Effective Prediction of Prostate Cancer Recurrence through the IQGAP1 Network
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
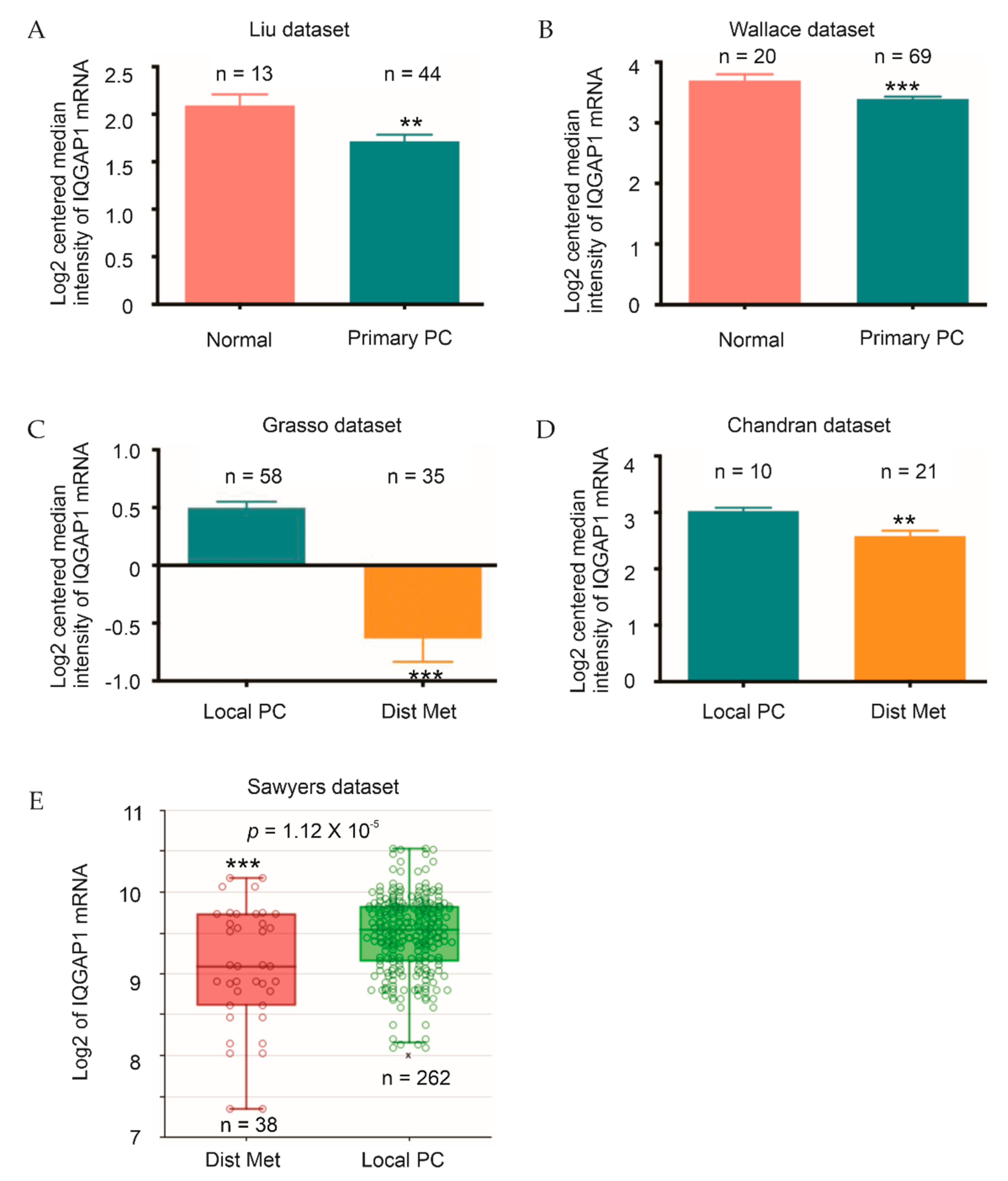
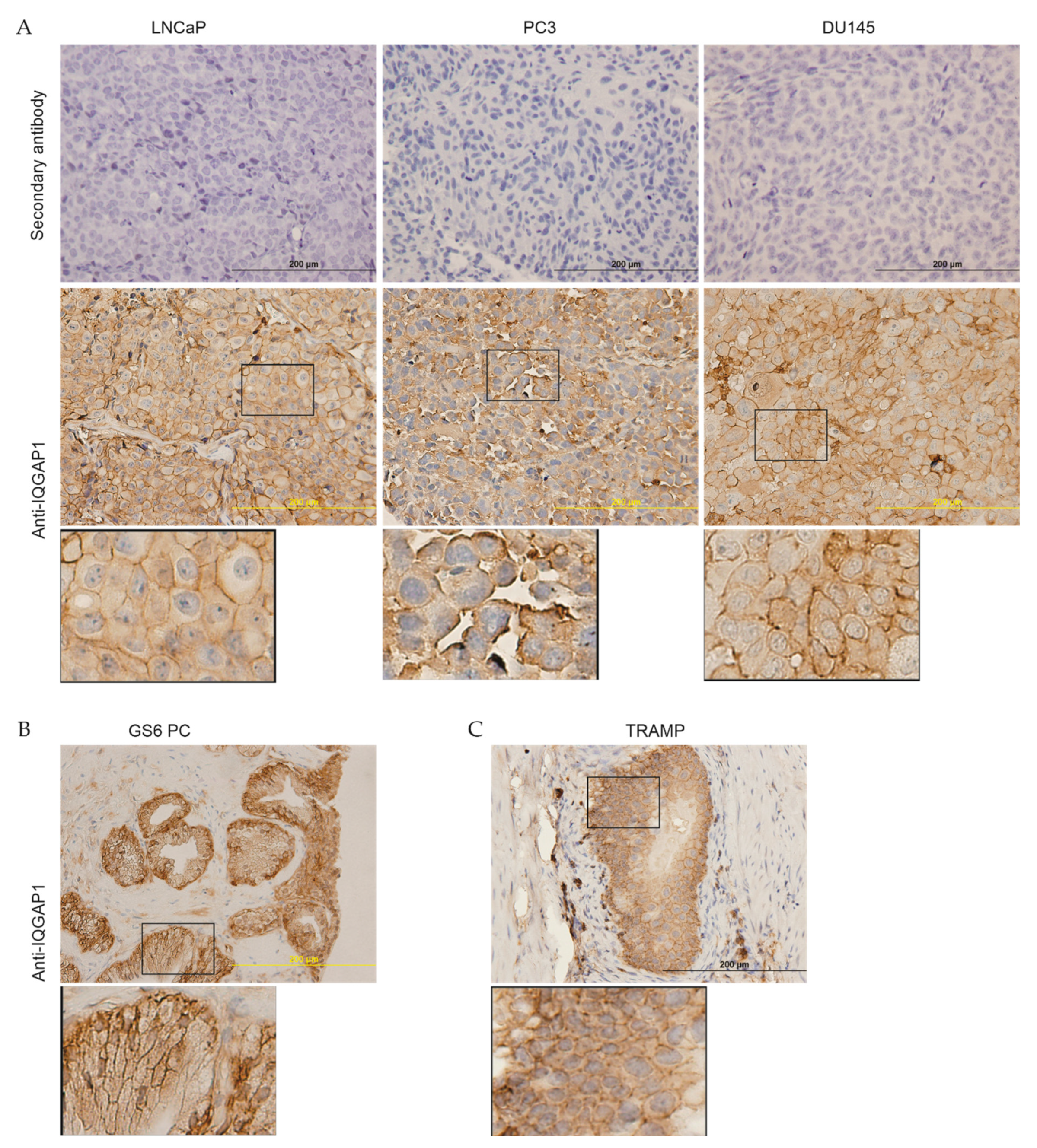
2.1. Downregulation of IQGAP1 following the Course of PC
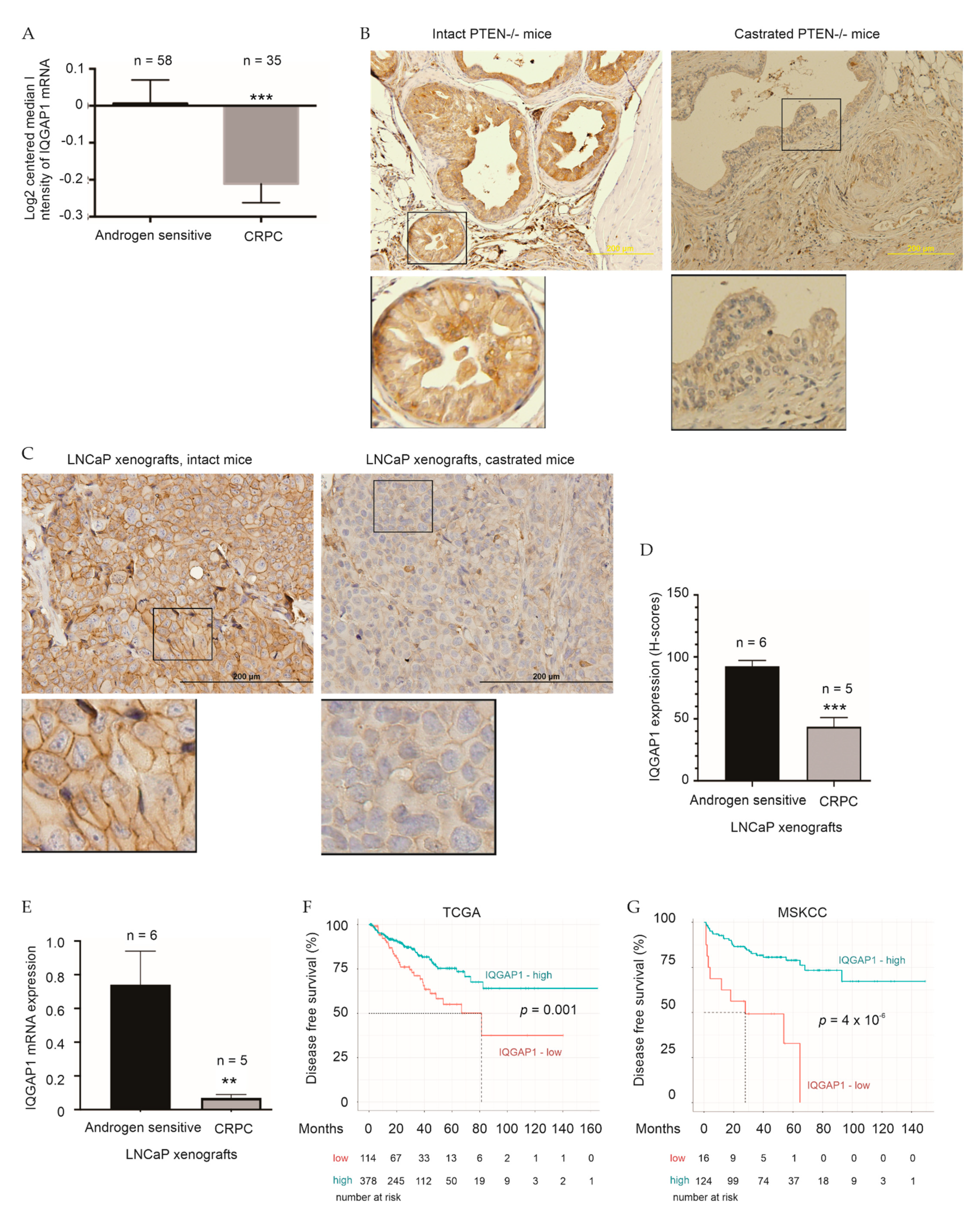
2.2. Association of IQGAP1 Downregulation with Therapy Resistance
2.3. Enrichment of Oncogenic Pathways within the IQGAP1 Network
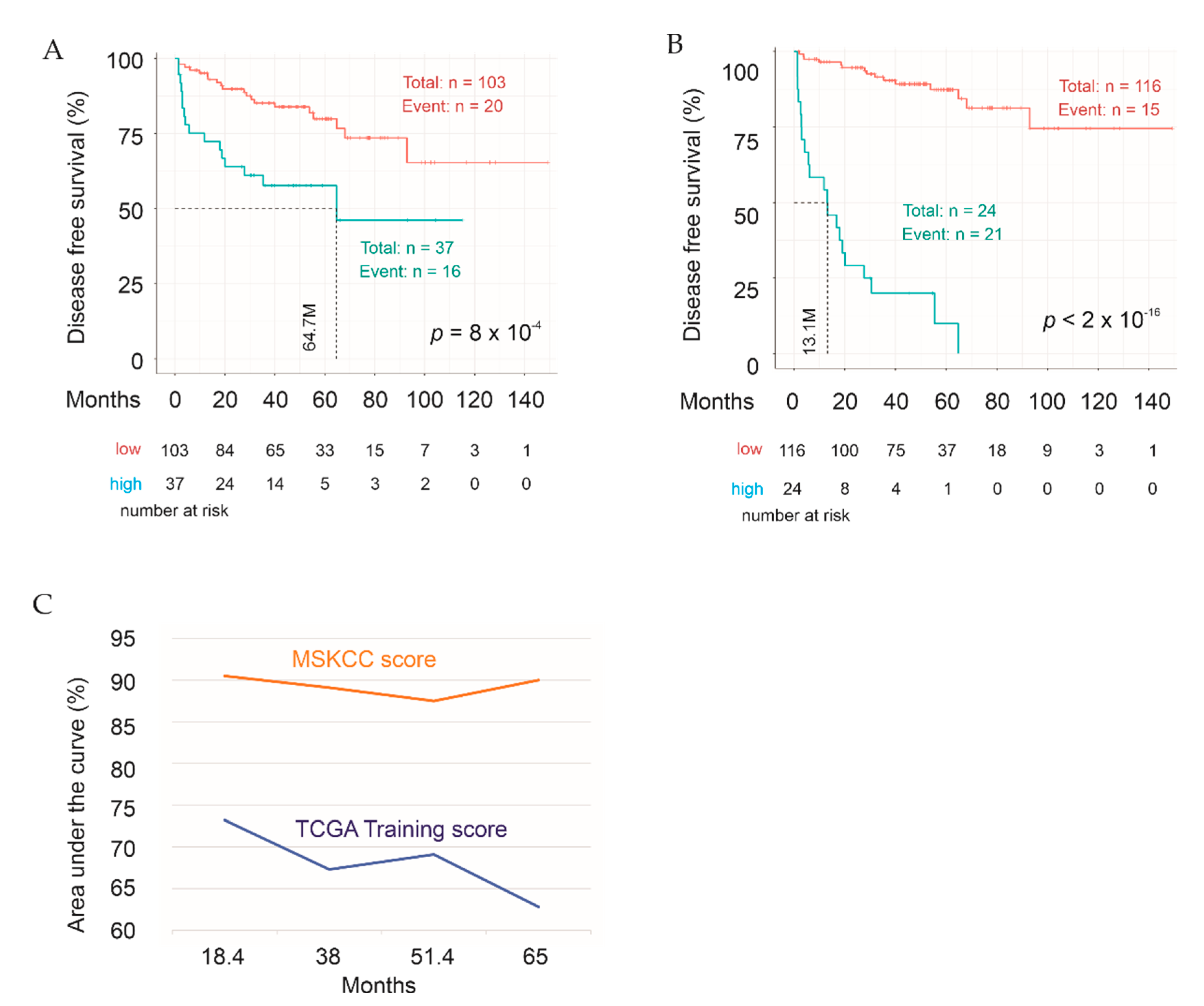
2.4. Construction of a Multigene Panel from the IQGAP1 Network in Predicting PC Recurrence following Prostatectomy
2.5. Testing Sig27gene
2.6. Validation of Sig27gene Using an Independent PC Cohort
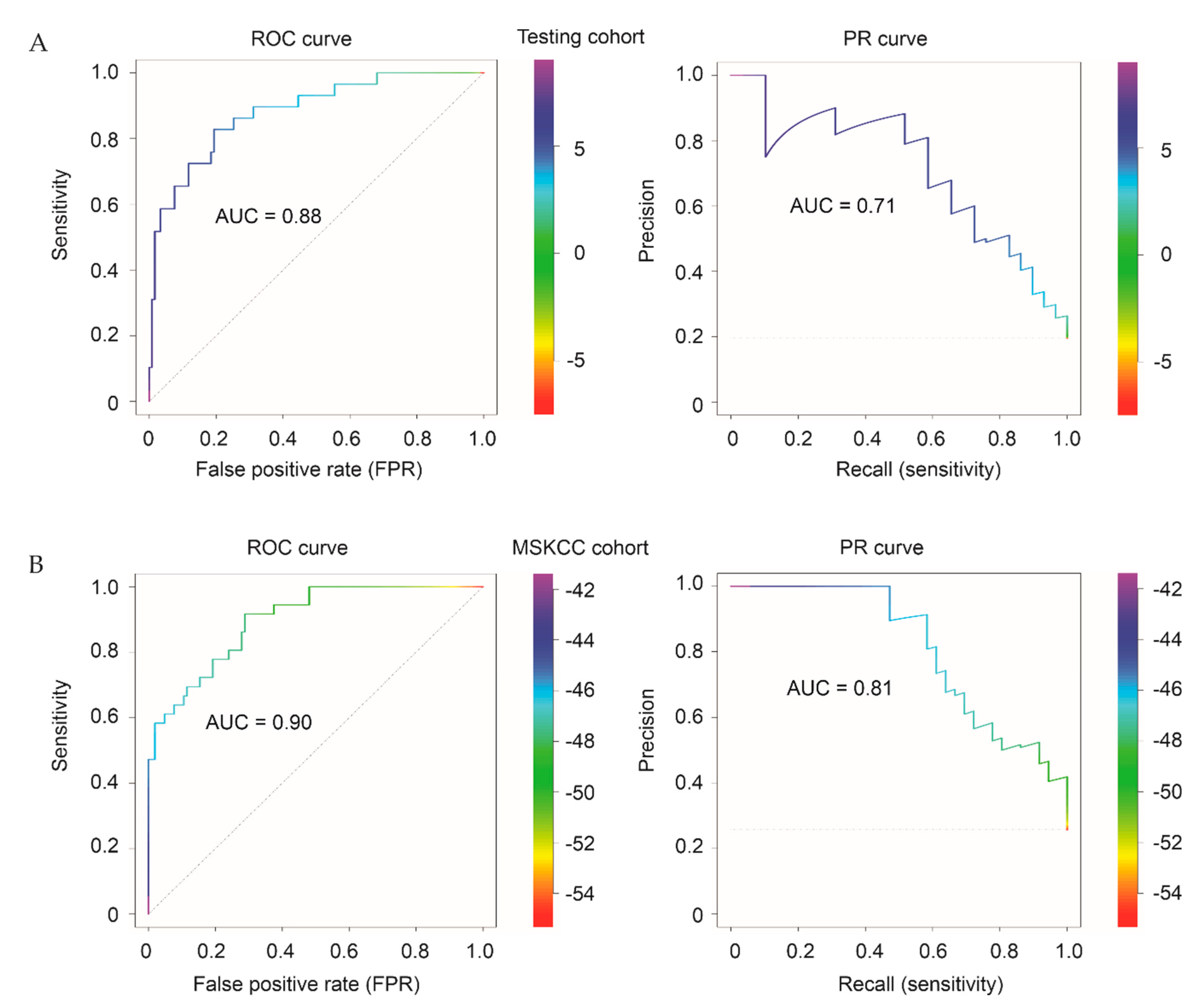
2.7. Evaluation of the Discriminative Performance of Sig27gene
2.8. Sig27gene as an Independent Risk Factor of PC Recurrence
2.9. Characterization of Sig27gene
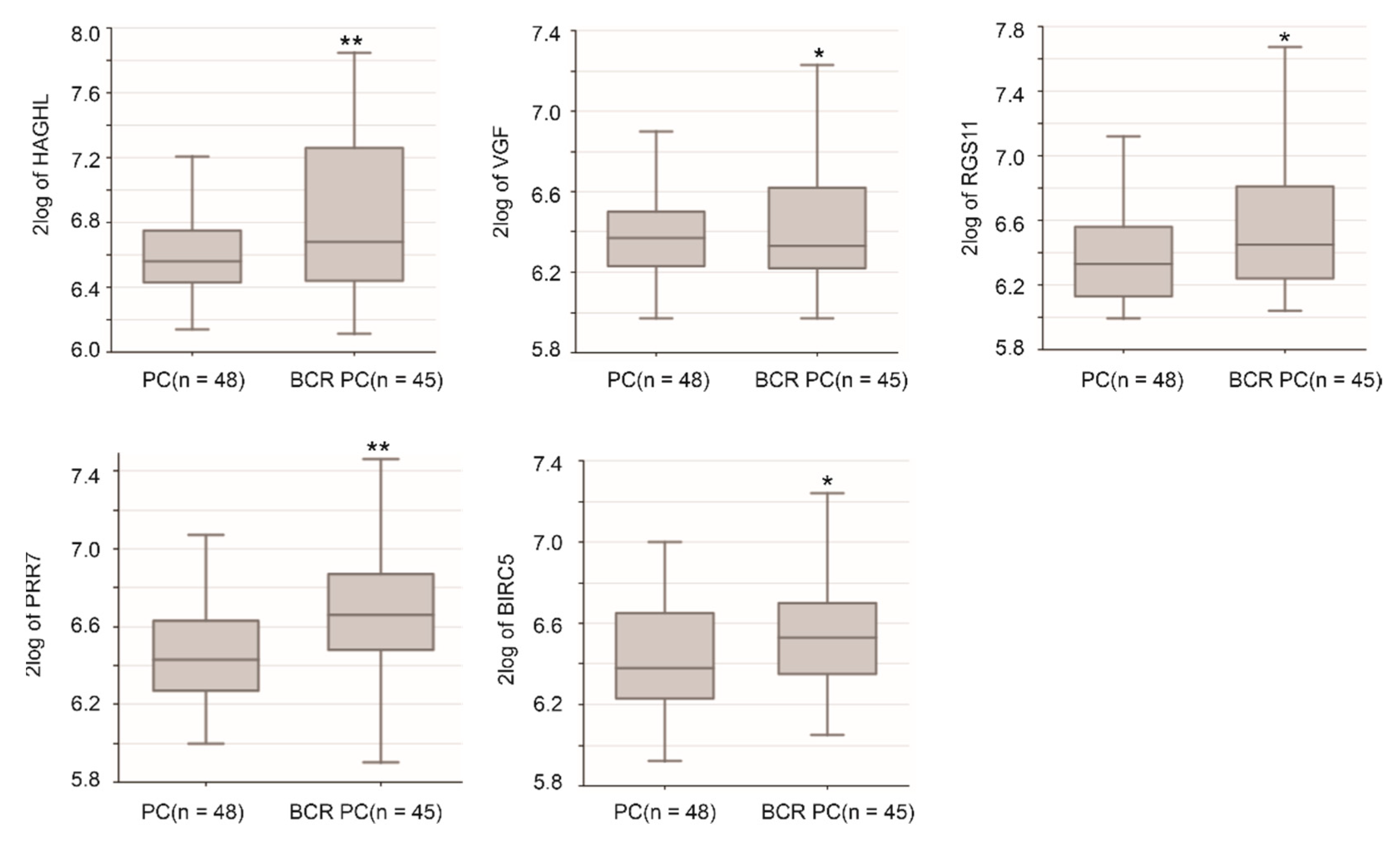
2.10. Potential Oncogenic Functions of Sig27gene Component Genes
3. Discussion
4. Materials and Methods
4.1. Collection of PC Tissues
4.2. Cell Culture
4.3. Formation of Xenograft Tumors
4.4. Generation of CRPC in Animal Models
4.5. Immunohistochemistry (IHC)
4.6. Analysis of IQGAP1 mRNA Expression
4.7. cBioPortal Database
4.8. Pathway Enrichment Analysis
4.9. Cutoff Point Estimation
4.10. Regression Analysis
4.11. Establishing of a Multigene Panel Predicting PC Biochemical Recurrence
4.12. Assignment of Signature Scores to Individual PCs
4.13. Examination of Gene Expression
4.14. Statistical Analysis
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in globocan 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef]
- Heidenreich, A.; Bastian, P.J.; Bellmunt, J.; Bolla, M.; Joniau, S.; van der Kwast, T.; Mason, M.; Matveev, V.; Wiegel, T.; Zattoni, F.; et al. Eau guidelines on prostate cancer. Part ii: Treatment of advanced, relapsing, and castration-resistant prostate cancer. Eur. Urol. 2014, 65, 467–479. [Google Scholar] [CrossRef]
- Egevad, L.; Delahunt, B.; Srigley, J.R.; Samaratunga, H. International society of urological pathology (isup) grading of prostate cancer—An isup consensus on contemporary grading. Acta Pathol. Microbiol. Immunol. Scand. 2016, 124, 433–435. [Google Scholar] [CrossRef]
- Gordetsky, J.; Epstein, J. Grading of prostatic adenocarcinoma: Current state and prognostic implications. Diagn. Pathol. 2016, 11, 25. [Google Scholar] [CrossRef]
- Epstein, J.I.; Zelefsky, M.J.; Sjoberg, D.D.; Nelson, J.B.; Egevad, L.; Magi-Galluzzi, C.; Vickers, A.J.; Parwani, A.V.; Reuter, V.E.; Fine, S.W.; et al. A contemporary prostate cancer grading system: A validated alternative to the gleason score. Eur. Urol. 2016, 69, 428–435. [Google Scholar] [CrossRef]
- Zaorsky, N.G.; Raj, G.V.; Trabulsi, E.J.; Lin, J.; Den, R.B. The dilemma of a rising prostate-specific antigen level after local therapy: What are our options? Semin. Oncol. 2013, 40, 322–336. [Google Scholar] [CrossRef]
- Shipley, W.U.; Seiferheld, W.; Lukka, H.R.; Major, P.P.; Heney, N.M.; Grignon, D.J.; Sartor, O.; Patel, M.P.; Bahary, J.P.; Zietman, A.L.; et al. Radiation with or without antiandrogen therapy in recurrent prostate cancer. N. Engl. J. Med. 2017, 376, 417–428. [Google Scholar] [CrossRef]
- Semenas, J.; Allegrucci, C.; Boorjian, S.A.; Mongan, N.P.; Persson, J.L. Overcoming drug resistance and treating advanced prostate cancer. Curr. Drug Targ. 2012, 13, 1308–1323. [Google Scholar] [CrossRef]
- Ojo, D.; Lin, X.; Wong, N.; Gu, Y.; Tang, D. Prostate cancer stem-like cells contribute to the development of castration-resistant prostate cancer. Cancers 2015, 7, 2290–2308. [Google Scholar] [CrossRef]
- De Bono, J.S.; Logothetis, C.J.; Molina, A.; Fizazi, K.; North, S.; Chu, L.; Chi, K.N.; Jones, R.J.; Goodman, O.B., Jr.; Saad, F.; et al. Abiraterone and increased survival in metastatic prostate cancer. N. Engl. J. Med. 2011, 364, 1995–2005. [Google Scholar] [CrossRef]
- Scher, H.I.; Fizazi, K.; Saad, F.; Taplin, M.E.; Sternberg, C.N.; Miller, K.; de Wit, R.; Mulders, P.; Chi, K.N.; Shore, N.D.; et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N. Engl. J. Med. 2012, 367, 1187–1197. [Google Scholar] [CrossRef]
- Chaturvedi, S.; Garcia, J.A. Novel agents in the management of castration resistant prostate cancer. J. Carcinogen. 2014, 13, 5. [Google Scholar]
- Drake, C.G. Prostate cancer as a model for tumour immunotherapy. Nat. Rev. Immunol. 2010, 10, 580–593. [Google Scholar] [CrossRef]
- Mei, W.; Gu, Y.; Jiang, Y.; Major, P.; Tang, D. Circulating cell-free DNA is a potential prognostic biomarker of metastatic castration-resistant prostate cancer for taxane therapy. AME Med. J. 2018, 3, 1–5. [Google Scholar] [CrossRef]
