NOTCH Activation via gp130/STAT3 Signaling Confers Resistance to Chemoradiotherapy
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
2.1. Inflammation Promotes CRT Resistance
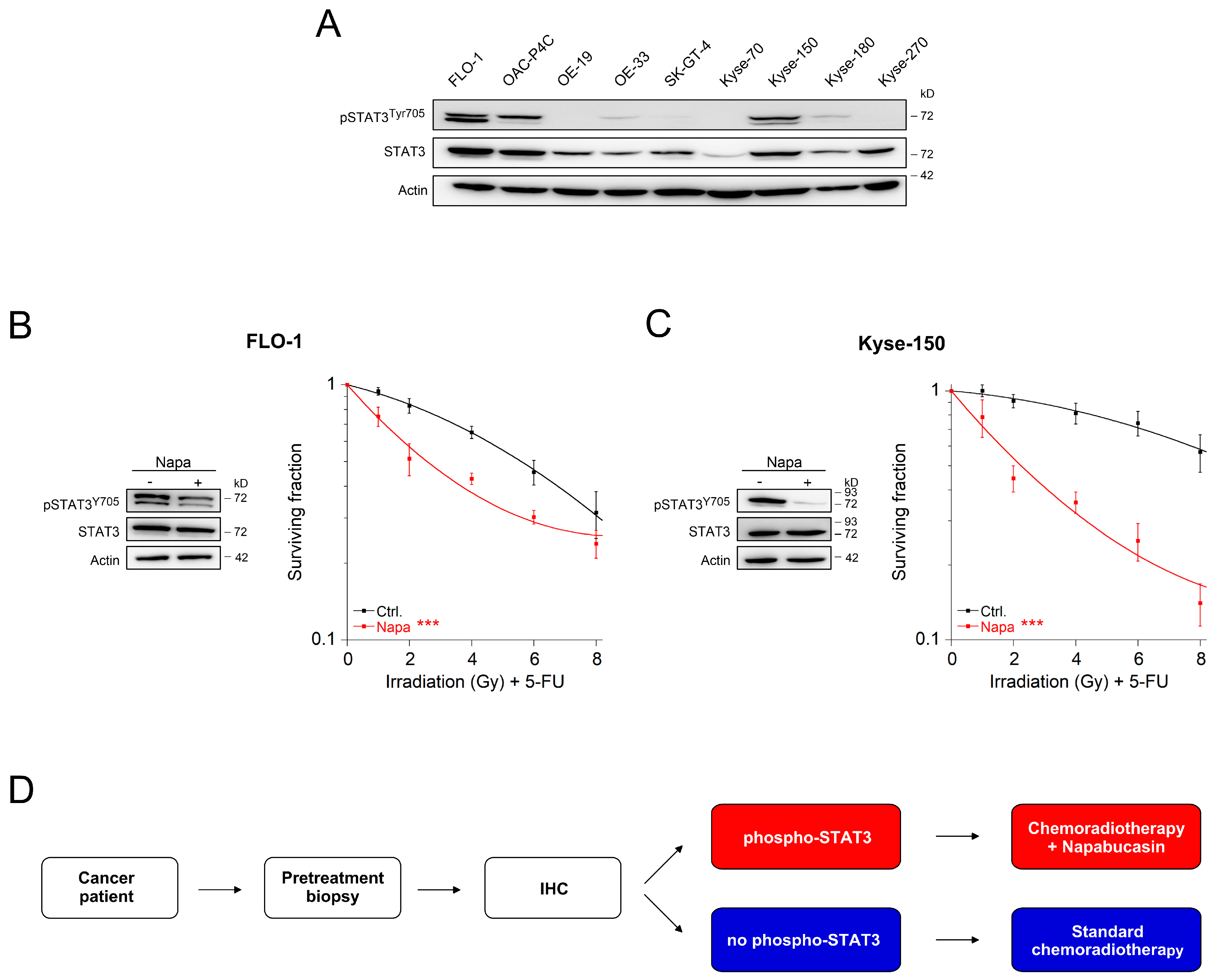
2.2. Inhibition of the gp130/STAT3 Axis as a Therapeutic Strategy
2.3. Resistance-Associated STAT3 Target Genes
2.4. Convergent STAT3 and NOTCH Signaling Cause CRT Resistance
3. Discussion
4. Materials and Methods
4.1. Cell Culture, RNA Interference, and Western Blot Analysis
4.2. Colony Formation Assay
4.3. STAT3 Activity and Expression of STAT3 Variants
4.4. Electrophoretic Mobility Shift Assay (EMSA)
4.5. Screening STAT3 Target Genes by RNA-Seq and Opposite Direction Analysis
4.6. Patients, Gene Expression Profiling and Survival Analysis
4.7. Mice
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sauer, R.; Becker, H.; Hohenberger, W.; Rödel, C.; Wittekind, C.; Fietkau, R.; Martus, P.; Tschmelitsch, J.; Hager, E.; Hess, C.F.; et al. Preoperative versus Postoperative Chemoradiotherapy for Rectal Cancer. N. Engl. J. Med. 2004, 351, 1731–1740. [Google Scholar] [CrossRef]
- Cunningham, D.; Atkin, W.; Lenz, H.-J.; Lynch, H.T.; Minsky, B.; Nordlinger, B.; Starling, N. Colorectal cancer. Lancet 2010, 375, 1030–1047. [Google Scholar] [CrossRef]
- Smith, J.J.; Garcia-Aguilar, J. Advances and Challenges in Treatment of Locally Advanced Rectal Cancer. J. Clin. Oncol. 2015, 33, 1797–1808. [Google Scholar] [CrossRef]
- Rödel, C.; Liersch, T.; Becker, H.; Fietkau, R.; Hohenberger, W.; Hothorn, T.; Graeven, U.; Arnold, D.; Lang-Welzenbach, M.; Raab, H.-R.; et al. Preoperative chemoradiotherapy and postoperative chemotherapy with fluorouracil and oxaliplatin versus fluorouracil alone in locally advanced rectal cancer: Initial results of the German CAO/ARO/AIO-04 randomised phase 3 trial. Lancet Oncol. 2012, 13, 679–687. [Google Scholar] [CrossRef]
- Lange, M.M.; Van De Velde, C.J.H. Urinary and sexual dysfunction after rectal cancer treatment. Nat. Rev. Urol. 2010, 8, 51–57. [Google Scholar] [CrossRef]
- Bryant, C.L.C.; Lunniss, P.J.; Knowles, C.H.; Thaha, M.; Chan, C.L.H. Anterior resection syndrome. Lancet Oncol. 2012, 13, e403–e408. [Google Scholar] [CrossRef]
- Fokas, E.; Liersch, T.; Fietkau, R.; Hohenberger, W.; Beißbarth, T.; Hess, C.; Becker, H.; Ghadimi, M.; Mrak, K.; Merkel, S.; et al. Tumor Regression Grading After Preoperative Chemoradiotherapy for Locally Advanced Rectal Carcinoma Revisited: Updated Results of the CAO/ARO/AIO-94 Trial. J. Clin. Oncol. 2014, 32, 1554–1562. [Google Scholar] [CrossRef]
