Subcellular Localization of uc.8+ as a Prognostic Biomarker in Bladder Cancer Tissue
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
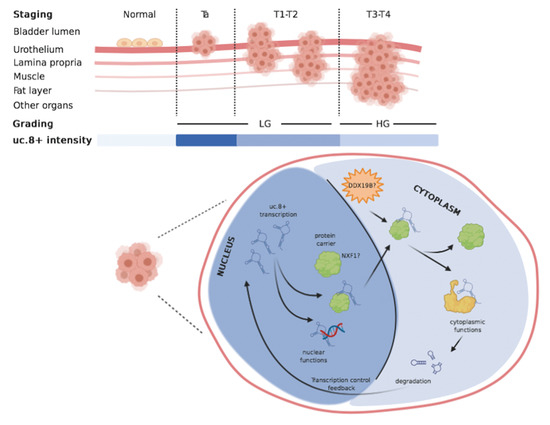
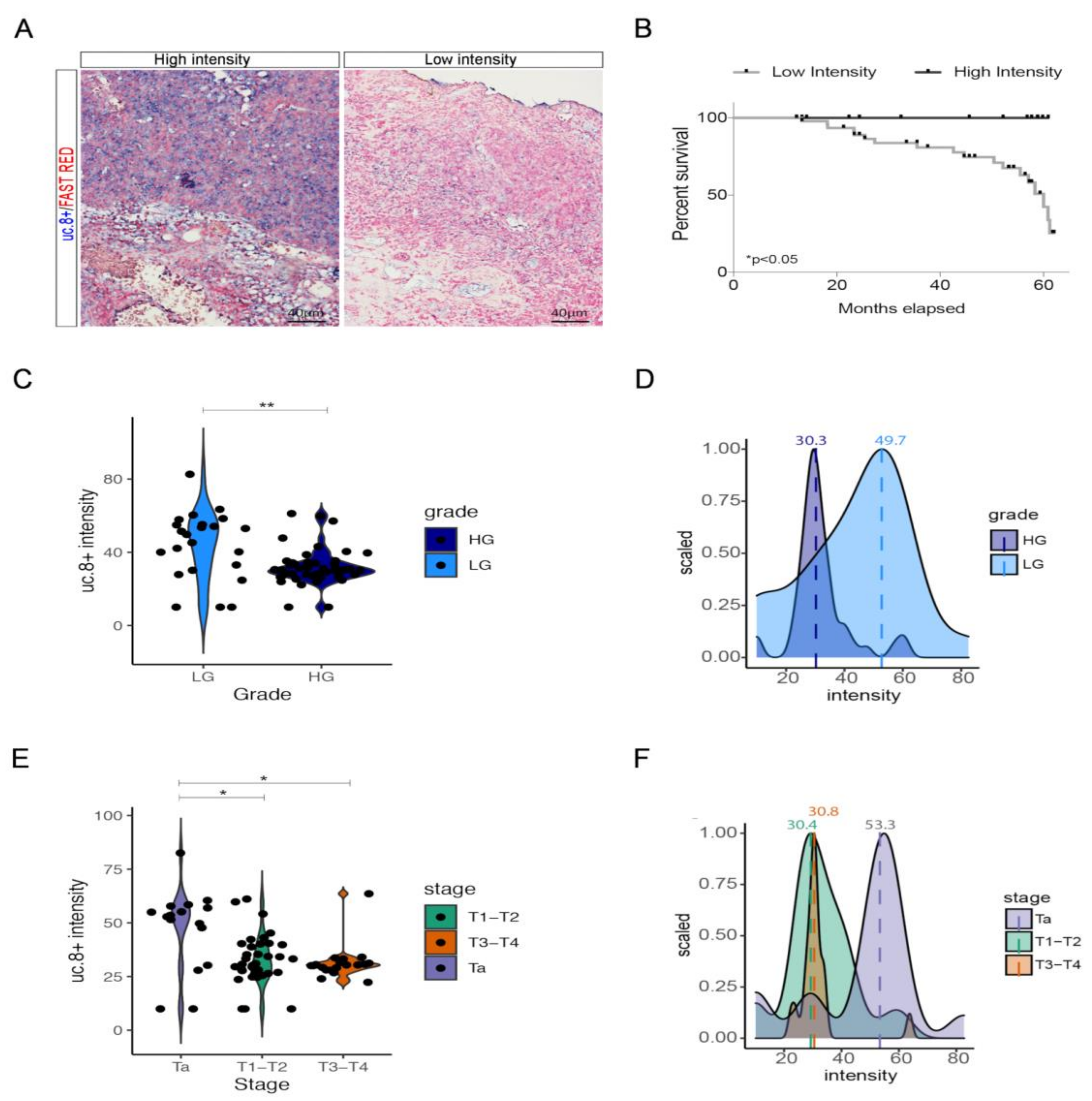
2.1. Association of uc.8+ Expression with Its Clinic-Pathologic Features and Survival of BlCa Patients
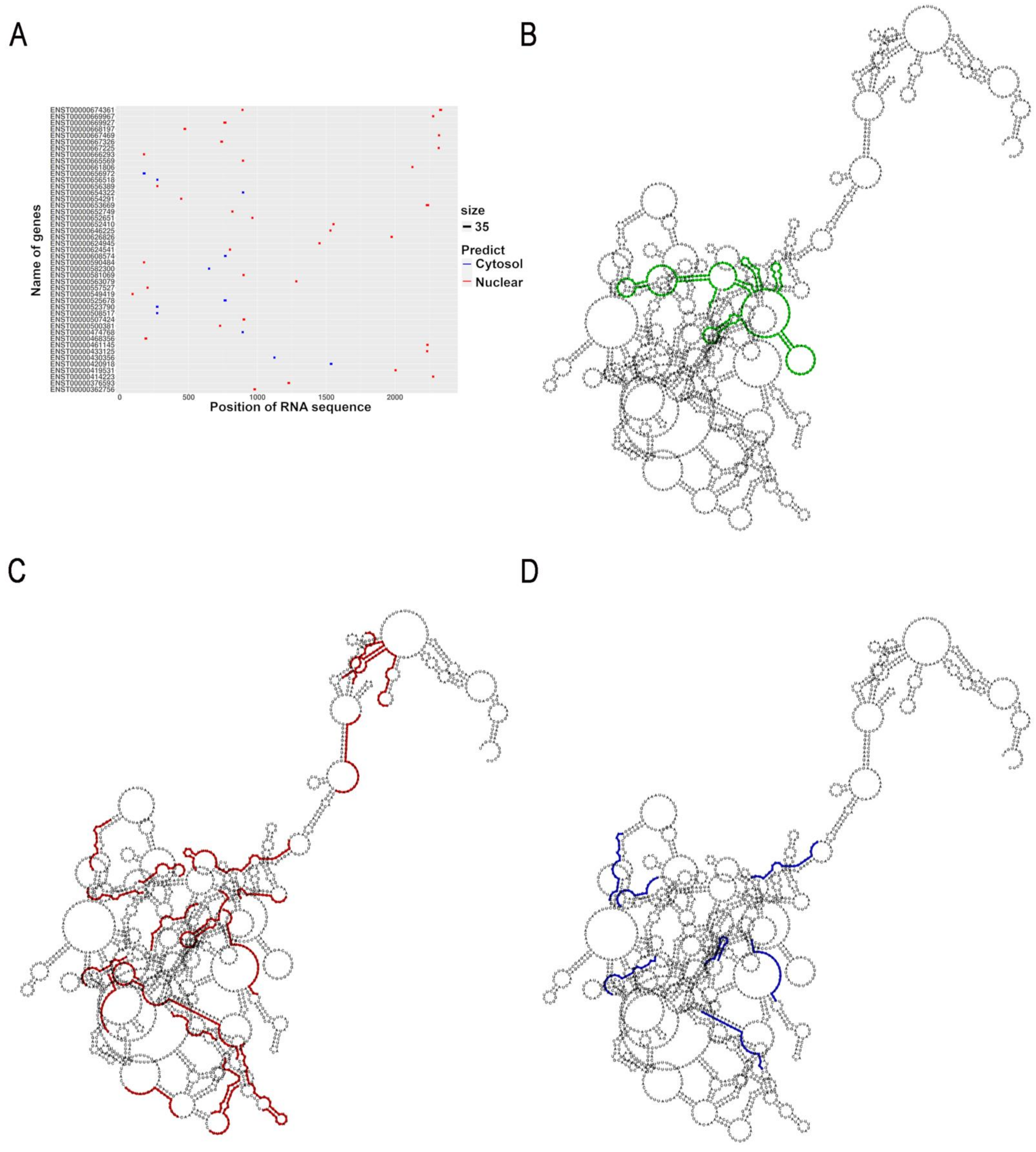
2.2. In Silico Analysis Indicates That uc.8+ Has Both Nuclear and Cytoplasmic Binding Partners
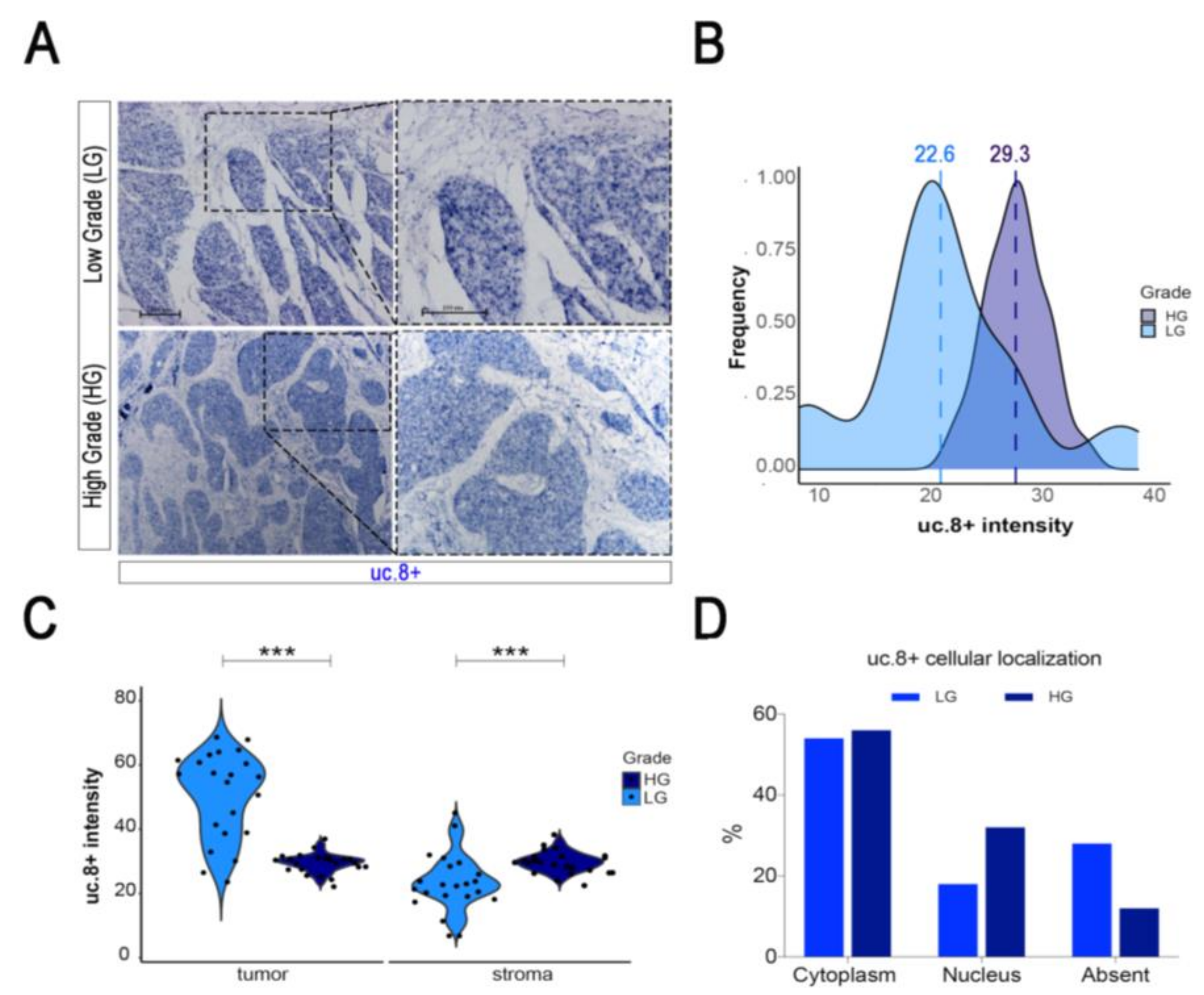
2.3. uc.8+ Expression and Subcellular Localization in BlCa Samples and Cell Lines
2.4. uc.8+ Localization in Bladder Cancer Cell Line
2.5. uc.8+ Expression and Histological Localization in Tumor-Surrounding Stroma
2.6. Prediction of uc.8+ Interactions in Bladder Cancer
2.7. DDX19B and NXF1 Involvement into Nuclear Export of uc.8+
3. Discussion
4. Materials and Methods
4.1. Clinical Characteristics of Bladder Cancer Patients
4.2. Ethics Statement
4.3. Tissue Microarray Building
4.4. In Situ RNA Hybridization on Paraffin Sections
4.5. Image Processing and Analysis
4.6. Transfection of siRNAs against uc.8+ in BlCa Cells
4.7. Cellular Fractionation
4.8. Nuclear Fractionation
4.9. Gene Expression Analysis
- XIST-F—5′TGATCCCATTGAAGATACCACGCTG3′;
- XIST-R—5′TGGCAACCCATCCAAGTAGATTAGC3′;
- rRNA-F—5′ACTGCTCGCCAGAACACTACGA3′;
- rRNA-R—5′GTCTTTACGTGGGTACTTGCGCT3′;
- snRNA-F—5′TCCCAGGGCGAGGCTTATCCATTG3′;
- snRNA-R—5′GCGAACGCAGTCCCCCACTACCAC3′;
- GAPDH-F—5′GTCAAGGCTGAGAACGGGAAGCT3′;
- GAPDH-R—5′GCCTTCTCCATGGTGGTGAAGA3′;
- uc.8+-F—5′-GGTCGCCATGGATATGACA3′;
- uc.8+-R—5′CACTGTGGCTTTAAACTCAGGA3′;
- Ddx19B-F—5′CGTCCATCCAAGATACAAGAGA3′;
- Ddx19B-R—5′ TTGGGCAATTAAGTTCTGTGG3′;
- NXF1-F—5′ATTTGGATCCATGGCGGACGAGGGGAAGT3′;
