A.; Goldberg, R.M.; Thibodeau, S.N.; et al. Prognostic Impact of Microsatellite Instability and DNA Ploidy in Human Colon Carcinoma Patients. Gastroenterology 2006, 131, 729–737. [Google Scholar] [CrossRef]
- Samowitz, W.S.; Curtin, K.; Ma, K.N.; Schaffer, D.; Coleman, L.W.; Leppert, M.; Slattery, M.L. Microsatellite instability in sporadic colon cancer is associated with an improved prognosis at the population level. Cancer Epidemiol. Biomark. Prev. 2001, 10, 917–923. [Google Scholar]
- Maoz, A.; Dennis, M.; Greenson, J.K. The Crohn’s-Like Lymphoid Reaction to Colorectal Cancer-Tertiary Lymphoid Structures with Immunologic and Potentially Therapeutic Relevance in Colorectal Cancer. Front. Immunol. 2019, 10, 1884. [Google Scholar] [CrossRef]
- Overman, M.J.; McDermott, R.; Leach, J.L.; Lonardi, S.; Lenz, H.-J.; Morse, M.A.; Desai, J.; Hill, A.; Axelson, M.; Moss, R.A.; et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): An open-label, multicentre, phase 2 study. Lancet Oncol. 2017, 18, 1182–1191. [Google Scholar] [CrossRef]
- Ribic, C.M.; Sargent, D.J.; Moore, M.J.; Thibodeau, S.N.; French, A.J.; Goldberg, R.M.; Hamilton, S.R.; Laurent-Puig, P.; Gryfe, R.; Shepherd, L.E.; et al. Tumor Microsatellite-Instability Status as a Predictor of Benefit from Fluorouracil-Based Adjuvant Chemotherapy for Colon Cancer. N. Engl. J. Med. 2003, 349, 247–257. [Google Scholar] [CrossRef]
- De Vos tot Nederveen Cappel, W.H.; Meulenbeld, H.J.; Kleibeuker, J.H.; Nagengast, F.M.; Menko, F.H.; Griffioen, G.; Cats, A.; Morreau, H.; Gelderblom, H.; Vasen, H.F.A. Survival after adjuvant 5-FU treatment for stage III colon cancer in hereditary nonpolyposis colorectal cancer. Int. J. Cancer 2004, 109, 468–471. [Google Scholar] [CrossRef] [PubMed]
- Sargent, D.J.; Marsoni, S.; Monges, G.; Thibodeau, S.N.; Labianca, R.; Hamilton, S.R.; French, A.J.; Kabat, B.; Foster, N.R.; Torri, V.; et al. Defective Mismatch Repair as a Predictive Marker for Lack of Efficacy of Fluorouracil-Based Adjuvant Therapy in Colon Cancer. J. Clin. Oncol. 2010, 28, 3219–3226. [Google Scholar] [CrossRef] [PubMed]
- Webber, E.M.; Kauffman, T.L.; O’Connor, E.; Goddard, K.A. Systematic review of the predictive effect of MSI status in colorectal cancer patients undergoing 5FU-based chemotherapy. BMC Cancer 2015, 15, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sepulveda, A.R.; Hamilton, S.R.; Allegra, C.J.; Grody, W.; Cushman-Vokoun, A.M.; Funkhouser, W.K.; Kopetz, S.E.; Lieu, C.; Lindor, N.M.; Minsky, B.D.; et al. Molecular Biomarkers for the Evaluation of Colorectal Cancer: Guideline From the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and American Society of Clinical Oncology. J. Mol. Diagn. 2017, 19, 187–225. [Google Scholar] [CrossRef]
- Shaikh, T.; Handorf, E.A.; Meyer, J.E.; Hall, M.J.; Esnaola, N.F. Mismatch Repair Deficiency Testing in Patients with Colorectal Cancer and Nonadherence to Testing Guidelines in Young Adults. JAMA Oncol. 2018, 4, e173580. [Google Scholar] [CrossRef]
- Greenson, J.K.; Huang, S.-C.; Herron, C.; Moreno, V.; Bonner, J.D.; Tomsho, L.P.; Ben-Izhak, O.; Cohen, H.I.; Trougouboff, P.; Bejhar, J.; et al. Pathologic Predictors of Microsatellite Instability in Colorectal Cancer. Am. J. Surg. Pathol. 2009, 33, 126–133. [Google Scholar] [CrossRef]
- Jenkins, M.A.; Hayashi, S.; O’Shea, A.-M.; Burgart, L.J.; Smyrk, T.C.; Shimizu, D.; Waring, P.M.; Ruszkiewicz, A.R.; Pollett, A.F.; Redston, M.; et al. Pathology Features in Bethesda Guidelines Predict Colorectal Cancer Microsatellite Instability: A Population-Based Study. Gastroenterology 2007, 133, 48–56. [Google Scholar] [CrossRef]
- Greenson, J.K.; Bonner, J.D.; Ben-Yzhak, O.; Cohen, H.I.; Miselevich, I.; Resnick, M.B.; Trougouboff, P.; Tomsho, L.D.; Kim, E.; Low, M.; et al. Phenotype of Microsatellite Unstable Colorectal Carcinomas: Well-Differentiated and Focally Mucinous Tumors and the Absence of Dirty Necrosis Correlate With Microsatellite Instability. Am. J. Surg. Pathol. 2003, 27, 563–570. [Google Scholar] [CrossRef]
- Walsh, M.D.; Cummings, M.C.; Buchanan, D.D.; Dambacher, W.M.; Arnold, S.; McKeone, D.; Byrnes, R.; Barker, M.A.; Leggett, B.A.; Gattas, M.; et al. Molecular, Pathologic, and Clinical Features of Early-Onset Endometrial Cancer: Identifying Presumptive Lynch Syndrome Patients. Clin. Cancer Res. 2008, 14, 1692–1700. [Google Scholar] [CrossRef]
- Alexander, J.; Watanabe, T.; Wu, T.-T.; Rashid, A.; Li, S.; Hamilton, S.R. Histopathological Identification of Colon Cancer with Microsatellite Instability. Am. J. Pathol. 2001, 158, 527–535. [Google Scholar] [CrossRef]