- Fokas, E.; Ströbel, P.; Fietkau, R.; Ghadimi, M.; Liersch, T.; Grabenbauer, G.G.; Hartmann, A.; Kaufmann, M.; Sauer, R.; Graeven, U.; et al. Tumor Regression Grading After Preoperative Chemoradiotherapy as a Prognostic Factor and Individual-Level Surrogate for Disease-Free Survival in Rectal Cancer. J. Natl. Cancer Inst. 2017, 109. [Google Scholar] [CrossRef]
- Begg, A.C.; Stewart, F.A.; Vens, C. Strategies to improve radiotherapy with targeted drugs. Nat. Rev. Cancer 2011, 11, 239–253. [Google Scholar] [CrossRef]
- Barker, H.E.; Paget, J.T.E.; Khan, A.A.; Harrington, K.J. The tumour microenvironment after radiotherapy: Mechanisms of resistance and recurrence. Nat. Rev. Cancer 2015, 15, 409–425. [Google Scholar] [CrossRef]
- Buckley, A.M.; Lynam-Lennon, N.; O’Neill, H.; O’Sullivan, J. Targeting hallmarks of cancer to enhance radiosensitivity in gastrointestinal cancers. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 298–313. [Google Scholar] [CrossRef]
- Grade, M.; Wolff, H.A.; Gaedcke, J.; Ghadimi, B.M. The molecular basis of chemoradiosensitivity in rectal cancer: Implications for personalized therapies. Langenbeck’s Arch. Surg. 2012, 397, 543–555. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yard, B.; Chie, E.K.; Adams, D.J.; Peacock, C.; Abazeed, M.E. Radiotherapy in the Era of Precision Medicine. Semin. Radiat. Oncol. 2015, 25, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Gerweck, L.E.; Wakimoto, H. At the Crossroads of Cancer Stem Cells, Radiation Biology, and Radiation Oncology. Cancer Res. 2016, 76, 994–998. [Google Scholar] [CrossRef]
- Garbers, C.; Aparicio-Siegmund, S.; Rose-John, S. The IL-6/gp130/STAT3 signaling axis: Recent advances towards specific inhibition. Curr. Opin. Immunol. 2015, 34, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Huynh, J.; Chand, A.L.; Gough, D.J.; Ernst, M. Therapeutically exploiting STAT3 activity in cancer—Using tissue repair as a road map. Nat. Rev. Cancer 2019, 19, 82–96. [Google Scholar] [CrossRef]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef]
- Yu, H.; Lee, H.; Herrmann, A.; Buettner, R.; Jove, R. Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat. Rev. Cancer 2014, 14, 736–746. [Google Scholar] [CrossRef]
- Spitzner, M.; Roesler, B.; Bielfeld, C.; Emons, G.; Gaedcke, J.; Wolff, H.A.; Rave-Fränk, M.; Kramer, F.; Beissbarth, T.; Kitz, J.; et al. STAT3 inhibition sensitizes colorectal cancer to chemoradiotherapyin vitroandin vivo. Int. J. Cancer 2014, 134, 997–1007. [Google Scholar] [CrossRef]
- Kang, S.; Narazaki, M.; Metwally, H.; Kishimoto, T. Historical overview of the interleukin-6 family cytokine. J. Exp. Med. 2020, 217, e20190347. [Google Scholar] [CrossRef]