- Schwartz, M. Rho signalling at a glance. J. Cell Sci. 2004, 117, 5457–5458. [Google Scholar] [CrossRef]
- White, C.D.; Brown, M.D.; Sacks, D.B. Iqgaps in cancer: A family of scaffold proteins underlying tumorigenesis. FEBS Lett. 2009, 583, 1817–1824. [Google Scholar] [CrossRef]
- Wang, S.; Watanabe, T.; Noritake, J.; Fukata, M.; Yoshimura, T.; Itoh, N.; Harada, T.; Nakagawa, M.; Matsuura, Y.; Arimura, N.; et al. Iqgap3, a novel effector of rac1 and cdc42, regulates neurite outgrowth. J. Cell Sci. 2007, 120, 567–577. [Google Scholar] [CrossRef]
- Dixon, M.J.; Gray, A.; Schenning, M.; Agacan, M.; Tempel, W.; Tong, Y.; Nedyalkova, L.; Park, H.W.; Leslie, N.R.; van Aalten, D.M.; et al. Iqgap proteins reveal an atypical phosphoinositide (api) binding domain with a pseudo c2 domain fold. J. Biol. Chem. 2012, 287, 22483–22496. [Google Scholar] [CrossRef]
- Xie, Y.; Yan, J.; Cutz, J.C.; Rybak, A.P.; He, L.; Wei, F.; Kapoor, A.; Schmidt, V.A.; Tao, L.; Tang, D. Iqgap2, a candidate tumour suppressor of prostate tumorigenesis. Biochim. Biophys. Acta 2012, 1822, 875–884. [Google Scholar] [CrossRef]
- Smith, J.M.; Hedman, A.C.; Sacks, D.B. Iqgaps choreograph cellular signaling from the membrane to the nucleus. Trends Cell Biol. 2015, 25, 171–184. [Google Scholar] [CrossRef]
- Xie, Y.; Kapoor, A.; Peng, H.; Cutz, J.C.; Tao, L.; Tang, D. Iqgap2 displays tumor suppression functions. J. Anal. Oncol. 2015, 4, 86–93. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, D.; Bojdani, E.; El-Naggar, A.K.; Vasko, V.; Xing, M. Iqgap1 plays an important role in the invasiveness of thyroid cancer. Clin. Cancer Res. 2010, 16, 6009–6018. [Google Scholar] [CrossRef]
- Jadeski, L.; Mataraza, J.M.; Jeong, H.W.; Li, Z.; Sacks, D.B. Iqgap1 stimulates proliferation and enhances tumorigenesis of human breast epithelial cells. J. Biol. Chem. 2008, 283, 1008–1017. [Google Scholar] [CrossRef]
- Hayashi, H.; Nabeshima, K.; Aoki, M.; Hamasaki, M.; Enatsu, S.; Yamauchi, Y.; Yamashita, Y.; Iwasaki, H. Overexpression of iqgap1 in advanced colorectal cancer correlates with poor prognosis-critical role in tumor invasion. Int. J. Cancer 2010, 126, 2563–2574. [Google Scholar] [CrossRef]
- Wang, X.X.; Wang, K.; Li, X.Z.; Zhai, L.Q.; Qu, C.X.; Zhao, Y.; Liu, Z.R.; Wang, H.Z.; An, Q.J.; Jing, L.W.; et al. Targeted knockdown of iqgap1 inhibits the progression of esophageal squamous cell carcinoma in vitro and in vivo. PLoS ONE 2014, 9, e96501. [Google Scholar] [CrossRef]
- Jin, X.; Liu, Y.; Liu, J.; Lu, W.; Liang, Z.; Zhang, D.; Liu, G.; Zhu, H.; Xu, N.; Liang, S. The overexpression of iqgap1 and beta-catenin is associated with tumor progression in hepatocellular carcinoma in vitro and in vivo. PLoS ONE 2015, 10, e0133770. [Google Scholar] [CrossRef]
- Dong, P.X.; Jia, N.; Xu, Z.J.; Liu, Y.T.; Li, D.J.; Feng, Y.J. Silencing of iqgap1 by shrna inhibits the invasion of ovarian carcinoma ho-8910pm cells in vitro. J. Exp. Clin. Cancer Res. 2008, 27, 77. [Google Scholar] [CrossRef][Green Version]
- Hensel, J.; Duex, J.E.; Owens, C.; Dancik, G.M.; Edwards, M.G.; Frierson, H.F.; Theodorescu, D. Patient mutation directed shrna screen uncovers novel bladder tumor growth suppressors. Mol. Cancer Res. 2015, 13, 1306–1315. [Google Scholar] [CrossRef]
- Liu, C.; Billadeau, D.D.; Abdelhakim, H.; Leof, E.; Kaibuchi, K.; Bernabeu, C.; Bloom, G.S.; Yang, L.; Boardman, L.; Shah, V.H.; et al. Iqgap1 suppresses tbetarii-mediated myofibroblastic activation and metastatic growth in liver. J. Clin. Investig. 2013, 123, 1138–1156. [Google Scholar] [CrossRef]
- Kaur, R.; Yuan, X.; Lu, M.L.; Balk, S.P. Increased pak6 expression in prostate cancer and identification of pak6 associated proteins. Prostate 2008, 68, 1510–1516. [Google Scholar] [CrossRef]
- Fram, S.; King, H.; Sacks, D.B.; Wells, C.M. A pak6-iqgap1 complex promotes disassembly of cell-cell adhesions. Cell. Mol. Life Sci. 2014, 71, 2759–2773. [Google Scholar] [CrossRef]
- Moon, H.; Ruelcke, J.E.; Choi, E.; Sharpe, L.J.; Nassar, Z.D.; Bielefeldt-Ohmann, H.; Parat, M.O.; Shah, A.; Francois, M.; Inder, K.L.; et al. Diet-induced hypercholesterolemia promotes androgen-independent prostate cancer metastasis via iqgap1 and caveolin-1. Oncotarget 2015, 6, 7438–7453. [Google Scholar] [CrossRef]
- Lin, X.; Gu, Y.; Kapoor, A.; Wei, F.; Aziz, T.; Ojo, D.; Jiang, Y.; Bonert, M.; Shayegan, B.; Yang, H.; et al. Overexpression of muc1 and genomic alterations in its network associate with prostate cancer progression. Neoplasia 2017, 19, 857–867. [Google Scholar] [CrossRef]