- NXF1-R—5′AATATGCGGCCGCTCACTTCATGAATGCCACTTCTGG3′.
4.10. LncRNA Subcellular Localization with Deep Learning
4.11. In Silico Data Analysis of Gene Expression and Protein-Ligand Interactions
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyazaki, J.; Nishiyama, H. Epidemiology of Urothelial Carcinoma. Int. J. Urol. 2017, 24, 730–734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witjes, J.A.; Lebret, T.; Compérat, E.M.; Cowan, N.C.; de Santis, M.; Bruins, H.M.; Hernández, V.; Espinós, E.L.; Dunn, J.; Rouanne, M.; et al. Updated 2016 EAU Guidelines on Muscle-Invasive and Metastatic Bladder Cancer. Eur. Urol. 2017, 71, 462–475. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Zhang, Y.; Li, H.; Ma, W. LncRNAs Act as Prognostic Biomarkers in Bladder Carcinoma: A Meta-Analysis. Heliyon 2019, 5, e02785. [Google Scholar] [CrossRef] [PubMed]
- Ilijazi, D.; Abufaraj, M.; Hassler, M.R.; Ertl, I.E.; D’Andrea, D.; Shariat, S.F. Waiting in the Wings: The Emerging Role of Molecular Biomarkers in Bladder Cancer. Expert Rev. Mol. Diagn. 2018, 18, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Katzman, S.; Kern, A.D.; Bejerano, G.; Fewell, G.; Fulton, L.; Wilson, R.K.; Salama, S.R.; Haussler, D. Human Genome Ultraconserved Elements Are Ultraselected. Science 2007, 317, 915. [Google Scholar] [CrossRef] [Green Version]
- Reneker, J.; Lyons, E.; Conant, G.C.; Pires, J.C.; Freeling, M.; Shyu, C.-R.; Korkin, D. Long Identical Multispecies Elements in Plant and Animal Genomes. Proc. Natl. Acad. Sci. USA 2012, 109, E1183–E1191. [Google Scholar] [CrossRef] [Green Version]
- Bejerano, G.; Pheasant, M.; Makunin, I.; Stephen, S.; Kent, W.J.; Mattick, J.S.; Haussler, D. Ultraconserved Elements in the Human Genome. Science 2004, 304, 1321. [Google Scholar] [CrossRef] [Green Version]