- Jones, S.; Jenkins, B.J. Recent insights into targeting the IL-6 cytokine family in inflammatory diseases and cancer. Nat. Rev. Immunol. 2018, 18, 773–789. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.; Goldschmitt, J.; Peschel, C.; Brakenhoff, J.P.G.; Kallen, K.-J.; Wollmer, A.; Grötzinger, J.; Rose-John, S. A bioactive designer cytokine for human hematopoietic progenitor cell expansion. Nat. Biotechnol. 1997, 15, 142–145. [Google Scholar] [CrossRef]
- Li, Y.; Rogoff, H.A.; Keates, S.; Gao, Y.; Murikipudi, S.; Mikule, K.; Leggett, D.; Li, W.; Pardee, A.B.; Li, C.J. Suppression of cancer relapse and metastasis by inhibiting cancer stemness. Proc. Natl. Acad. Sci. USA 2015, 112, 1839–1844. [Google Scholar] [CrossRef] [PubMed]
- Jonker, D.J.; Nott, L.; Yoshino, T.; Gill, S.; Shapiro, J.; Ohtsu, A.; Zalcberg, J.; Vickers, M.M.; Wei, A.C.; Gao, Y.; et al. Napabucasin versus placebo in refractory advanced colorectal cancer: A randomised phase 3 trial. Lancet Gastroenterol. Hepatol. 2018, 3, 263–270. [Google Scholar] [CrossRef]
- Wahl, D.R.; Dresser, J.; Wilder-Romans, K.; Parsels, J.D.; Zhao, S.G.; Davis, M.; Zhao, L.; Kachman, M.; Wernisch, S.; Burant, C.F.; et al. Glioblastoma Therapy Can Be Augmented by Targeting IDH1-Mediated NADPH Biosynthesis. Cancer Res. 2017, 77, 960–970. [Google Scholar] [CrossRef]
- Pennathur, A.; Gibson, M.K.; Jobe, B.; Luketich, J.D. Oesophageal carcinoma. Lancet 2013, 381, 400–412. [Google Scholar] [CrossRef]
- Yoshimura, A.; Naka, T.; Kubo, M. SOCS proteins, cytokine signalling and immune regulation. Nat. Rev. Immunol. 2007, 7, 454–465. [Google Scholar] [CrossRef]
- Kendziorra, E.; Ahlborn, K.; Ebner, R.; Ghadimi, B.; Pukrop, T.; Ried, T.; Grade, M.; Spitzner, M.; Rave-Fränk, M.; Emons, G.; et al. Silencing of the Wnt transcription factor TCF4 sensitizes colorectal cancer cells to (chemo-) radiotherapy. Carcinogenesis 2011, 32, 1824–1831. [Google Scholar] [CrossRef]
- Emons, G.; Spitzner, M.; Reineke, S.; Möller, J.; Auslander, N.; Kramer, F.; Hu, Y.; Beissbarth, T.; Wolff, H.A.; Rave-Fränk, M.; et al. Chemoradiotherapy Resistance in Colorectal Cancer Cells is Mediated by Wnt/β-catenin Signaling. Mol. Cancer Res. 2017, 15, 1481–1490. [Google Scholar] [CrossRef]
- Henricks, L.M.; Lunenburg, C.A.T.C.; De Man, F.M.; Meulendijks, D.; Frederix, G.W.J.; Kienhuis, E.; Creemers, G.-J.; Baars, A.; Dezentjé, V.; Imholz, A.L.T.; et al. DPYD genotype-guided dose individualisation of fluoropyrimidine therapy in patients with cancer: A prospective safety analysis. Lancet Oncol. 2018, 19, 1459–1467. [Google Scholar] [CrossRef]
- Gunda, V.; Souchek, J.; Abrego, J.; Shukla, S.K.; Goode, G.D.; Vernucci, E.; Dasgupta, A.; Chaika, N.V.; King, R.J.; Li, S.; et al. MUC1-Mediated Metabolic Alterations Regulate Response to Radiotherapy in Pancreatic Cancer. Clin. Cancer Res. 2017, 23, 5881–5891. [Google Scholar] [CrossRef] [PubMed]