- Wong, N.; Major, P.; Kapoor, A.; Wei, F.; Yan, J.; Aziz, T.; Zheng, M.; Jayasekera, D.; Cutz, J.C.; Chow, M.J.; et al. Amplification of muc1 in prostate cancer metastasis and crpc development. Oncotarget 2016, 7, 83115–83133. [Google Scholar] [CrossRef]
- Liu, P.; Ramachandran, S.; Ali Seyed, M.; Scharer, C.D.; Laycock, N.; Dalton, W.B.; Williams, H.; Karanam, S.; Datta, M.W.; Jaye, D.L.; et al. Sex-determining region y box 4 is a transforming oncogene in human prostate cancer cells. Cancer Res. 2006, 66, 4011–4019. [Google Scholar] [CrossRef]
- Wallace, T.A.; Prueitt, R.L.; Yi, M.; Howe, T.M.; Gillespie, J.W.; Yfantis, H.G.; Stephens, R.M.; Caporaso, N.E.; Loffredo, C.A.; Ambs, S. Tumor immunobiological differences in prostate cancer between african-american and european-american men. Cancer Res. 2008, 68, 927–936. [Google Scholar] [CrossRef]
- Grasso, C.S.; Wu, Y.M.; Robinson, D.R.; Cao, X.; Dhanasekaran, S.M.; Khan, A.P.; Quist, M.J.; Jing, X.; Lonigro, R.J.; Brenner, J.C.; et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012, 487, 239–243. [Google Scholar] [CrossRef]
- Chandran, U.R.; Ma, C.; Dhir, R.; Bisceglia, M.; Lyons-Weiler, M.; Liang, W.; Michalopoulos, G.; Becich, M.; Monzon, F.A. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer 2007, 7, 64. [Google Scholar] [CrossRef]
- Taylor, B.S.; Schultz, N.; Hieronymus, H.; Gopalan, A.; Xiao, Y.; Carver, B.S.; Arora, V.K.; Kaushik, P.; Cerami, E.; Reva, B.; et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010, 18, 11–22. [Google Scholar] [CrossRef]
- Horoszewicz, J.S.; Leong, S.S.; Kawinski, E.; Karr, J.P.; Rosenthal, H.; Chu, T.M.; Mirand, E.A.; Murphy, G.P. Lncap model of human prostatic carcinoma. Cancer Res. 1983, 43, 1809–1818. [Google Scholar]
- Kaighn, M.E.; Narayan, K.S.; Ohnuki, Y.; Lechner, J.F.; Jones, L.W. Establishment and characterization of a human prostatic carcinoma cell line (pc-3). Investig. Urol. 1979, 17, 16–23. [Google Scholar]
- Thomas, C.; Zoubeidi, A.; Kuruma, H.; Fazli, L.; Lamoureux, F.; Beraldi, E.; Monia, B.P.; MacLeod, A.R.; Thuroff, J.W.; Gleave, M.E. Transcription factor stat5 knockdown enhances androgen receptor degradation and delays castration-resistant prostate cancer progression in vivo. Mol. Cancer Ther. 2011, 10, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Lin, X.; Kapoor, A.; He, L.; Wei, F.; Gu, Y.; Mei, W.; Zhao, K.; Yang, H.; Tang, D. Fam84b promotes prostate tumorigenesis through a network alteration. Ther. Adv. Med. Oncol. 2019, 11, 1758835919846372. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Mei, W.; Gu, Y.; Lin, X.; He, L.; Zeng, H.; Wei, F.; Wan, X.; Yang, H.; Major, P.; et al. Construction of a set of novel and robust gene expression signatures predicting prostate cancer recurrence. Mol. Oncol. 2018, 12, 1559–1578. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Kamarudin, A.N.; Cox, T.; Kolamunnage-Dona, R. Time-dependent roc curve analysis in medical research: Current methods and applications. BMC Med. Res. Methodol. 2017, 17, 53. [Google Scholar] [CrossRef]
- Brott, T.; Marler, J.R.; Olinger, C.P.; Adams, H.P., Jr.; Tomsick, T.; Barsan, W.G.; Biller, J.; Eberle, R.; Hertzberg, V.; Walker, M. Measurements of acute cerebral infarction: Lesion size by computed tomography. Stroke 1989, 20, 871–875. [Google Scholar] [CrossRef]
- Ozenne, B.; Subtil, F.; Maucort-Boulch, D. The precision--recall curve overcame the optimism of the receiver operating characteristic curve in rare diseases. J. Clin. Epidemiol. 2015, 68, 855–859. [Google Scholar] [CrossRef]
- Shen, R.; Olshen, A.B.; Ladanyi, M. Integrative clustering of multiple genomic data types using a joint latent variable model with application to breast and lung cancer subtype analysis. Bioinformatics 2009, 25, 2906–2912. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell 2015, 163, 1011–1025. [Google Scholar]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. Gepia2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucl. Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [PubMed]
- Ross-Adams, H.; Lamb, A.D.; Dunning, M.J.; Halim, S.; Lindberg, J.; Massie, C.M.; Egevad, L.A.; Russell, R.; Ramos-Montoya, A.; Vowler, S.L.; et al. Integration of copy number and transcriptomics provides risk stratification in prostate cancer: A discovery and validation cohort study. EBioMedicine 2015, 2, 1133–1144. [Google Scholar] [CrossRef] [PubMed]
- Knezevic, D.; Goddard, A.D.; Natraj, N.; Cherbavaz, D.B.; Clark-Langone, K.M.; Snable, J.; Watson, D.; Falzarano, S.M.; Magi-Galluzzi, C.; Klein, E.A.; et al. Analytical validation of the oncotype dx prostate cancer assay—A clinical rt-pcr assay optimized for prostate needle biopsies. BMC Genom. 2013, 14, 690. [Google Scholar] [CrossRef]