- Buckowitz, A.; Knaebel, H.-P.; Benner, A.; Bläker, H.; Gebert, J.; Kienle, P.; Doeberitz, M.V.K.; Kloor, M. Microsatellite instability in colorectal cancer is associated with local lymphocyte infiltration and low frequency of distant metastases. Br. J. Cancer 2005, 92, 1746–1753. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.S.; Bird, M.J.; Jorissen, R.N.; Yu, Y.L.; Walker, F.; Zhang, H.H.; Nice, E.C.; Burgess, A.W. Nonsense Mediated Decay Resistant Mutations Are a Source of Expressed Mutant Proteins in Colon Cancer Cell Lines with Microsatellite Instability. PLoS ONE 2010, 5, e16012. [Google Scholar] [CrossRef] [PubMed]
- Willis, J.A.; Reyes-Uribe, L.; Chang, K.; Lipkin, S.M.; Vilar, E. Immune Activation in Mismatch Repair–Deficient Carcinogenesis: More Than Just Mutational Rate. Clin. Cancer Res. 2020, 26, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Umar, A.; Boland, C.R.; Terdiman, J.P.; Syngal, S.; De La Chapelle, A.; Rüschoff, J.; Fishel, R.; Lindor, N.M.; Burgart, L.J.; Hamelin, R.; et al. Revised Bethesda Guidelines for Hereditary Nonpolyposis Colorectal Cancer (Lynch Syndrome) and Microsatellite Instability. J. Natl. Cancer Inst. 2004, 96, 261–268. [Google Scholar] [CrossRef]
- Hyde, A.; Fontaine, D.; Stuckless, S.; Green, R.; Pollett, A.; Simms, M.; Sipahimalani, P.; Parfrey, P.; Younghusband, B. A Histology-Based Model for Predicting Microsatellite Instability in Colorectal Cancers. Am. J. Surg. Pathol. 2010, 34, 1820–1829. [Google Scholar] [CrossRef]
- Fujiyoshi, K.; Yamaguchi, T.; Kakuta, M.; Takahashi, A.; Arai, Y.; Yamada, M.; Yamamoto, G.; Ohde, S.; Takao, M.; Horiguchi, S.-I.; et al. Predictive model for high-frequency microsatellite instability in colorectal cancer patients over 50 years of age. Cancer Med. 2017, 6, 1255–1263. [Google Scholar] [CrossRef]
- Roman, R.; Verdu, M.; Calvo, M.; Vidal, A.; Sanjuan, X.; Jimeno, M.; Salas, A.; Autonell, J.; Trias, I.; González, M.; et al. Microsatellite instability of the colorectal carcinoma can be predicted in the conventional pathologic examination. A prospective multicentric study and the statistical analysis of 615 cases consolidate our previously proposed logistic regression model. Virchows Arch. 2010, 456, 533–541. [Google Scholar] [CrossRef]
- Bessa, X.; Alenda, C.; Paya, A.; Álvarez, C.; Iglesias, M.; Seoane, A.; Dedeu, J.M.; Abulí, A.; Ilzarbe, L.; Navarro, G.; et al. Validation Microsatellite Path Score in a Population-Based Cohort of Patients with Colorectal Cancer. J. Clin. Oncol. 2011, 29, 3374–3380. [Google Scholar] [CrossRef]
- Brazowski, E.; Rozen, P.; Pel, S.; Samuel, Z.; Solar, I.; Rosner, G. Can a gastrointestinal pathologist identify microsatellite instability in colorectal cancer with reproducibility and a high degree of specificity? Fam. Cancer 2012, 11, 249–257. [Google Scholar] [CrossRef]
- Bera, K.; Schalper, K.A.; Rimm, D.L.; Velcheti, V.; Madabhushi, A. Artificial intelligence in digital pathology—New tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 2019, 16, 703–715. [Google Scholar] [CrossRef] [PubMed]
- Acs, B.; Rantalainen, M.; Hartman, J. Artificial intelligence as the next step towards precision pathology. J. Intern. Med. 2020, 288, 62–81. [Google Scholar] [CrossRef] [PubMed]
- Kather, J.N.; Pearson, A.T.; Halama, N.; Jäger, D.; Krause, J.; Loosen, S.H.; Marx, A.; Boor, P.; Tacke, F.; Neumann, U.P.; et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 2019, 25, 1054–1056. [Google Scholar] [CrossRef] [PubMed]
- Rashidi, H.H.; Tran, N.K.; Betts, E.V.; Howell, L.P.; Green, R. Artificial Intelligence and Machine Learning in Pathology: The Present Landscape of Supervised Methods. Acad. Pathol. 2019, 6, 2374289519873088. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Mobadersany, P.; Yousefi, S.; Amgad, M.; Gutman, D.A.; Barnholtz-Sloan, J.S.; Vega, J.E.V.; Brat, D.J.; Cooper, L.A. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc. Natl. Acad. Sci. USA 2018, 115, e2970–e2979. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Wolf, L.; Hassner, T.; Maoz, I. Face recognition in unconstrained videos with matched background similarity. In Proceedings of the CVPR 2011, Colorado Springs, CO, USA, 20–25 June 2011; pp. 529–534. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 1–9. [Google Scholar]
- Bizzego, A.; Bussola, N.; Chierici, M.; Maggio, V.; Francescatto, M.; Cima, L.; Cristoforetti, M.; Jurman, G.; Furlanello, C. Evaluating reproducibility of AI algorithms in digital pathology with DAPPER. PLoS Comput. Biol. 2019, 15, e1006269. [Google Scholar] [CrossRef]