- McCole, R.B.; Fonseka, C.Y.; Koren, A.; Wu, C. Abnormal Dosage of Ultraconserved Elements Is Highly Disfavored in Healthy Cells but Not Cancer Cells. PLoS Genet. 2014, 10, e1004646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira Zambalde, E.; Mathias, C.; Rodrigues, A.C.; de Souza Fonseca Ribeiro, E.M.; Fiori Gradia, D.; Calin, G.A.; Carvalho de Oliveira, J. Highlighting Transcribed Ultraconserved Regions in Human Diseases. Wiley Interdiscip. Rev. RNA 2020, 11, e1567. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-L. Linking Long Noncoding RNA Localization and Function. Trends Biochem. Sci. 2016, 41, 761–772. [Google Scholar] [CrossRef]
- Ulitsky, I.; Bartel, D.P. LincRNAs: Genomics, Evolution, and Mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef] [Green Version]
- Terreri, S.; Durso, M.; Colonna, V.; Romanelli, A.; Terracciano, D.; Ferro, M.; Perdonà, S.; Castaldo, L.; Febbraio, F.; de Nigris, F.; et al. New Cross-Talk Layer between Ultraconserved Non-Coding RNAs, MicroRNAs and Polycomb Protein YY1 in Bladder Cancer. Genes 2016, 7, 127. [Google Scholar] [CrossRef]
- Olivieri, M.; Ferro, M.; Terreri, S.; Durso, M.; Romanelli, A.; Avitabile, C.; de Cobelli, O.; Messere, A.; Bruzzese, D.; Vannini, I.; et al. Long Non-Coding RNA Containing Ultraconserved Genomic Region 8 Promotes Bladder Cancer Tumorigenesis. Oncotarget 2016, 7, 20636–20654. [Google Scholar] [CrossRef]
- Gudenas, B.L.; Wang, L. Prediction of LncRNA Subcellular Localization with Deep Learning from Sequence Features. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmitt, C.; von Kobbe, C.; Bachi, A.; Panté, N.; Rodrigues, J.P.; Boscheron, C.; Rigaut, G.; Wilm, M.; Séraphin, B.; Carmo-Fonseca, M.; et al. Dbp5, a DEAD-Box Protein Required for MRNA Export, Is Recruited to the Cytoplasmic Fibrils of Nuclear Pore Complex via a Conserved Interaction with CAN/Nup159p. EMBO J. 1999, 18, 4332–4347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cobianchi, F.; Biamonti, G.; Maconi, M.; Riva, S. Human HnRNP Protein A1: A Model Polypeptide for a Structural and Genetic Investigation of a Broad Family of RNA Binding Proteins. Genetica 1994, 94, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Williams, K.R.; Szer, W. Purification and Domain Structure of Core HnRNP Proteins A1 and A2 and Their Relationship to Single-Stranded DNA-Binding Proteins. J. Biol. Chem. 1986, 261, 11266–11273. [Google Scholar] [CrossRef]
- Mayeda, A.; Helfman, D.M.; Krainer, A.R. Modulation of Exon Skipping and Inclusion by Heterogeneous Nuclear Ribonucleoprotein A1 and Pre-MRNA Splicing Factor SF2/ASF. Mol. Cell Biol. 1993, 13, 2993–3001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herold, A.; Suyama, M.; Rodrigues, J.P.; Braun, I.C.; Kutay, U.; Carmo-Fonseca, M.; Bork, P.; Izaurralde, E. TAP (NXF1) Belongs to a Multigene Family of Putative RNA Export Factors with a Conserved Modular Architecture. Mol. Cell Biol. 2000, 20, 8996–9008. [Google Scholar] [CrossRef] [Green Version]
- Kang, Y.; Bogerd, H.P.; Yang, J.; Cullen, B.R. Analysis of the RNA Binding Specificity of the Human Tap Protein, a Constitutive Transport Element-Specific Nuclear RNA Export Factor. Virology 1999, 262, 200–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben-Yishay, R.; Mor, A.; Shraga, A.; Ashkenazy-Titelman, A.; Kinor, N.; Schwed-Gross, A.; Jacob, A.; Kozer, N.; Kumar, P.; Garini, Y.; et al. Imaging within Single NPCs Reveals NXF1’s Role in MRNA Export on the Cytoplasmic Side of the Pore. J. Cell Biol. 2019, 218, 2962–2981. [Google Scholar] [CrossRef] [Green Version]