- Klein, E.A.; Cooperberg, M.R.; Magi-Galluzzi, C.; Simko, J.P.; Falzarano, S.M.; Maddala, T.; Chan, J.M.; Li, J.; Cowan, J.E.; Tsiatis, A.C.; et al. A 17-gene assay to predict prostate cancer aggressiveness in the context of gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. Eur. Urol. 2014, 66, 550–560. [Google Scholar] [CrossRef] [PubMed]
- Cuzick, J.; Swanson, G.P.; Fisher, G.; Brothman, A.R.; Berney, D.M.; Reid, J.E.; Mesher, D.; Speights, V.O.; Stankiewicz, E.; Foster, C.S.; et al. Prognostic value of an rna expression signature derived from cell cycle proliferation genes in patients with prostate cancer: A retrospective study. Lancet Oncol. 2011, 12, 245–255. [Google Scholar] [CrossRef]
- Oderda, M.; Cozzi, G.; Daniele, L.; Sapino, A.; Munegato, S.; Renne, G.; De Cobelli, O.; Gontero, P. Cell-cycle progression-score might improve the current risk assessment in newly diagnosed prostate cancer patients. Urology 2017, 102, 73–78. [Google Scholar] [CrossRef]
- Albala, D.; Kemeter, M.J.; Febbo, P.G.; Lu, R.; John, V.; Stoy, D.; Denes, B.; McCall, M.; Shindel, A.W.; Dubeck, F. Health economic impact and prospective clinical utility of oncotype dx(r) genomic prostate score. Rev. Urol. 2016, 18, 123–132. [Google Scholar]
- Cullen, J.; Rosner, I.L.; Brand, T.C.; Zhang, N.; Tsiatis, A.C.; Moncur, J.; Ali, A.; Chen, Y.; Knezevic, D.; Maddala, T.; et al. A biopsy-based 17-gene genomic prostate score predicts recurrence after radical prostatectomy and adverse surgical pathology in a racially diverse population of men with clinically low- and intermediate-risk prostate cancer. Eur. Urol. 2015, 68, 123–131. [Google Scholar] [CrossRef]
- Cooperberg, M.R.; Simko, J.P.; Cowan, J.E.; Reid, J.E.; Djalilvand, A.; Bhatnagar, S.; Gutin, A.; Lanchbury, J.S.; Swanson, G.P.; Stone, S.; et al. Validation of a cell-cycle progression gene panel to improve risk stratification in a contemporary prostatectomy cohort. J. Clin. Oncol. 2013, 31, 1428–1434. [Google Scholar] [CrossRef]
- Seifert, M.; Peitzsch, C.; Gorodetska, I.; Borner, C.; Klink, B.; Dubrovska, A. Network-based analysis of prostate cancer cell lines reveals novel marker gene candidates associated with radioresistance and patient relapse. PLoS Comput. Biol. 2019, 15, e1007460. [Google Scholar] [CrossRef]
- Hwang, W.; Chiu, Y.F.; Kuo, M.H.; Lee, K.L.; Lee, A.C.; Yu, C.C.; Chang, J.L.; Huang, W.C.; Hsiao, S.H.; Lin, S.E.; et al. Expression of neuroendocrine factor vgf in lung cancer cells confers resistance to egfr kinase inhibitors and triggers epithelial-to-mesenchymal transition. Cancer Res. 2017, 77, 3013–3026. [Google Scholar] [CrossRef] [PubMed]
- Pudova, E.A.; Lukyanova, E.N.; Nyushko, K.M.; Mikhaylenko, D.S.; Zaretsky, A.R.; Snezhkina, A.V.; Savvateeva, M.V.; Kobelyatskaya, A.A.; Melnikova, N.V.; Volchenko, N.N.; et al. Differentially expressed genes associated with prognosis in locally advanced lymph node-negative prostate cancer. Front. Gen. 2019, 10, 730. [Google Scholar] [CrossRef]
- Chang, Y.F.; Huang, Y.Q.; Wu, K.M.; Jou, A.F.; Shih, N.Y.; Ho, J.A. Diagnosing the rgs11 lung cancer biomarker: The integration of competitive immunoassay and isothermal nucleic acid exponential amplification reaction. Anal. Chem. 2019, 91, 3327–3335. [Google Scholar] [CrossRef]
- Yang, F.; Chen, X.; Li, X.; Chen, J.; Tang, Y.; Cai, Y.; Wang, Y.; Chen, Z.; Li, L.; Li, R.; et al. Long intergenic non-protein coding rna 1089 suppresses cell proliferation and metastasis in gastric cancer by regulating mirna-27a-3p/epithelial-mesenchymal transition (emt) axis. Cancer Manag. Res. 2020, 12, 5587–5596. [Google Scholar] [CrossRef] [PubMed]
- Barisone, G.A.; Ngo, T.; Tran, M.; Cortes, D.; Shahi, M.H.; Nguyen, T.V.; Perez-Lanza, D.; Matayasuwan, W.; Diaz, E. Role of mxd3 in proliferation of daoy human medulloblastoma cells. PLoS ONE 2012, 7, e38508. [Google Scholar] [CrossRef] [PubMed]
- Koike, H.; Morikawa, Y.; Sekine, Y.; Matsui, H.; Shibata, Y.; Suzuki, K. Survivin is associated with cell proliferation and has a role in 1a,25-dihydroxyvitamin d3 induced cell growth inhibition in prostate cancer. J. Urol. 2011, 185, 1497–1503. [Google Scholar] [CrossRef]
- Cao, L.; Li, C.; Shen, S.; Yan, Y.; Ji, W.; Wang, J.; Qian, H.; Jiang, X.; Li, Z.; Wu, M.; et al. Oct4 increases birc5 and ccnd1 expression and promotes cancer progression in hepatocellular carcinoma. BMC Cancer 2013, 13, 82. [Google Scholar] [CrossRef]