- Nagpal, K.; Foote, D.; Liu, Y.; Chen, P.-H.C.; Wulczyn, E.; Tan, F.; Olson, N.; Smith, G.; Mohtashamian, A.; Wren, J.H.; et al. Development and validation of a deep learning algorithm for improving Gleason scoring of prostate cancer. NPJ Digit. Med. 2019, 2, 1–10. [Google Scholar] [CrossRef]
- Bychkov, D.; Linder, N.; Turkki, R.; Nordling, S.; Kovanen, P.E.; Verrill, C.; Walliander, M.; Lundin, M.; Haglund, C.; Lundin, J. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Kather, J.N.; Krisam, J.; Charoentong, P.; Luedde, T.; Herpel, E.; Weis, C.-A.; Gaiser, T.; Marx, A.; Valous, N.A.; Ferber, D.; et al. Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study. PLoS Med. 2019, 16, e1002730. [Google Scholar] [CrossRef] [PubMed]
- Skrede, O.-J.; De Raedt, S.; Kleppe, A.; Hveem, T.S.; Liestøl, K.; Maddison, J.; Askautrud, H.A.; Pradhan, M.; Nesheim, J.A.; Albregtsen, F.; et al. Deep learning for prediction of colorectal cancer outcome: A discovery and validation study. Lancet 2020, 395, 350–360. [Google Scholar] [CrossRef]
- Pagès, F.; Mlecnik, B.; Marliot, F.; Bindea, G.; Ou, F.-S.; Bifulco, C.; Lugli, A.; Zlobec, I.; Rau, T.T.; Berger, M.D.; et al. International validation of the consensus Immunoscore for the classification of colon cancer: A prognostic and accuracy study. Lancet 2018, 391, 2128–2139. [Google Scholar] [CrossRef]
- Coudray, N.; Ocampo, P.S.; Sakellaropoulos, T.; Narula, N.; Snuderl, M.; Fenyo, D.; Moreira, A.L.; Razavian, N.; Tsirigos, A. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat. Med. 2018, 24, 1559–1567. [Google Scholar] [CrossRef] [PubMed]
- Yip, S.S.F.; Sha, L.; Osinski, B.L.; Ho, I.Y.; Tan, T.L.; Willis, C.; Weiss, H.; Beaubier, N.; Mahon, B.M.; Taxter, T.J. Multi-field-of-view deep learning model predicts nonsmall cell lung cancer programmed death-ligand 1 status from whole-slide hematoxylin and eosin images. J. Pathol. Inform. 2019, 10, 24. [Google Scholar] [CrossRef] [PubMed]
- Couture, H.D.; Williams, L.A.; Geradts, J.; Nyante, S.J.; Butler, E.N.; Marron, J.S.; Perou, C.M.; Troester, M.A.; Niethammer, M. Image analysis with deep learning to predict breast cancer grade, ER status, histologic subtype, and intrinsic subtype. NPJ Breast Cancer 2018, 4, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sirinukunwattana, K.; Domingo, E.; Richman, S.D.; Redmond, K.L.; Blake, A.; Verrill, C.; Leedham, S.J.; Chatzipli, A.; Hardy, C.; Whalley, C.M.; et al. Image-based consensus molecular subtype (imCMS) classification of colorectal cancer using deep learning. Gut 2020. [Google Scholar] [CrossRef]
- Steu, S.; Baucamp, M.; Von Dach, G.; Bawohl, M.; Dettwiler, S.; Storz, M.; Moch, H.; Schraml, P. A procedure for tissue freezing and processing applicable to both intra-operative frozen section diagnosis and tissue banking in surgical pathology. Virchows Arch. 2008, 452, 305–312. [Google Scholar] [CrossRef]
- Sprung, R.W.; Brock, J.W.C.; Tanksley, J.P.; Li, M.; Washington, M.K.; Slebos, R.J.C.; Liebler, D.C. Equivalence of Protein Inventories Obtained from Formalin-fixed Paraffin-embedded and Frozen Tissue in Multidimensional Liquid Chromatography-Tandem Mass Spectrometry Shotgun Proteomic Analysis. Mol. Cell. Proteom. 2009, 8, 1988–1998. [Google Scholar] [CrossRef]
- Yeh, Y.-C.; Nitadori, J.-I.; Kadota, K.; Yoshizawa, A.; Rekhtman, N.; Moreira, A.L.; Sima, C.S.; Rusch, V.W.; Adusumilli, P.S.; Travis, W.D. Using frozen section to identify histological patterns in stage I lung adenocarcinoma of ≤3 cm: Accuracy and interobserver agreement. Histopathology 2015, 66, 922–938. [Google Scholar] [CrossRef]
- Ratnavelu, N.D.; Brown, A.P.; Mallett, S.; Scholten, R.J.; Patel, A.; Founta, C.; Galaal, K.; Cross, P.; Naik, R. Intraoperative frozen section analysis for the diagnosis of early stage ovarian cancer in suspicious pelvic masses. Cochrane Database Syst. Rev. 2016, 2016, 010360. [Google Scholar] [CrossRef]
- Mantel, H.T.; Westerkamp, A.C.; Sieders, E.; Peeters, P.M.J.G.; De Jong, K.P.; Boer, M.T.; de Kleine, R.H.; Gouw, A.S.H.; Porte, R.J. Intraoperative frozen section analysis of the proximal bile ducts in hilar cholangiocarcinoma is of limited value. Cancer Med. 2016, 5, 1373–1380. [Google Scholar] [CrossRef]
- Kather, J.N.; Heij, L.R.; Grabsch, H.I.; Loeffler, C.; Echle, A.; Muti, H.S.; Krause, J.; Niehues, J.M.; Sommer, K.A.J.; Bankhead, P.; et al. Pan-cancer image-based detection of clinically actionable genetic alterations. Nat. Rev. Cancer 2020, 1, 789–799. [Google Scholar] [CrossRef]