- Calin, G.A.; Liu, C.; Ferracin, M.; Hyslop, T.; Spizzo, R.; Sevignani, C.; Fabbri, M.; Cimmino, A.; Lee, E.J.; Wojcik, S.E.; et al. Ultraconserved Regions Encoding NcRNAs Are Altered in Human Leukemias and Carcinomas. Cancer Cell 2007, 12, 215–229. [Google Scholar] [CrossRef]
- Terracciano, D.; Ferro, M.; Terreri, S.; Lucarelli, G.; D’Elia, C.; Musi, G.; de Cobelli, O.; Mirone, V.; Cimmino, A. Urinary Long Noncoding RNAs in Nonmuscle-Invasive Bladder Cancer: New Architects in Cancer Prognostic Biomarkers. Transl. Res. 2017, 184, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Cabili, M.N.; Dunagin, M.C.; McClanahan, P.D.; Biaesch, A.; Padovan-Merhar, O.; Regev, A.; Rinn, J.L.; Raj, A. Localization and Abundance Analysis of Human LncRNAs at Single-Cell and Single-Molecule Resolution. Genome Biol. 2015, 16, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rueden, C.T.; Eliceiri, K.W. Visualization Approaches for Multidimensional Biological Image Data. BioTechniques 2007, 43, S31–S36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Werner, M.S.; Ruthenburg, A.J. Nuclear Fractionation Reveals Thousands of Chromatin-Tethered Noncoding RNAs Adjacent to Active Genes. Cell Rep. 2015, 12, 1089–1098. [Google Scholar] [CrossRef] [Green Version]
- Pandya-Jones, A.; Black, D.L. Co-Transcriptional Splicing of Constitutive and Alternative Exons. RNA 2009, 15, 1896–1908. [Google Scholar] [CrossRef] [Green Version]
- Wuarin, J.; Schibler, U. Physical Isolation of Nascent RNA Chains Transcribed by RNA Polymerase II: Evidence for Cotranscriptional Splicing. Mol. Cell Biol. 1994, 14, 7219–7225. [Google Scholar] [CrossRef] [Green Version]