- Ali, H.E.A.; Lung, P.Y.; Sholl, A.B.; Gad, S.A.; Bustamante, J.J.; Ali, H.I.; Rhim, J.S.; Deep, G.; Zhang, J.; Abd Elmageed, Z.Y. Dysregulated gene expression predicts tumor aggressiveness in african-american prostate cancer patients. Sci. Rep. 2018, 8, 16335. [Google Scholar] [CrossRef]
- Halvorsen, A.R.; Helland, A.; Fleischer, T.; Haug, K.M.; Grenaker Alnaes, G.I.; Nebdal, D.; Syljuasen, R.G.; Touleimat, N.; Busato, F.; Tost, J.; et al. Differential DNA methylation analysis of breast cancer reveals the impact of immune signaling in radiation therapy. Int. J. Cancer 2014, 135, 2085–2095. [Google Scholar] [CrossRef]
- Shi, X.; Huo, J.; Gao, X.; Cai, H.; Zhu, W. A newly identified lncrna h1fx-as1 targets dact1 to inhibit cervical cancer via sponging mir-324-3p. Cancer Cell Int. 2020, 20, 358. [Google Scholar] [CrossRef]
- Hochwagen, A.; Tham, W.H.; Brar, G.A.; Amon, A. The fk506 binding protein fpr3 counteracts protein phosphatase 1 to maintain meiotic recombination checkpoint activity. Cell 2005, 122, 861–873. [Google Scholar] [CrossRef]
- Dong, P.; Yu, B.; Pan, L.; Tian, X.; Liu, F. Identification of key genes and pathways in triple-negative breast cancer by integrated bioinformatics analysis. Biomed. Res. Int. 2018, 2018, 2760918. [Google Scholar] [CrossRef]
- Mei, J.; Xing, Y.; Lv, J.; Gu, D.; Pan, J.; Zhang, Y.; Liu, J. Construction of an immune-related gene signature for prediction of prognosis in patients with cervical cancer. Int. Immunopharmacol. 2020, 88, 106882. [Google Scholar] [CrossRef]
- Kang, M.J.; Heo, S.K.; Song, E.J.; Kim, D.J.; Han, S.Y.; Han, J.H.; Kim, B.Y.; Park, J.H. Activation of nod1 and nod2 induces innate immune responses of prostate epithelial cells. Prostate 2012, 72, 1351–1358. [Google Scholar] [CrossRef]
- Castano-Rodriguez, N.; Kaakoush, N.O.; Mitchell, H.M. Pattern-recognition receptors and gastric cancer. Front. Immunol. 2014, 5, 336. [Google Scholar]
- Balakrishnan, A.; Penachioni, J.Y.; Lamba, S.; Bleeker, F.E.; Zanon, C.; Rodolfo, M.; Vallacchi, V.; Scarpa, A.; Felicioni, L.; Buck, M.; et al. Molecular profiling of the "plexinome" in melanoma and pancreatic cancer. Hum. Mutat. 2009, 30, 1167–1174. [Google Scholar] [CrossRef] [PubMed]
- Wen, H.; Lei, Y.; Eun, S.Y.; Ting, J.P. Plexin-a4-semaphorin 3a signaling is required for toll-like receptor- and sepsis-induced cytokine storm. J. Exp. Med. 2010, 207, 2943–2957. [Google Scholar] [CrossRef]
- Tsun, Z.Y.; Bar-Peled, L.; Chantranupong, L.; Zoncu, R.; Wang, T.; Kim, C.; Spooner, E.; Sabatini, D.M. The folliculin tumor suppressor is a gap for the ragc/d gtpases that signal amino acid levels to mtorc1. Mol. Cell 2013, 52, 495–505. [Google Scholar] [CrossRef]
- Puertollano, R.; Ferguson, S.M.; Brugarolas, J.; Ballabio, A. The complex relationship between tfeb transcription factor phosphorylation and subcellular localization. EMBO J. 2018, 37. [Google Scholar] [CrossRef]
- Jiang, Y.; Zheng, X.; Jiao, D.; Chen, P.; Xu, Y.; Wei, H.; Qian, Y. Peptidase inhibitor 15 as a novel blood diagnostic marker for cholangiocarcinoma. EBioMedicine 2019, 40, 422–431. [Google Scholar] [CrossRef]
- Stacey, S.N.; Helgason, H.; Gudjonsson, S.A.; Thorleifsson, G.; Zink, F.; Sigurdsson, A.; Kehr, B.; Gudmundsson, J.; Sulem, P.; Sigurgeirsson, B.; et al. New basal cell carcinoma susceptibility loci. Nat. Commun. 2015, 6, 6825. [Google Scholar] [CrossRef]
- Pennati, M.; Lopergolo, A.; Profumo, V.; De Cesare, M.; Sbarra, S.; Valdagni, R.; Zaffaroni, N.; Gandellini, P.; Folini, M. Mir-205 impairs the autophagic flux and enhances cisplatin cytotoxicity in castration-resistant prostate cancer cells. Biochem. Pharmacol. 2014, 87, 579–597. [Google Scholar] [CrossRef]
- Nagelkerke, A.; Mujcic, H.; Bussink, J.; Wouters, B.G.; van Laarhoven, H.W.; Sweep, F.C.; Span, P.N. Hypoxic regulation and prognostic value of lamp3 expression in breast cancer. Cancer 2011, 117, 3670–3681. [Google Scholar] [CrossRef]
- Vasmatzis, G.; Kosari, F.; Murphy, S.J.; Terra, S.; Kovtun, I.V.; Harris, F.R.; Zarei, S.; Smadbeck, J.B.; Johnson, S.H.; Gaitatzes, A.G.; et al. Large chromosomal rearrangements yield biomarkers to distinguish low-risk from intermediate- and high-risk prostate cancer. Mayo Clin. Proc. 2019, 94, 27–36. [Google Scholar] [CrossRef]
- Buckwalter, J.M.; Chan, W.; Shuman, L.; Wildermuth, T.; Ellis-Mohl, J.; Walter, V.; Warrick, J.I.; Wu, X.R.; Kaag, M.; Raman, J.D.; et al. Characterization of histone deacetylase expression within in vitro and in vivo bladder cancer model systems. Int. J. Mol. Sci. 2019, 20, 2599. [Google Scholar] [CrossRef]