- Cao, R.; Yang, F.; Ma, S.-C.; Liu, L.; Zhao, Y.; Li, Y.; Wu, D.-H.; Wang, T.; Lu, W.-J.; Cai, W.-J.; et al. Development and interpretation of a pathomics-based model for the prediction of microsatellite instability in Colorectal Cancer. Theranostics 2020, 10, 11080–11091. [Google Scholar] [CrossRef] [PubMed]
- Schmauch, B.; Romagnoni, A.; Pronier, E.; Saillard, C.; Maillé, P.; Calderaro, J.; Kamoun, A.; Sefta, M.; Toldo, S.; Zaslavskiy, M.; et al. A deep learning model to predict RNA-Seq expression of tumours from whole slide images. Nat. Commun. 2020, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Osinski, B.L.; Taxter, T.J.; Perera, J.; Lau, D.J.; Khan, A.A. Adversarial deep learning for microsatellite instability prediction from histopathology slides. In Proceedings of the 1st Conference on Medical Imaging with Deep Learning (MIDL 2018), Amsterdam, The Netherlands, 4–6 July 2018. [Google Scholar]
- Hong, R.; Liu, W.; DeLair, D.; Razavian, N.; Fenyö, D. Predicting Endometrial Cancer Subtypes and Molecular Features from Histopathology Images Using Multi-resolution Deep Learning Models. bioRxiv 2020. [Google Scholar] [CrossRef]
- Echle, A.; Grabsch, H.I.; Quirke, P.; van den Brandt, P.A.; West, N.P.; Hutchins, G.G.; Heij, L.R.; Tan, X.; Richman, S.D.; Krause, J.; et al. Clinical-Grade Detection of Microsatellite Instability in Colorectal Tumors by Deep Learning. Gastroenterology 2020, 159, 1406–1416.e11. [Google Scholar] [CrossRef]
- Gatalica, Z.; Snyder, C.; Maney, T.; Ghazalpour, A.; Holterman, D.A.; Xiao, N.; Overberg, P.; Rose, I.; Basu, G.D.; Vranic, S.; et al. Programmed Cell Death 1 (PD-1) and Its Ligand (PD-L1) in Common Cancers and Their Correlation with Molecular Cancer Type. Cancer Epidemiol. Biomark. Prev. 2014, 23, 2965–2970. [Google Scholar] [CrossRef]
- Wang, H.; Wang, X.; Xu, L.; Zhang, J.; Cao, H. Analysis of the transcriptomic features of microsatellite instability subtype colon cancer. BMC Cancer 2019, 19, 605. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Zhang, X.; Zhou, X.; Lin, M.; Sun, J. ShuffleNet: An Extremely Efficient Convolutional Neural Network for Mobile Devices. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, MA, USA, 18–23 June 2018; pp. 6848–6856. [Google Scholar]
- Quellec, G.; Cazuguel, G.; Cochener, B.; Lamard, M. Multiple-Instance Learning for Medical Image and Video Analysis. IEEE Rev. Biomed. Eng. 2017, 10, 213–234. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Lu, W.; Yang, F.; Liu, L.; Dong, Z.; Tang, W.; Chang, J.; Huan, W.; Huang, K.; Yao, J. Microsatellite Instability Prediction of Uterine Corpus Endometrial Carcinoma Based on H&E Histology Whole-Slide Imaging. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 1289–1292. [Google Scholar] [CrossRef]
- Khan, A.A. Generalizable and Interpretable Deep Learning Framework for Predicting MSI from Histopathology Slide Images. U.S. Patent 20190347557, 14 November 2019. [Google Scholar]
- Hellmann, M.D.; Kim, T.-W.; Lee, C.; Goh, B.-C.; Miller, W.; Oh, D.-Y.; Jamal, R.; Chee, C.-E.; Chow, L.; Gainor, J.; et al. Phase Ib study of atezolizumab combined with cobimetinib in patients with solid tumors. Ann. Oncol. 2019, 30, 1134–1142. [Google Scholar] [CrossRef]
- Chalabi, M.; Fanchi, L.F.; Dijkstra, K.K.; Berg, J.G.V.D.; Aalbers, A.G.; Sikorska, K.; Lopez-Yurda, M.; Grootscholten, C.; Beets, G.L.; Snaebjornsson, P.; et al. Neoadjuvant immunotherapy leads to pathological responses in MMR-proficient and MMR-deficient early-stage colon cancers. Nat. Med. 2020, 26, 566–576. [Google Scholar] [CrossRef]
- Le, D.T.; Kim, T.W.; Van Cutsem, E.; Geva, R.; Jäger, D.; Hara, H.; Burge, M.; O’Neil, B.; Kavan, P.; Yoshino, T.; et al. Phase II Open-Label Study of Pembrolizumab in Treatment-Refractory, Microsatellite Instability–High/Mismatch Repair–Deficient Metastatic Colorectal Cancer: KEYNOTE-164. J. Clin. Oncol. 2020, 38, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Postow, M.; Sidlow, R.; Hellmann, M.D. Immune-Related Adverse Events Associated with Immune Checkpoint Blockade. N. Engl. J. Med. 2018, 378, 158–168. [Google Scholar] [CrossRef]
- Das, S.; Johnson, D.B. Immune-related adverse events and anti-tumor efficacy of immune checkpoint inhibitors. J. Immunother. Cancer 2019, 7, 1–11. [Google Scholar] [CrossRef]