- Moeller, B.J.; Cao, Y.; Li, C.Y.; Dewhirst, M.W. Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: Role of reoxygenation, free radicals, and stress granules. Cancer Cell 2004, 5, 429–441. [Google Scholar] [CrossRef]
- Kopan, R.; Ilagan, M.X.G. The Canonical Notch Signaling Pathway: Unfolding the Activation Mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef]
- Ntziachristos, P.; Lim, J.S.; Sage, J.; Aifantis, I. From Fly Wings to Targeted Cancer Therapies: A Centennial for Notch Signaling. Cancer Cell 2014, 25, 318–334. [Google Scholar] [CrossRef]
- Decker, T.; Kovarik, P.; Meinke, A. GAS Elements: A Few Nucleotides with a Major Impact on Cytokine-Induced Gene Expression. J. Interf. Cytokine Res. 1997, 17, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Sasse, J.; Hemmann, U.; Schwartz, C.; Schniertshauer, U.; Heesel, B.; Landgraf, C.; Schneider-Mergener, J.; Heinrich, P.C.; Horn, F. Mutational analysis of acute-phase response factor/Stat3 activation and dimerization. Mol. Cell. Biol. 1997, 17, 4677–4686. [Google Scholar] [CrossRef]
- Hurtado, C.; Safarova, A.; Smith, G.; Chung, R.; Bruyneel, A.A.N.; Gomez-Galeno, J.; Oswald, F.; Larson, C.J.; Cashman, J.R.; Ruiz-Lozano, P.; et al. Disruption of NOTCH signaling by a small molecule inhibitor of the transcription factor RBPJ. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Kamakura, S.; Oishi, K.; Yoshimatsu, T.; Nakafuku, M.; Masuyama, N.; Gotoh, Y. Hes binding to STAT3 mediates crosstalk between Notch and JAK–STAT signalling. Nat. Cell Biol. 2004, 6, 547–554. [Google Scholar] [CrossRef]
- Greten, F.R.; Grivennikov, S.I. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity 2019, 51, 27–41. [Google Scholar] [CrossRef]
- Ganesh, K.; Wu, C.; O’Rourke, K.P.; Szeglin, B.C.; Zheng, Y.; Sauve, C.G.; Adileh, M.; Wasserman, I.; Marco, M.R.; Kim, A.S.; et al. A rectal cancer organoid platform to study individual responses to chemoradiation. Nat. Med. 2019, 25, 1607–1614. [Google Scholar] [CrossRef]
- Kersten, K.; De Visser, K.; Van Miltenburg, M.H.; Jonkers, J. Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol. Med. 2016, 9, 137–153. [Google Scholar] [CrossRef]
- Nagaraju, G.P.; Farran, B.; Farren, M.; Chalikonda, G.; Wu, C.; Lesinski, G.B.; El-Rayes, B.F. Napabucasin (BBI 608), a potent chemoradiosensitizer in rectal cancer. Cancer 2020, 126, 3360–3371. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef]
- Ebbing, E.A.; Van Der Zalm, A.P.; Steins, A.; Creemers, A.; Hermsen, S.; Rentenaar, R.; Klein, M.; Waasdorp, C.; Hooijer, G.K.J.; Meijer, S.L.; et al. Stromal-derived interleukin 6 drives epithelial-to-mesenchymal transition and therapy resistance in esophageal adenocarcinoma. Proc. Natl. Acad. Sci. USA 2019, 116, 2237–2242. [Google Scholar] [CrossRef]