- Heaton, J. Ian Goodfellow, Yoshua Bengio, and Aaron Courville: Deep Learning. Genet. Program. Evolvable Mach. 2018, 19, 305–307. [Google Scholar] [CrossRef] [Green Version]
- Giudice, G.; Sánchez-Cabo, F.; Torroja, C.; Lara-Pezzi, E. ATtRACT-a Database of RNA-Binding Proteins and Associated Motifs. Database 2016, 2016. [Google Scholar] [CrossRef] [Green Version]
- Krüger, J.; Rehmsmeier, M. RNAhybrid: MicroRNA Target Prediction Easy, Fast and Flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, R.; Bernhart, S.H.; Zu Siederdissen, C.H.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef]
- Grossman, R.L.; Heath, A.P.; Ferretti, V.; Varmus, H.E.; Lowy, D.R.; Kibbe, W.A.; Staudt, L.M. Toward a Shared Vision for Cancer Genomic Data. N. Engl. J. Med. 2016, 375, 1109–1112. [Google Scholar] [CrossRef]
- Huber, W.; Carey, V.J.; Gentleman, R.; Anders, S.; Carlson, M.; Carvalho, B.S.; Bravo, H.C.; Davis, S.; Gatto, L.; Girke, T.; et al. Orchestrating High-Throughput Genomic Analysis with Bioconductor. Nat. Methods 2015, 12, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Mounir, M.; Lucchetta, M.; Silva, T.C.; Olsen, C.; Bontempi, G.; Chen, X.; Noushmehr, H.; Colaprico, A.; Papaleo, E. New Functionalities in the TCGAbiolinks Package for the Study and Integration of Cancer Data from GDC and GTEx. PLoS Comput. Biol. 2019, 15, e1006701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgan, M.; Obenchain, V.; Hester, J.; Pagès, H. SummarizedExperiment: SummarizedExperiment Container. Bioconductor version: Release (3.11). Bioconductor Package Maintainer. 2020. Available online: http://bioconductor.riken.jp/packages/3.6/bioc/html/ExperimentHub.html (accessed on 3 February 2021).