- Johannet, P.; Coudray, N.; Jour, G.; Donnelly, D.M.; Bajaj, S.; Moran, U.; Dawood, Z.; Nomikou, S.; Kim, R.H.; Pavlick, A.C.; et al. Using machine learning algorithms to predict response and toxicity to immune checkpoint inhibitors (ICIs) in melanoma patients. J. Clin. Oncol. 2019, 37, 2581. [Google Scholar] [CrossRef]
- Barrera, C.V.; Velu, P.; Bera, K.; Wang, X.; Prasanna, P.; Khunger, M.; Khunger, A.; Velcheti, V.; Romero, E.; Madabhushi, A. Computer-extracted features relating to spatial arrangement of tumor infiltrating lymphocytes to predict response to nivolumab in non-small cell lung cancer (NSCLC). J. Clin. Oncol. 2018, 35, 12115. [Google Scholar] [CrossRef]
- Samstein, R.M.; Lee, C.; Shoushtari, A.N.; Hellmann, M.D.; Shen, R.; Janjigian, Y.Y.; Barron, D.A.; Zehir, A.; Jordan, E.J.; Omuro, A.; et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019, 51, 202–206. [Google Scholar] [CrossRef]
- Schrock, A.; Ouyang, C.; Sandhu, J.; Sokol, E.; Jin, D.; Ross, J.; Miller, V.; Lim, D.; Amanam, I.; Chao, J.; et al. Tumor mutational burden is predictive of response to immune checkpoint inhibitors in MSI-high metastatic colorectal cancer. Ann. Oncol. 2019, 30, 1096–1103. [Google Scholar] [CrossRef]
- Mandal, R.; Samstein, R.M.; Lee, K.-W.; Havel, J.J.; Wang, H.; Krishna, C.; Sabio, E.Y.; Makarov, V.; Kuo, F.; Blecua, P.; et al. Genetic diversity of tumors with mismatch repair deficiency influences anti–PD-1 immunotherapy response. Science 2019, 364, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Bai, X.; Wang, J.; Tang, X.-R.; Wu, D.-H.; Du, S.; Du, X.-J.; Zhang, Y.-W.; Zhu, H.-B.; Fang, Y.; et al. Combination of TMB and CNA Stratifies Prognostic and Predictive Responses to Immunotherapy Across Metastatic Cancer. Clin. Cancer Res. 2019, 25, 7413–7423. [Google Scholar] [CrossRef] [PubMed]
- Tran, E.; Ahmadzadeh, M.; Lu, Y.-C.; Gros, A.; Turcotte, S.; Robbins, P.F.; Gartner, J.J.; Zheng, Z.; Li, Y.F.; Ray, S.; et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 2015, 350, 1387–1390. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.N.; Llosa, N.J.; Cottrell, T.R.; Siegel, N.; Fan, H.; Suri, P.; Chan, H.Y.; Guo, H.; Oke, T.; Awan, A.H.; et al. Persistent mutant oncogene specific T cells in two patients benefitting from anti-PD-1. J. Immunother. Cancer 2019, 7, 40. [Google Scholar] [CrossRef]
- Llosa, N.J.; Luber, B.; Tam, A.J.; Smith, K.N.; Siegel, N.; Awan, A.H.; Fan, H.; Oke, T.; Zhang, J.; Domingue, J.; et al. Intratumoral Adaptive Immunosuppression and Type 17 Immunity in Mismatch Repair Proficient Colorectal Tumors. Clin. Cancer Res. 2019, 25, 5250–5259. [Google Scholar] [CrossRef]
- Saltz, J.H.; Gupta, R.; Hou, L.; Kurc, T.; Singh, P.; Nguyen, V.; Samaras, D.; Shroyer, K.R.; Zhao, T.; Batiste, R.; et al. Spatial Organization and Molecular Correlation of Tumor-Infiltrating Lymphocytes Using Deep Learning on Pathology Images. Cell Rep. 2018, 23, 181–193.e7. [Google Scholar] [CrossRef]
- Lu, Z.; Xu, S.; Shao, W.; Wu, Y.; Zhang, J.; Han, Z.; Feng, Q.; Huang, K. Deep-Learning–Based Characterization of Tumor-Infiltrating Lymphocytes in Breast Cancers From Histopathology Images and Multiomics Data. JCO Clin. Cancer Inform. 2020, 4, 480–490. [Google Scholar] [CrossRef]
- Huyghe, N.; Baldin, P.; Eynde, M.V.D. Immunotherapy with immune checkpoint inhibitors in colorectal cancer: What is the future beyond deficient mismatch-repair tumours? Gastroenterol. Rep. 2019, 8, 11–24. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Huebner, L.J.; Finnes, H.D.; Muranyi, A.; Clements, J.; Singh, S.; Hubbard, J.M.; McWilliams, R.R.; Shanmugam, K.; Sinicrope, F.A. Intratumoral CD3+ and CD8+ T-Cell Densities in Patients with DNA Mismatch Repair–Deficient Metastatic Colorectal Cancer Receiving Programmed Cell Death-1 Blockade. JCO Precis. Oncol. 2019, 3, 1–7. [Google Scholar] [CrossRef]
- Miao, Y.-R.; Zhang, Q.; Lei, Q.; Luo, M.; Xie, G.; Wang, H.; Guo, A.-Y. ImmuCellAI: A Unique Method for Comprehensive T-Cell Subsets Abundance Prediction and its Application in Cancer Immunotherapy. Adv. Sci. 2020, 7, 1902880. [Google Scholar] [CrossRef]
- Wiesweg, M.; Mairinger, F.; Reis, H.; Goetz, M.; Walter, R.F.H.; Hager, T.; Metzenmacher, M.; Eberhardt, W.; McCutcheon, A.; Köster, J.; et al. Machine learning-based predictors for immune checkpoint inhibitor therapy of non-small-cell lung cancer. Ann. Oncol. 2019, 30, 655–657. [Google Scholar] [CrossRef] [PubMed]