- Januchowski, R.; Sterzynska, K.; Zawierucha, P.; Rucinski, M.; Swierczewska, M.; Partyka, M.; Bednarek-Rajewska, K.; Brazert, M.; Nowicki, M.; Zabel, M.; et al. Microarray-based detection and expression analysis of new genes associated with drug resistance in ovarian cancer cell lines. Oncotarget 2017, 8, 49944–49958. [Google Scholar] [CrossRef]
- Zhu, Q.L.; Zhan, D.M.; Chong, Y.K.; Ding, L.; Yang, Y.G. Mir-652-3p promotes bladder cancer migration and invasion by targeting kcnn3. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8806–8812. [Google Scholar]
- Zhao, S.; Geybels, M.S.; Leonardson, A.; Rubicz, R.; Kolb, S.; Yan, Q.; Klotzle, B.; Bibikova, M.; Hurtado-Coll, A.; Troyer, D.; et al. Epigenome-wide tumor DNA methylation profiling identifies novel prognostic biomarkers of metastatic-lethal progression in men diagnosed with clinically localized prostate cancer. Clin. Cancer Res. 2017, 23, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Kapoor, A.; Gu, Y.; Chow, M.J.; Xu, H.; Major, P.; Tang, D. Assessment of biochemical recurrence of prostate cancer (review). Int. J. Oncol. 2019, 55, 1194–1212. [Google Scholar] [CrossRef]
- Worst, T.S.; Previti, C.; Nitschke, K.; Diessl, N.; Gross, J.C.; Hoffmann, L.; Frey, L.; Thomas, V.; Kahlert, C.; Bieback, K.; et al. Mir-10a-5p and mir-29b-3p as extracellular vesicle-associated prostate cancer detection markers. Cancers 2019, 12, 43. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Li, T.; Kapoor, A.; Major, P.; Tang, D. Contactin 1: An important and emerging oncogenic protein promoting cancer progression and metastasis. Genes 2020, 11, 874. [Google Scholar] [CrossRef]
- Osman, M.A.; Antonisamy, W.J.; Yakirevich, E. Iqgap1 control of centrosome function defines distinct variants of triple negative breast cancer. Oncotarget 2020, 11, 2493–2511. [Google Scholar] [CrossRef]
- Zhao, H.; Xie, C.; Lin, X.; Zhao, Y.; Han, Y.; Fan, C.; Zhang, X.; Du, J.; Han, Y.; Han, Q.; et al. Coexpression of iq-domain gtpase-activating protein 1 (iqgap1) and dishevelled (dvl) is correlated with poor prognosis in non-small cell lung cancer. PLoS ONE 2014, 9, e113713. [Google Scholar] [CrossRef]
- Hayashi, T.; Fujita, K.; Matsushita, M.; Nonomura, N. Main inflammatory cells and potentials of anti-inflammatory agents in prostate cancer. Cancers 2019, 11, 1153. [Google Scholar] [CrossRef]
- Wu, C.T.; Chen, W.C.; Chen, M.F. The response of prostate cancer to androgen deprivation and irradiation due to immune modulation. Cancers 2018, 11, 20. [Google Scholar] [CrossRef]
- Korolchuk, V.I.; Saiki, S.; Lichtenberg, M.; Siddiqi, F.H.; Roberts, E.A.; Imarisio, S.; Jahreiss, L.; Sarkar, S.; Futter, M.; Menzies, F.M.; et al. Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 2011, 13, 453–460. [Google Scholar] [CrossRef]
- Ojo, D.; Lin, X.; Wu, Y.; Cockburn, J.; Bane, A.; Tang, D. Polycomb complex protein bmi1 confers resistance to tamoxifen in estrogen receptor positive breast cancer. Cancer Lett. 2018, 426, 4–13. [Google Scholar] [CrossRef]
- He, L.; Fan, C.; Kapoor, A.; Ingram, A.J.; Rybak, A.P.; Austin, R.C.; Dickhout, J.; Cutz, J.C.; Scholey, J.; Tang, D. Alpha-mannosidase 2c1 attenuates pten function in prostate cancer cells. Nat. Commun. 2011, 2, 307. [Google Scholar] [CrossRef]
- He, L.; Ingram, A.; Rybak, A.P.; Tang, D. Shank-interacting protein-like 1 promotes tumorigenesis via pten inhibition in human tumor cells. J. Clin. Investig. 2010, 120, 2094–2108. [Google Scholar] [CrossRef]
- Yan, J.; Ojo, D.; Kapoor, A.; Lin, X.; Pinthus, J.H.; Aziz, T.; Bismar, T.A.; Wei, F.; Wong, N.; De Melo, J.; et al. Neural cell adhesion protein cntn1 promotes the metastatic progression of prostate cancer. Cancer Res. 2016, 76, 1603–1614. [Google Scholar] [CrossRef]
- Wong, N.; Gu, Y.; Kapoor, A.; Lin, X.; Ojo, D.; Wei, F.; Yan, J.; de Melo, J.; Major, P.; Wood, G.; et al. Upregulation of fam84b during prostate cancer progression. Oncotarget 2017. [Google Scholar] [CrossRef]
- Gu, Y.; Chow, M.J.; Kapoor, A.; Mei, W.; Jiang, Y.; Yan, J.; De Melo, J.; Seliman, M.; Yang, H.; Cutz, J.C.; et al. Biphasic alteration of butyrylcholinesterase (bche) during prostate cancer development. Transl. Oncol. 2018, 11, 1012–1022. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cbio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cbioportal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An integrated tcga pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 2018, 173, 400–416.e411. [Google Scholar] [CrossRef]


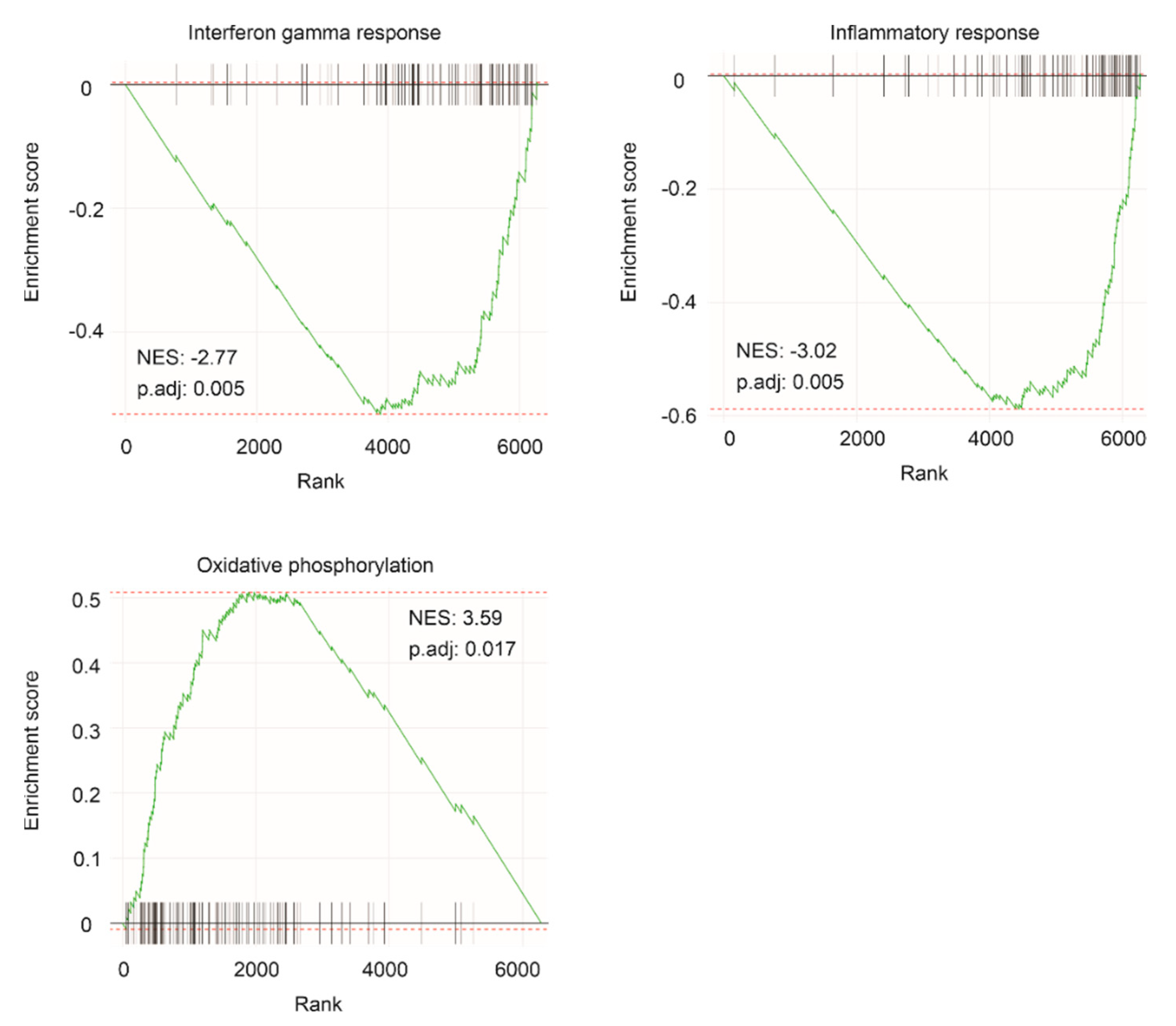




| Pathway | P.Adj | ES | NES | Size |
|---|---|---|---|---|
| INFγ response | 0.004662 | −0.53228 | −2.77404 | 92 |