- Kawada, M.; Seno, H.; Uenoyama, Y.; Sawabu, T.; Kanda, N.; Fukui, H.; Shimahara, Y.; Chiba, T. Signal Transducers and Activators of Transcription 3 Activation Is Involved in Nuclear Accumulation of β-Catenin in Colorectal Cancer. Cancer Res. 2006, 66, 2913–2917. [Google Scholar] [CrossRef]
- Morikawa, T.; Baba, Y.; Yamauchi, M.; Kuchiba, A.; Nosho, K.; Shima, K.; Tanaka, N.; Huttenhower, C.; Frank, D.A.; Fuchs, C.S.; et al. STAT3 Expression, Molecular Features, Inflammation Patterns, and Prognosis in a Database of 724 Colorectal Cancers. Clin. Cancer Res. 2011, 17, 1452–1462. [Google Scholar] [CrossRef]
- Chen, M.-F.; Chang, Y.-H.; Lin, P.-Y.; Chen, P.-T.; Chen, W.-C.; Lee, K.-D. The role of DNA methyltransferase 3b in esophageal squamous cell carcinoma. Cancer 2011, 118, 4074–4089. [Google Scholar] [CrossRef]
- Zhang, N.; Zhang, M.; Wang, Z.; Gao, W.; Sun, Z.-G. Activated STAT3 Could Reduce Survival in Patients with Esophageal Squamous Cell Carcinoma by Up-regulating VEGF and Cyclin D1 Expression. J. Cancer 2020, 11, 1859–1868. [Google Scholar] [CrossRef]
- Weng, M.T.; Tsao, P.N.; Lin, H.L.; Tung, C.C.; Change, M.C.; Chang, Y.T.; Wong, J.M.; Wei, S.C. Hes1 Increases the Invasion Ability of Colorectal Cancer Cells via the STAT3-MMP14 Pathway. PLoS ONE 2015, 10, e0144322. [Google Scholar] [CrossRef]
- Yang, J.; Xing, H.; Lu, D.; Wang, J.; Li, B.; Tang, J.; Gu, F.; Hong, L. Role of Jagged1/STAT3 signalling in platinum-resistant ovarian cancer. J. Cell. Mol. Med. 2019, 23, 4005–4018. [Google Scholar] [CrossRef]
- Bui, Q.T.; Im, J.H.; Jeong, S.B.; Kim, Y.-M.; Lim, S.C.; Kim, B.; Kang, K.W. Essential role of Notch4/STAT3 signaling in epithelial–mesenchymal transition of tamoxifen-resistant human breast cancer. Cancer Lett. 2017, 390, 115–125. [Google Scholar] [CrossRef]
- Yahyanejad, S.; Theys, J.; Vooijs, M. Targeting Notch to overcome radiation resistance. Oncotarget 2015, 7, 7610–7628. [Google Scholar] [CrossRef]
- Speiser, J.; Foreman, K.; Drinka, E.; Godellas, C.; Perez, C.; Salhadar, A.; Erşahin, Ç.; Rajan, P. Notch-1 and Notch-4 Biomarker Expression in Triple-Negative Breast Cancer. Int. J. Surg. Pathol. 2011, 20, 137–143. [Google Scholar] [CrossRef]
- Jin, M.-M.; Ye, Y.; Qian, Z.-D.; Zhang, Y. Notch signaling molecules as prognostic biomarkers for non-small cell lung cancer. Oncol. Lett. 2015, 10, 3252–3260. [Google Scholar] [CrossRef]
- Hu, J.; Yu, J.; Gan, J.; Song, N.; Shi, L.; Liu, J.; Zhang, Z.; Du, J. Notch1/2/3/4 are prognostic biomarker and correlated with immune infiltrates in gastric cancer. Aging 2020, 12, 2595–2609. [Google Scholar] [CrossRef]
- Marijnen, C.A.M. Organ preservation in rectal cancer: Have all questions been answered? Lancet Oncol. 2015, 16, e13–e22. [Google Scholar] [CrossRef]