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential Expression Analysis of Multifactor RNA-Seq Experiments with Respect to Biological Variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef] [Green Version]
- Kassambara, A. Ggpubr: “ggplot2” Based Publication Ready Plots. R Package Version 0.2.4. GitHub. 2019. Available online: https://github.com/ (accessed on 3 February 2021).
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]



| All Patients | Intensity-Score Subgroups | ||||
|---|---|---|---|---|---|
| n = 73 | Low Intensity Score (<35) n = 46 | High Intensity Score (≥35) n = 27 | Chi-Square | p-Value | |
| Age at diagnosis | 0.440 | 0.735 | |||
| <60 years | 17 | 11 | 6 | ||
| ≥60 years | 56 | 35 | 21 | ||
| Sex | 0.006 | 0.936 | |||
| Male | 61 | 37 | 24 | ||
| Female | 12 | 9 | 3 | ||
| Grade | 16.394 | 5.14 × 10−5 | |||
| High | 50 | 39 | 11 | ||
| Low | 23 | 7 | 16 | ||
| Stage | 18.34 | 1.04 × 10−4 | |||
| Ta | 16 | 4 | 12 | ||
| T1–T2 | 37 | 24 | 13 | ||
| T3–T4 | 20 | 18 | 2 | ||
| Metastasis | 8.967 | 2.75 × 10−3 | |||
| absent | 59 | 33 | 26 | ||
| present | 14 | 13 | 1 | ||
| Therapy | 10.084 | 0.007 | |||
| no | 51 | 27 | 24 | ||
| yes | 22 | 19 | 3 | ||
| Localization | 3.582 | 0.058 | |||
| cytoplasm | 29 | 21 | 8 | ||
| nucleus | 26 | 15 | 11 | ||
| nucleus-cytoplasm | 18 | 7 | 11 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Terreri, S.; Mancinelli, S.; Ferro, M.; Vitale, M.C.; Perdonà, S.; Castaldo, L.; Gigantino, V.; Mercadante, V.; De Cecio, R.; Aquino, G.; et al. Subcellular Localization of uc.8+ as a Prognostic Biomarker in Bladder Cancer Tissue. Cancers 2021, 13, 681. https://doi.org/10.3390/cancers13040681
Terreri S, Mancinelli S, Ferro M, Vitale MC, Perdonà S, Castaldo L, Gigantino V, Mercadante V, De Cecio R, Aquino G, et al. Subcellular Localization of uc.8+ as a Prognostic Biomarker in Bladder Cancer Tissue. Cancers. 2021; 13(4):681. https://doi.org/10.3390/cancers13040681
Chicago/Turabian StyleTerreri, Sara, Sara Mancinelli, Matteo Ferro, Maria Concetta Vitale, Sisto Perdonà, Luigi Castaldo, Vincenzo Gigantino, Vincenzo Mercadante, Rossella De Cecio, Gabriella Aquino, and et al. 2021. "Subcellular Localization of uc.8+ as a Prognostic Biomarker in Bladder Cancer Tissue" Cancers 13, no. 4: 681. https://doi.org/10.3390/cancers13040681
APA StyleTerreri, S., Mancinelli, S., Ferro, M., Vitale, M. C., Perdonà, S., Castaldo, L., Gigantino, V., Mercadante, V., De Cecio, R., Aquino, G., Montella, M., Angelini, C., Del Prete, E., Aprile, M., Ciaramella, A., Liguori, G. L., Costa, V., Calin, G. A., La Civita, E., ... Cimmino, A. (2021). Subcellular Localization of uc.8+ as a Prognostic Biomarker in Bladder Cancer Tissue. Cancers, 13(4), 681. https://doi.org/10.3390/cancers13040681