| Apical junction | 0.004662 | −0.44445 | −2.25531 | 78 |
| Inflammatory response | 0.004662 | −0.58813 | −3.02081 | 84 |
| Il2_Stat5 signaling | 0.004662 | −0.4212 | −2.14177 | 80 |
| Complement | 0.004662 | −0.42784 | −2.17532 | 81 |
| Allograft rejection | 0.004662 | −0.55785 | −2.83455 | 82 |
| KRAS signaling UP | 0.004662 | −0.62089 | −3.10098 | 73 |
| UV response DN | 0.004662 | −0.52657 | −2.58735 | 67 |
| TNFα signaling via NFκB | 0.004662 | −0.56906 | −2.80937 | 70 |
| Epithelial mesenchymal transition | 0.004662 | −0.64204 | −3.10811 | 63 |
| Estrogen response early | 0.004662 | −0.50057 | −2.37491 | 59 |
| INFα response | 0.004662 | −0.47271 | −2.12179 | 45 |
| IL6 Jak Stat3 signaling | 0.004662 | −0.60951 | −2.574 | 33 |
| Estrogen response late | 0.007924 | −0.42778 | −1.91355 | 44 |
| Coagulation | 0.007924 | −0.46215 | −2.02948 | 41 |
| Myc targets V2 | 0.011409 | 0.481626 | 2.48835 | 32 |
| Fatty acid metabolism | 0.011897 | 0.30286 | 1.71898 | 46 |
| Apoptosis | 0.011897 | −0.36188 | −1.74708 | 62 |
| Hypoxia | 0.011897 | −0.37568 | −1.79535 | 60 |
| DNA repair | 0.013372 | 0.37213 | 2.35074 | 69 |
| Myc targets V1 | 0.016632 | 0.41489 | 2.90611 | 102 |
| Mitotic spindle | 0.016632 | −0.32065 | −1.71067 | 103 |
| Myogenesis | 0.017405 | −0.37748 | −1.71929 | 49 |
| Oxidative phosphorylation | 0.017405 | 0.508354 | 3.58833 | 113 |
| TGFβ signaling | 0.017405 | −0.44961 | −1.80892 | 28 |
| Gene | Locus | Log2 Ratio 1 | p-Value | q-Value |
|---|---|---|---|---|
| HAGHL | 16p13.3 | 1.72 | 3.54 × 10−17 *** | 1.00 × 10−15 *** |
| LCN12 | 9q34.3 | 1.34 | 6.75 × 10−18 *** | 2.39 × 10−16 *** |
| DCST2 | 1q21.3 | 1.28 | 6.10 × 10−16 *** | 1.14 × 10−14 *** |
| VGF | 7q22.1 | 1.2 | 5.54 × 10−8 *** | 1.53 × 10−7 *** |
| RGS11 | 16p13.3 | 1.18 | 9.68 × 10−12 *** | 6.41 × 10−11 *** |
| PRR7 | 5q35.3 | 1.17 | 1.65 × 10−16 *** | 3.82 × 10−15 *** |
| LINC01089 | 12q24.31 | 1.1 | 1.43 × 10−15 *** | 2.37 × 10−14 *** |
| MXD3 | 5q35.3 | 1.07 | 1.83 × 10−26 *** | 3.50 × 10−23 *** |
| BIRC5 | 17q25.3 | 1.04 | 1.70 × 10−10 *** | 8.74 × 10−10 *** |
| LTC4S | 5q35.3 | 1.03 | 1.68 × 10−11 *** | 1.06 × 10−10 *** |
| H1FX-AS1 | 3q21.3 | 1.03 | 2.83 × 10−17 *** | 8.46 × 10−16 *** |
| FPR3 | 19q13.41 | −1 | 1.12 × 10−10 *** | 5.96 × 10−10 *** |
| RAB30 | 11q14.1 | −1 | 8.65 × 10−12 *** | 5.79 × 10−11 *** |
| RIPOR2 | 6p22.3 | −1.01 | 3.04 × 10−5 *** | 6.67 × 10−5 *** |
| NOD2 | 16q12.1 | −1.01 | 1.05 × 10−8 *** | 3.93 × 10−8 *** |
| PLXNA4 | 7q32.3 | −1.02 | 4.74 × 10−11 *** | 2.71 × 10−10 *** |
| RRAGC | 1p34.3 | −1.03 | 1.33 × 10−11 *** | 8.61 × 10−11 *** |
| TF × 10C | 7q31.2 | −1.06 | 5.65 × 10−13 *** | 4.84 × 10−12 *** |
| PI15 | 8q21.13 | −1.08 | 5.34 × 10−4 *** | 9.88 × 10−4 *** |
| ZFHX4 | 8q21.13 | −1.08 | 1.92 × 10−8 *** | 6.90 × 10−8 *** |
| LAMP3 | 3q27.1 | −1.21 | 7.78 × 10−10 *** | 3.55 × 10−9 *** |
| HDAC9 | 7p21.1 | −1.24 | 1.07 × 10−7 *** | 3.83 × 10−7 *** |
| MCTP1 | 5q15 | −1.29 | 1.27 × 10−11 *** | 8.23 × 10−11 *** |
| KCNN3 | 1q21.3 | −1.4 | 5.58 × 10−10 *** | 2.73 × 10−9 *** |
| PCDHB8 | 5q31.3 | −1.42 | 1.39 × 10−8 *** | 5.09 × 10−8 *** |
| PCDHGB2 | 5q31.3 | −1.66 | 4.86 × 10−9 *** | 1.93 × 10−8 *** |
| PCDHGA5 | 5q31.3 | −1.94 | 6.00 × 10−10 *** | 2.79 × 10−9 *** |
| Factors | Univariate Cox Analysis | Multivariate Cox Analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p-Value | HR | 95% CI | p-Value | |
| Sig27gene | 2.72 | 2.27–3.25 | <2 × 10−16 *** | 2.32 | 1.88–2.88 | 5.74 × 10−15 *** |
| Age 1 | 1.02 | 0.99–1.05 | 0.189 | 0.99 | 0.96–1.02 | 0.402 |
| WHO IV 2 | 9.76 | 1.28–74.61 | 0.02817 * | 3.46 | 0.43–27.83 | 0.244 |
| WHO V 2 | 21.38 | 2.96–154.52 | 0.00241 ** | 5.11 | 0.66–39.64 | 0.199 |
| Margin 1 3 | 2.30 | 1.52–3.48 | 8.1 × 10−5 *** | 1.28 | 0.81–2.03 | 0.294 |
| Tstage 1 4 | 3.69 | 2.08–6.52 | 7.54 × 10−6 *** | 1.34 | 0.70–2.69 | 0.352 |
| Gene | Details | HR a | 95% CI | p-Value |
|---|---|---|---|---|
| HAGHL c | Hydroxyacylglutathione Hydrolase Like | 1.002 | 1.001–1.003 | 3.6 × 10−6 *** |
| LCN12 c | Lipocalin 12 | 1.009 | 1.004–1.015 | 8.83 × 10−4 *** |
| DCST2 c | DC-STAMP Domain Containing 2 | 1.01 | 1.005–1.015 | 3.61 × 10−5 *** |
| VGF c | VGF Nerve Growth Factor Inducible | 1.003 | 1.001–1.004 | 4.17 × 10−6 *** |
| RGS11 | Regulator of G Protein Signaling 11 | 1.001 | 1.001–1.001 | 4.76 × 10−7 *** |
| PRR7 c | Proline Rich 7, Synaptic | 1.005 | 1.003–1.007 | 1.27 × 10−7 *** |
| LINC01089 c | Long Intergenic Non-Protein Coding RNA 1089 | 1.001 | 1–1.001 | 2.66 × 10−5 *** |
| MXD3 c | MAX Dimerization Protein 3 | 1.004 | 1.002–1.005 | 2.05 × 10−7 *** |