- Bao, X.; Shi, R.; Zhao, T.; Wang, Y.; Anastasov, N.; Rosemann, M.; Fang, W. Integrated analysis of single-cell RNA-seq and bulk RNA-seq unravels tumour heterogeneity plus M2-like tumour-associated macrophage infiltration and aggressiveness in TNBC. Cancer Immunol. Immunother. 2020, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Trebeschi, S.; Drago, S.; Birkbak, N.; Kurilova, I.; Cǎlin, A.; Pizzi, A.D.; Lalezari, F.; Lambregts, D.; Rohaan, M.; Parmar, C.; et al. Predicting response to cancer immunotherapy using noninvasive radiomic biomarkers. Ann. Oncol. 2019, 30, 998–1004. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Limkin, E.J.; Vakalopoulou, M.; Dercle, L.; Champiat, S.; Han, S.R.; Verlingue, L.; Brandao, D.; Lancia, A.; Ammari, S.; et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: An imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018, 19, 1180–1191. [Google Scholar] [CrossRef]
- Park, C.; Na, K.J.; Choi, H.; Ock, C.-Y.; Ha, S.; Kim, M.; Park, S.; Keam, B.; Kim, T.M.; Paeng, J.C.; et al. Tumor immune profiles noninvasively estimated by FDG PET with deep learning correlate with immunotherapy response in lung adenocarcinoma. Theranostics 2020, 10, 10838–10848. [Google Scholar] [CrossRef]
- Briggs, S.; Tomlinson, I.P.M. Germline and somatic polymerase ϵ and δ mutations define a new class of hypermutated colorectal and endometrial cancers. J. Pathol. 2013, 230, 148–153. [Google Scholar] [CrossRef]
- Giannakis, M.; Mu, X.J.; Shukla, S.A.; Qian, Z.R.; Cohen, O.; Nishihara, R.; Bahl, S.; Cao, Y.; Amin-Mansour, A.; Yamauchi, M.; et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep. 2016, 17, 1206. [Google Scholar] [CrossRef]
- Domingo, E.; Freeman-Mills, L.; Rayner, E.; Glaire, M.A.; Briggs, S.; Vermeulen, L.; Fessler, E.; Medema, J.P.; Boot, A.; Morreau, H.; et al. Somatic POLE proofreading domain mutation, immune response, and prognosis in colorectal cancer: A retrospective, pooled biomarker study. Lancet Gastroenterol. Hepatol. 2016, 1, 207–216. [Google Scholar] [CrossRef]
- Gong, J.; Wang, C.; Lee, P.P.; Chu, P.; Fakih, M. Response to PD-1 Blockade in Microsatellite Stable Metastatic Colorectal Cancer Harboring aPOLEMutation. J. Natl. Compr. Cancer Netw. 2017, 15, 142–147. [Google Scholar] [CrossRef]
- Picard, E.; Verschoor, C.P.; Ma, G.W.; Pawelec, G. Relationships Between Immune Landscapes, Genetic Subtypes and Responses to Immunotherapy in Colorectal Cancer. Front. Immunol. 2020, 11, 369. [Google Scholar] [CrossRef]
- Rodon, J.; Soria, J.-C.; Berger, R.; Miller, W.H.; Rubin, E.; Kugel, A.; Tsimberidou, A.; Saintigny, P.; Ackerstein, A.; Braña, I.; et al. Genomic and transcriptomic profiling expands precision cancer medicine: The WINTHER trial. Nat. Med. 2019, 25, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Salipante, S.J.; Scroggins, S.M.; Hampel, H.L.; Turner, E.H.; Pritchard, C.C. Microsatellite Instability Detection by Next Generation Sequencing. Clin. Chem. 2014, 60, 1192–1199. [Google Scholar] [CrossRef]
- Middha, S.; Zhang, L.; Nafa, K.; Jayakumaran, G.; Wong, D.; Kim, H.R.; Sadowska, J.; Berger, M.F.; Delair, D.F.; Shia, J.; et al. Reliable Pan-Cancer Microsatellite Instability Assessment by Using Targeted Next-Generation Sequencing Data. JCO Precis. Oncol. 2017, 2017, 1–17. [Google Scholar] [CrossRef]
- Willis, J.; Lefterova, M.I.; Artyomenko, A.; Kasi, P.M.; Nakamura, Y.; Mody, K.; Catenacci, D.V.; Fakih, M.; Barbacioru, C.; Zhao, J.; et al. Validation of Microsatellite Instability Detection Using a Comprehensive Plasma-Based Genotyping Panel. Clin. Cancer Res. 2019, 25, 7035–7045. [Google Scholar] [CrossRef]
- Fan, S.; Li, X.; Cui, X.; Zheng, L.; Ren, X.; Ma, W.; Ye, Z. Computed Tomography-Based Radiomic Features Could Potentially Predict Microsatellite Instability Status in Stage II Colorectal Cancer: A Preliminary Study. Acad. Radiol. 2019, 26, 1633–1640. [Google Scholar] [CrossRef]
- Pernicka, J.S.G.; Gagniere, J.; Chakraborty, J.; Yamashita, R.; Nardo, L.; Creasy, J.M.; Petkovska, I.; Do, R.R.K.; Bates, D.D.B.; Paroder, V.; et al. Radiomics-based prediction of microsatellite instability in colorectal cancer at initial computed tomography evaluation. Abdom. Radiol. 2019, 44, 3755–3763. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, Q.; Zhao, Y.; Liu, Y.; Chen, A.; Li, X.; Wu, T.; Li, J.; Guo, Y.; Liu, A. Radiomics Analysis of Iodine-Based Material Decomposition Images with Dual-Energy Computed Tomography Imaging for Preoperatively Predicting Microsatellite Instability Status in Colorectal Cancer. Front. Oncol. 2019, 9, 1250. [Google Scholar] [CrossRef]
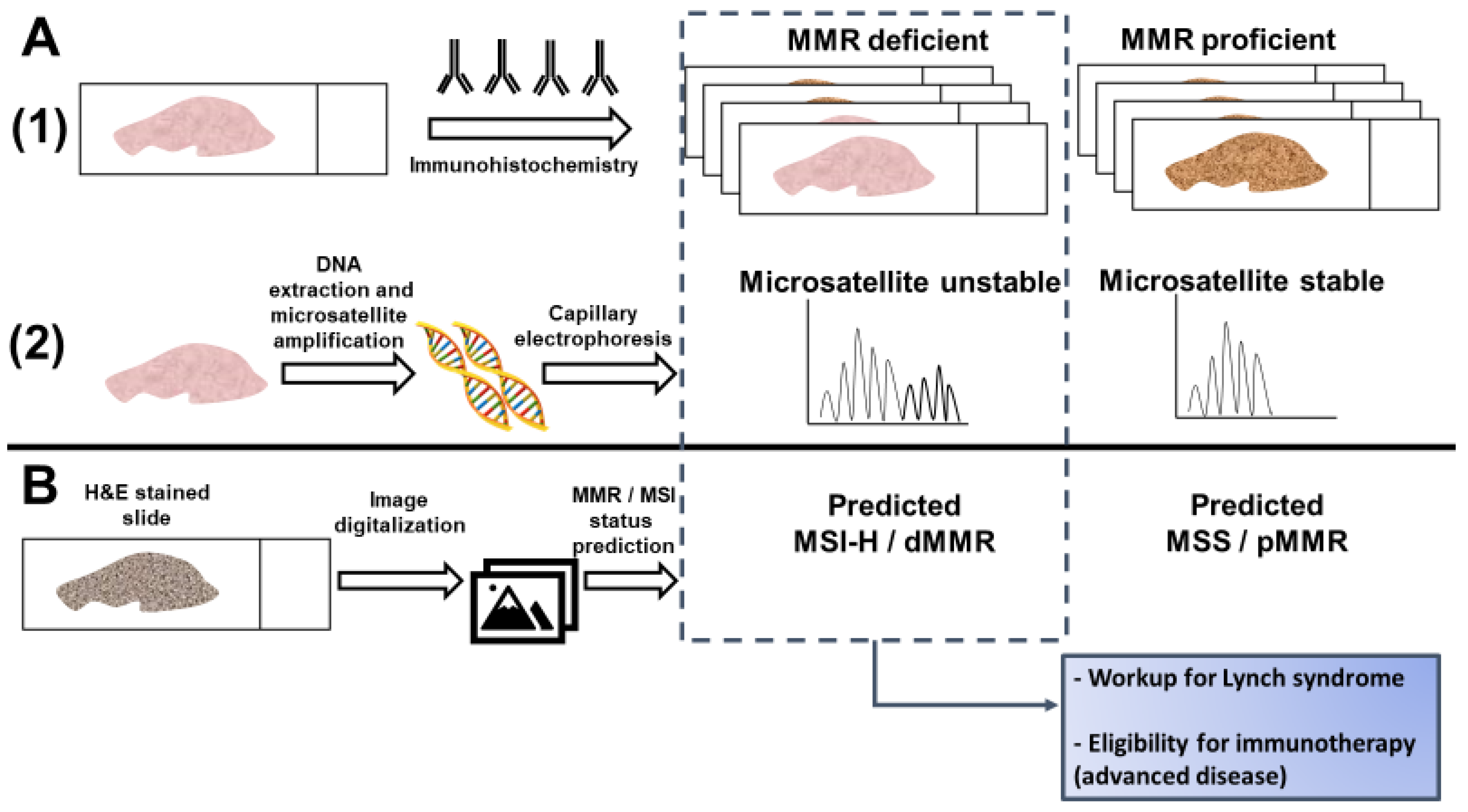
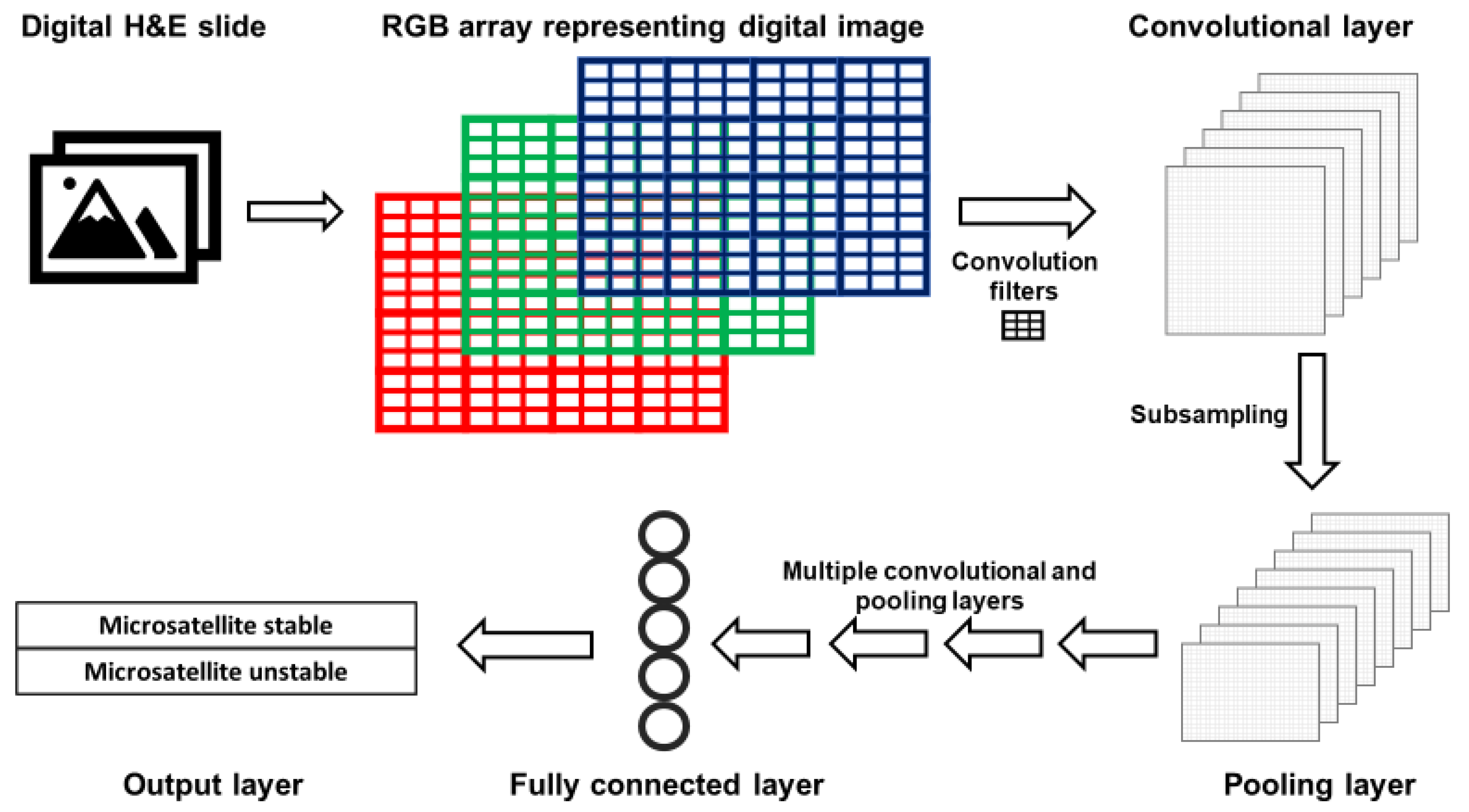
| A | |||||
| Variable | Sensitivity % (95% CI) | Specificity % (95% CI) | Odds Ratio, Univariate (95% CI) | Odds Ratio, Multivariate (95% CI) | |
