Clinicopathological Correlates of γδ T Cell Infiltration in Triple-Negative Breast Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
2.1. Patient and Tumor Characteristics
2.2. Association of Tumor-Infiltrating Lymphocytes with TNBC Biological Features
2.3. In Situ γδ T Cell Infiltration Analysis
2.4. Association of In Situ γδ T Cell Infiltration with Clinicopathological Features
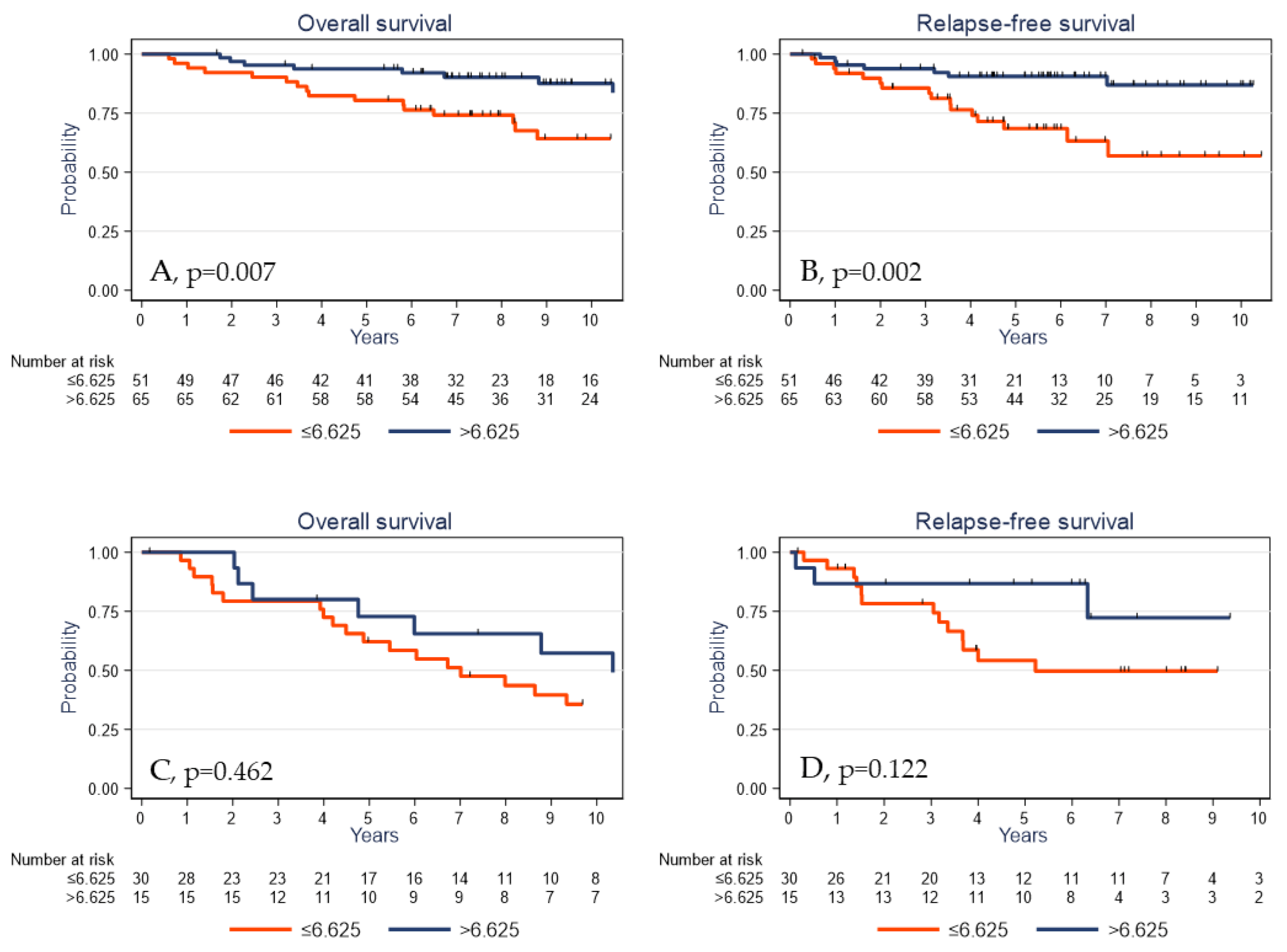
2.5. Survival Analyses
3. Discussion
4. Materials and Methods
4.1. Objectives
4.2. Patients and Tumor Samples
4.3. Tissue Microarray
4.4. Immunohistochemistry
4.5. TIL Assessment
4.6. Tissue Processing and DNA Extraction
4.7. Molecular Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AR | Androgen receptor |
| BC | Breast cancer |
| BRCA1 | Breast cancer type 1 susceptibility gene |
| CK 5/6 | Cytokeratin 5/6 |
| DC | Dendritic cells |
| DNA | Deoxyribonucleic acid |
| EGFR | Epidermal growth factor receptor |
| ER | Estrogen receptor |
| FOXA1 | Forkhead box protein A1 |
| HES | Hematoxylin-Eosin-Saffron |
| HER2 | Human epidermal growth factor receptor 2 |
| HR | Hazard ratio |
| IHC | Immunohistochemistry |
| NK | Natural killer |
| OS | Overall survival |
| PCR | Polymerase chain reaction |
| PD-1 | Programmed cell death 1 |
| PD-L1 | Programmed cell death ligand 1 |
| PR | Progesterone receptor |
| REMARK | Reporting recommendations for tumor marker prognostic studies |
| RFS | Relapse-free survival |
| SC | Stromal cells |
| TC | Tumor cells |
| TCR | T cell receptor |
| TIL | Tumor infiltrating lymphocyte |
| TMA | Tissue microarray |
| TNBC | Triple-negative breast cancer |
References
- Dent, R.; Hanna, W.M.; Trudeau, M.; Rawlinson, E.; Sun, P.; Narod, S.A. Pattern of metastatic spread in triple-negative breast cancer. Breast Cancer Res. Treat. 2009, 115, 423–428. [Google Scholar] [CrossRef]