| BIRC5 | Baculoviral IAP Repeat Containing 5 (survivin) | 1.002 | 1.002–1.003 | 2.78 × 10−7 *** |
| LTC4S c | Leukotriene C4 Synthase | 1.017 | 1.011–1.023 | 3.12 × 10−8 *** |
| H1FX-AS1 c | H1−10 Antisense RNA 1 | 1.006 | 1.004–1.009 | 2.28 × 10−7 *** |
| FPR3 | Formyl Peptide Receptor 3 | 1.002 | 1.001–1.004 | 0.00434 ** |
| RAB30 | member RAS Oncogene Family | 1.001 | 0.998–1.004 | 0.444 |
| RIPOR2 | Atypical inhibitor of the small G protein RhoA | 1 | 1–1 | 0.0464 * |
| NOD2 c | Nucleotide Binding Oligomerization Domain Containing 2 | 1.01 | 1.004–1.016 | 9.95 × 10−4 *** |
| PLXNA4 | Plexin A4 | 1.004 | 1.001–1.006 | 0.0177 * |
| RRAGC | Ras Related GTP Binding C | 1 | 0.998–1.003 | 0.768 |
| TFEC | Transcription Factor EC | 1.005 | 1.002–1.008 | 0.0194 * |
| PI15 | Peptidase Inhibitor 15 | 1 | 1–1 | 0.12 |
| ZFHX4 | Zinc Finger Homeobox 4 | 1.001 | 1–1.002 | 0.0385 * |
| LAMP3 b | Lysosomal Associated Membrane Protein 3 | 1.001 | 1–1.001 | 0.0616 |
| HDAC9 | Histone Deacetylase 9 | 1 | 0.9998–1 | 0.585 |
| MCTP1 | Multiple C2 And Transmembrane Domain Containing 1 | 1.003 | 1–1.005 | 0.0497 * |
| KCNN3 | Potassium Calcium-Activated Channel Subfamily N Member 3 | 1.005 | 0.997–1.009 | 0.0668 |
| PCDHB8 | Protocadherin Beta 8 | 1 | 0.9991–1.002 | 0.523 |
| PCDHGB2 | Protocadherin Gamma Subfamily B2 | 1.001 | 1–1.002 | 0.00166 ** |
| PCDHGA5 | Protocadherin Gamma Subfamily A5 | 1 | 1–1.001 | 0.0109 * |
| Gene | Oncogenic Role in PC | Oncogenic Role in Others | Refs |
|---|---|---|---|
| HAGHL | unknown | unknown | NA |
| LCN12 | unknown | unknown | NA |
| DCST2 | unknown | unknown | NA |
| VGF | facilitation of radioresistance in prostate cancer cells | promotion of resistance to tyrosine kinase inhibitors in lung cancer | [60,61] |
| RGS11 | association with TMPRSS2-ERG | a biomarker of lung cancer | [62,63] |
| PRR7 | unknown | unknown | NA |
| LINC01089 | unknown | suppression of cell proliferation | [64] |
| MXD3 | unknown | promotion of medullobastoma | [65] |
| BIRC5 | promotion of prostate cancer | promotion of cancer progression and metastasis | [66,67] |
| LTC4S | predicting prostate cancer progression | a component gene of a immune signature of breast cancer | [68,69] |
| H1FX-AS1 | unknown | reverse association with cervical cancer prognosis | [70] |
| FPR3 | unknown | sustain meiotic recombination checkpoint actions | [71] |
| RAB30 | unknown | association of good prognosis in triple negative breast cancer | [72] |
| RIPOR2 | unknown | association of immune cell infiltration and thus inhibition of cervical cancer | [73] |
| NOD2 | induction of innate immune responses of prostate cancer | immunosuppression of tumorigenesis in gastric cancer | [74,75] |
| PLXNA4 | unknown | inhibition of tumor cell migration and contribution to innate immunity in working with Toll-like receptor | [76,77] |
| RRAGC | unknown | regulation (activation and inactivation) of mTOC1 activation | [78] |
| TFEC | unknown | regulation of lysosome biogenesis and mTOR activation | [79] |
| PI15 | methylation of its CpGs occurs in metastatic PCs | biomarker of cholangiocarcinoma | [80] |
| ZFHX4 | unknown | a susceptibility locus of cutaneous basal cell carcinoma | [81] |
| LAMP3 b | evidence suggests its association with resistance to platinum in PC | a hypoxia-induced gene associated with aggressive breast cancer | [82,83] |
| HDAC9 | mutations detected in PC | increases in expression in bladder cancer | [84,85] |
| MCTP1 | unknown | downregulation in paclitaxel-resistant ovarian cancer cells | [86] |
| KCNN3 | unknown | suppression of bladder cancer cell migration and invasion | [87] |
| PCDHB8 | unknown | unknown | NA |
| PCDHGB2 | unknown | unknown | NA |
| PCDHGA5 | unknown | unknown | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, Y.; Lin, X.; Kapoor, A.; Li, T.; Major, P.; Tang, D. Effective Prediction of Prostate Cancer Recurrence through the IQGAP1 Network. Cancers 2021, 13, 430. https://doi.org/10.3390/cancers13030430
Gu Y, Lin X, Kapoor A, Li T, Major P, Tang D. Effective Prediction of Prostate Cancer Recurrence through the IQGAP1 Network. Cancers. 2021; 13(3):430. https://doi.org/10.3390/cancers13030430
Chicago/Turabian StyleGu, Yan, Xiaozeng Lin, Anil Kapoor, Taosha Li, Pierre Major, and Damu Tang. 2021. "Effective Prediction of Prostate Cancer Recurrence through the IQGAP1 Network" Cancers 13, no. 3: 430. https://doi.org/10.3390/cancers13030430
APA StyleGu, Y., Lin, X., Kapoor, A., Li, T., Major, P., & Tang, D. (2021). Effective Prediction of Prostate Cancer Recurrence through the IQGAP1 Network. Cancers, 13(3), 430. https://doi.org/10.3390/cancers13030430