| Host response features | |||||
| Tumor-infiltrating lymphocytes (TILs) | 70 [26] | 76 [26] | 7.4 (5.4–10.3) [26] | 3.8 (2.5–5.6) [26] | |
| 72 (64–78) [27] | 82 (80–85) [27] | 9.1 (5.9–14.1) [27] | |||
| 21 [30] | 97 [30] | 9.8 (3.5–28.5) [30] | |||
| 60 (50–70) [36] | 78 (76–79) [36] | 5.2 (3.2–8.5) [36] | 3.7 (2.0–6.8) [36] | ||
| Crohn’s-like lymphocytic reaction (CLR) | 68 [26] | 54 [26] | 2.5 (1.8–3.4) [26] | 2.3 (1.6–3.5) [26] | |
| 56 (48–63) [27] | 77 (74–80) [27] | 1.9 (1.2–2.9) [27] | |||
| 49 [30] | 64 [30] | 1.7 (0.9–3.2) [30] | |||
| 69 (59–78) [36] | 45 (44–46) [36] | 1.8 (1.1–3.0) [36] | 1.1 (0.6–2.0) [36] | ||
| Peritumoral lymphocytic reaction | 86 (77–92) [36] | 42 (40–42) [36] | 4.3 (2.3–8.3) [36] | 3.7 (1.6–8.6) [36] | |
| Stromal plasma cells | 78 (68–86) [36] | 48 (46–48) [36] | 3.2 (1.9–5.6) [36] | 2.1 (1.1–4.1) [36] | |
| Tumor characteristics | |||||
| Mucinous morphology * | 53 [26] | 80 [26] | 4.6 (3.4–6.3) [26] | 1.7 (1.1–2.7) [26] | |
| 28 (21–35) [27] ** | 91 (89–93) [27] ** | 2.8 (1.7–4.8) [27] ** | |||
| 22 [30] | 93 [30] | 3.7 (1.7–8.0) [30] | |||
| 51 (41–61) [36] | 78 (77–79) [36] | 3.7 (2.3–6.0) [36] | 4.71 (2.1–10.7) [36] | ||
| 2.13 (1.3–3.4) [37] | |||||
| Medullary morphology (10–70%) | 25 [30] | 97 [30] | 12.5 (4.6–35.9) [30] | ||
| Grade † | 64 [26] † | 81 [26] † | 7.4 (5.4–10.1) [26] † | 3.4 (2.2–5.2) [26] † | |
| 38 (31–46) [27] | 82 (79–84) [27] | 1.9 (1.2–3.1) [27] | |||
| 38 [30] | 87 [30] | 4.0 (2.2–7.3) [30] | |||
| 17 (10–26) [36] | 90 (89–91) [36] | 1.8 (0.9–3.5) [36] | |||
| 32 (23–43) [36] † | 77 (76–78) [36] † | 1.6 (0.9–2.6) [36] † | |||
| Signet ring cells | 4.3 (2.2–8.7) [26] | ||||
| 13 [30] | 95 [30] | 2.7 (1.1–6.8) [30] | |||
| Lack of dirty or garland necrosis | 59 [26] | 79 [26] | 5.4 (3.9–7.4) [26] | 1.8 (1.1–2.8) [26] | |
| 26 (18–35) [36] | 89 (88–90) [36] | 2.7 (1.5–4.7) [36] | 1.4 (0.7–3.0) [36] | ||
| Cribriform pattern | 13 [30] | 72 [30] | 0.4 (0.2–0.8) [30] | ||
| Histologic heterogeneity | 4.4 (3.0–6.4) [26] | ||||
| 55 (45–65) [36] | 69 (68–70) [36] | 2.7 (1.7–4.4) [36] | |||
| Clinical/Molecular Features | |||||
| Age <50 years | 2.2 (1.3–3.8) [26] | 3.1 (1.5–6.5) [26] | |||
| 52 (44–60) [27] | 59 (56–62) [27] | 1.9 (1.3–2.9) [27] | |||
| 21 (13–29) [36] | 89 (88–90) [36] | 2.0 (1.1–3.7) [36] | 3.8 (1.8–8.0) [36] | ||
| Female | 1.4 (1.0–1.9) [26] | ||||
| 51 (41–62) [36] | 63 (62–64) [36] | 1.8 (1.1–2.8) [36] | 1.3 (0.7–2.2) [36] | ||
| 1.56 (1.0–2.4) [37] | |||||
| Size > or equal to 60 mm | 2.75 (1.8–4.2) [37] | ||||
| Anatomic site (right sided/proximal) | 70 [26] | 63 [26] | 4.1 (2.9–5.7) [26] | 2.2 (1.5–3.3) [26] | |
| 74 (67–81) [27] | 70 (67–73) [27] | 4.7 (3.1–7.3) [27] | |||
| 79 (70–87) [36] | 63 (61–63) [36] | 6.4 (3.6–11.2) [36] | 5.08 (2.7–9.6) [36] | ||
| 3.76 (2.4–5.9) [37] | |||||
| BRAF mutant | 13.33 (8.0–22.2) [37] | ||||
| B | |||||
| Model | Model Variables | Sensitivity (%) | Specificity (%) | Positive/Negative Predictive Value (%) | AUC or Accuracy (95% CI) |
| Greenson et al. ** [26] | TIL/HPF, well or poorly differentiated, age < 50, CLR, R-sided, lack of dirty necrosis, any mucinous differentiation | 92 ** | 46 ** | AUC 0.850 ** | |
| MsPath [27] | Age < 50, proximal location, mucinous/signet ring/undifferentiated, poorly differentiated, CLR, TILs | 93 | 55 | AUC 0.890 (0.83– 0.94) | |
| PREDICT [36] | R-sided, mucinous component, age < 50 years, TILs, peritumoral reaction, increased stromal plasma cells | 96.9 | 76.6 | 35.2/99.5 | AUC 0.924 |
| Fujiyoshi et al. [37] | Female, mucinous component, tumor size > or equal to 60 mm, proximal location, BRAF mutation | 76 | 77 | AUC 0.856 (0.806 –0.905) | |
| RERTest6 ** [38] | Proximal location, expansive growth pattern, CLR, solid pattern %, mucinous pattern %, cribriform pattern | 78.01 ** | 93.39 ** | 51.8/97.9 ** | Accuracy 0.921 ** |
| CNN and Additional Methods | Other CNNs Evaluated | Training Cohort | Test Cohort(s) with AUC (95% CI) or Accuracy | External Validation Cohort(s) with AUC (95% CI) |
|---|---|---|---|---|
| ResNet-18 Whole-slide image classified per majority of image tiles [43] | AlexNet, VGG-19, InceptionV3, SqueezeNet | TCGA CRC FFPE | TGCA CRC FFPEAUC 0.77 (0.62–0.87) | DACHS CRC FFPE AUC 0.84 (0.72–0.92) |
| TCGA CRC frozen | TCGA CRC frozen AUC 0.84 (0.73–0.91) | DACHS CRC FFPE 0.61 (0.50–0.73) | ||
| TCGA gastric FFPE | TCGA gastric FFPE AUC 0.81 (0.69–0.90) | DACHS CRC FFPE AUC 0.60 (0.48–0.69) KCCH gastric FFPE AUC 0.69 (0.52–0.82) | ||