- Van Der Valk, M.J.M.; Hilling, D.; Bastiaannet, E.; Kranenbarg, E.M.-K.; Beets, G.L.; Figueiredo, N.L.; Habr-Gama, A.; Perez, R.; Renehan, A.G.; Van De Velde, C.J.H.; et al. Long-term outcomes of clinical complete responders after neoadjuvant treatment for rectal cancer in the International Watch & Wait Database (IWWD): An international multicentre registry study. Lancet 2018, 391, 2537–2545. [Google Scholar] [CrossRef]
- Smith, J.J.; Strombom, P.; Chow, O.S.; Roxburgh, C.S.; Lynn, P.; Eaton, A.; Widmar, M.; Ganesh, K.; Yaeger, R.; Cercek, A.; et al. Assessment of a Watch-and-Wait Strategy for Rectal Cancer in Patients with a Complete Response after Neoadjuvant Therapy. JAMA Oncol. 2019, 5, e185896. [Google Scholar] [CrossRef]
- Smith, J.J.; Paty, P.B.; Garcia-Aguilar, J. Watch and Wait in Rectal Cancer or More Wait and See? JAMA Surg. 2020, 155, 657. [Google Scholar] [CrossRef]
- Shimada, Y.; Imamura, M.; Wagata, T.; Yamaguchi, N.; Tobe, T. Characterization of 21 newly established esophageal cancer cell lines. Cancer 1992, 69, 277–284. [Google Scholar] [CrossRef]
- Franken, N.A.P.; Rodermond, H.M.; Stap, J.; Haveman, J.; Van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef] [PubMed]
- Petersen, J.; Staab, J.; Bader, O.; Buhl, T.; Ivetic, A.; Meyer, T. Identification of a distinct subset of disease-associated gain-of-function missense mutations in the STAT1 coiled-coil domain as system mutants. Mol. Immunol. 2019, 114, 30–40. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koerdel, K.; Spitzner, M.; Meyer, T.; Engels, N.; Krause, F.; Gaedcke, J.; Conradi, L.-C.; Haubrock, M.; Beißbarth, T.; Leha, A.; et al. NOTCH Activation via gp130/STAT3 Signaling Confers Resistance to Chemoradiotherapy. Cancers 2021, 13, 455. https://doi.org/10.3390/cancers13030455
Koerdel K, Spitzner M, Meyer T, Engels N, Krause F, Gaedcke J, Conradi L-C, Haubrock M, Beißbarth T, Leha A, et al. NOTCH Activation via gp130/STAT3 Signaling Confers Resistance to Chemoradiotherapy. Cancers. 2021; 13(3):455. https://doi.org/10.3390/cancers13030455
Chicago/Turabian StyleKoerdel, Kristin, Melanie Spitzner, Thomas Meyer, Niklas Engels, Florian Krause, Jochen Gaedcke, Lena-Christin Conradi, Martin Haubrock, Tim Beißbarth, Andreas Leha, and et al. 2021. "NOTCH Activation via gp130/STAT3 Signaling Confers Resistance to Chemoradiotherapy" Cancers 13, no. 3: 455. https://doi.org/10.3390/cancers13030455
APA StyleKoerdel, K., Spitzner, M., Meyer, T., Engels, N., Krause, F., Gaedcke, J., Conradi, L.-C., Haubrock, M., Beißbarth, T., Leha, A., Johnsen, S. A., Ghadimi, B. M., Rose-John, S., Grade, M., & Wienands, J. (2021). NOTCH Activation via gp130/STAT3 Signaling Confers Resistance to Chemoradiotherapy. Cancers, 13(3), 455. https://doi.org/10.3390/cancers13030455