| TCGA uterine FFPE | TCGA uterine FFPE AUC 0.75 (0.63–0.83) | |||
| ShuffleNet [67] | AlexNet, InceptionV3, ResNet-18, DenseNet201 | TCGA CRC | TCGA CRC AUC 0.805 | DACHS CRC AUC 0.89 (0.88–0.92) |
| ResNet-18 Whole slide image classified using two multiple instance learning pipelines integrated into an ensemble classifier [68] | none | TCGA CRC frozen | TCGA CRC frozen AUC 0.885 | Asian CRC AUC FFPE 0.650 |
| TCGA CRC frozen with 10% Asian CRC FFPE | Asian CRC FFPEAUC 0.850 | |||
| TCGA CRC frozen with 70% Asian CRC FFPE | Asian CRC FFPEAUC 0.926 | |||
| Custom multilayer perceptron (HE2RNA) applied after feature extraction by ResNet-50; with and without transcriptomic representation of histology [69] | none | TCGA CRC FFPE, with transcriptomic representation and 20% of training cohort | TCGA CRC FFPEAUC ~0.80 * | |
| TCGA CRC FFPE, using >80% of training cohort | TCGA CRC FFPEAUC ~0.80 * | |||
| Inception-V3 with and without adversarial learning [70] | VGG-19, ResNet-50 | TCGA CRC | TCGA CRC Accuracy 98.3% | TCGA endometrial Accuracy 53.7% |
| TCGA CRC and endometrial | TCGA CRC Accuracy 72.3% TCGA endometrial Accuracy 84.2% | TCGA gastric Accuracy 34.9% | ||
| TCGA CRC and endometrial with adversarial learning | TCGA CRC Accuracy 85.0%TCGA endometrial Accuracy 94.6% | TCGA gastric Accuracy 57.4% | ||
| InceptionResNetV1 [71] | InceptionV1-3, InceptionResnetV1-2, Panoptes1-4 (multibranch custom InceptionResnet) | TCGA and CPTAC endometrial carcinoma | TCGA and CPTAC endometrial carcinoma AUC 0.827 (0.705–0.948) | |
| ShuffleNet [72] | none | MSIDETECT CRC (color normalized) | MSIDETECT CRC AUC 0.92 (0.90–0.93) | YCR-BCIP CRC surgical samples AUC 0.96 (0.93–0.98) |
| YCR-BCIP CRC biopsy samples AUC 0.78 (0.75–0.81) | ||||
| YCR-BCIP CRC biopsy samples | YCR-BCIP CRC biopsy samples |
| Advantages | Limitations | Future Directions | |
|---|---|---|---|
| Classification accuracy |
|
|
|
| Generalizability | Excellent performance on well curated cohorts that are similar to training data | Performance not robust to differing patient and tissue characteristics | Increase availability of datasets for global and local model refinement |
| Accessibility | Potential to expand access to settings without pathology experts or molecular testing, including via cellular devices |
| Design dedicated CNNs for settings with reduced access to healthcare |
| Clinical endpoint prediction | Very good classification of MSI | No direct prediction of clinical endpoints | Shift from surrogate marker classification to clinical endpoint prediction |
| Patient selection for immunotherapy | Decreased accuracy of MSI classification in metastatic disease, where immunotherapy is approved |
| |
| Identification of Lynch Syndrome | Inability to distinguish between somatic and germline etiology of MSI, such that confirmatory testing is required |
| |
| Comparison with next generation sequencing (NGS) | Rapid and cost-effective after initial investment | Cannot currently reliably detect KRAS or BRAF mutations, tumor mutational burden and other clinically actionable alterations |
|
| Cost effectiveness | Long term savings on molecular assays | Initial investment required in hardware and software for digital pathology | Expand use of and access to digital pathology |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hildebrand, L.A.; Pierce, C.J.; Dennis, M.; Paracha, M.; Maoz, A. Artificial Intelligence for Histology-Based Detection of Microsatellite Instability and Prediction of Response to Immunotherapy in Colorectal Cancer. Cancers 2021, 13, 391. https://doi.org/10.3390/cancers13030391
Hildebrand LA, Pierce CJ, Dennis M, Paracha M, Maoz A. Artificial Intelligence for Histology-Based Detection of Microsatellite Instability and Prediction of Response to Immunotherapy in Colorectal Cancer. Cancers. 2021; 13(3):391. https://doi.org/10.3390/cancers13030391
Chicago/Turabian StyleHildebrand, Lindsey A., Colin J. Pierce, Michael Dennis, Munizay Paracha, and Asaf Maoz. 2021. "Artificial Intelligence for Histology-Based Detection of Microsatellite Instability and Prediction of Response to Immunotherapy in Colorectal Cancer" Cancers 13, no. 3: 391. https://doi.org/10.3390/cancers13030391
APA StyleHildebrand, L. A., Pierce, C. J., Dennis, M., Paracha, M., & Maoz, A. (2021). Artificial Intelligence for Histology-Based Detection of Microsatellite Instability and Prediction of Response to Immunotherapy in Colorectal Cancer. Cancers, 13(3), 391. https://doi.org/10.3390/cancers13030391