- Dent, R.; Trudeau, M.; Pritchard, K.I.; Hanna, W.M.; Kahn, H.K.; Sawka, C.A.; Lickley, L.A.; Rawlinson, E.; Sun, P.; Narod, S.A. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin. Cancer Res. 2007, 13, 4429–4434. [Google Scholar] [CrossRef] [Green Version]
- Carey, L.A.; Dees, E.C.; Sawyer, L.; Gatti, L.; Moore, D.T.; Collichio, F.; Ollila, D.W.; Sartor, C.I.; Graham, M.L.; Perou, C.M. The triple negative paradox: Primary tumor chemosensitivity of breast cancer subtypes. Clin. Cancer Res. 2007, 13, 2329–2334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prat, A.; Adamo, B.; Cheang, M.C.; Anders, C.K.; Carey, L.A.; Perou, C.M. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist 2013, 18, 123–133. [Google Scholar] [CrossRef] [Green Version]
- Cheang, M.C.; Voduc, D.; Bajdik, C.; Leung, S.; McKinney, S.; Chia, S.K.; Perou, C.M.; Nielsen, T.O. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin. Cancer Res. 2008, 14, 1368–1376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef] [Green Version]
- Masuda, H.; Baggerly, K.A.; Wang, Y.; Zhang, Y.; Gonzalez-Angulo, A.M.; Meric-Bernstam, F.; Valero, V.; Lehmann, B.D.; Pietenpol, J.A.; Hortobagyi, G.N.; et al. Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clin. Cancer. Res. 2013, 19, 5533–5540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehmann, B.D.; Jovanovic, B.; Chen, X.; Estrada, M.V.; Johnson, K.N.; Shyr, Y.; Moses, H.L.; Sanders, M.E.; Pietenpol, J.A. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS ONE 2016, 11, e0157368. [Google Scholar] [CrossRef]
- Ellis, H.; Ma, C.X. PI3K Inhibitors in Breast Cancer Therapy. Curr. Oncol. Rep. 2019, 21, 110. [Google Scholar] [CrossRef]
- Sobral-Leite, M.; Salomon, I.; Opdam, M.; Kruger, D.T.; Beelen, K.J.; van der Noort, V.; van Vlierberghe, R.L.P.; Blok, E.J.; Giardiello, D.; Sanders, J.; et al. Cancer-immune interactions in ER-positive breast cancers: PI3K pathway alterations and tumor-infiltrating lymphocytes. Breast Cancer Res. 2019, 21, 90. [Google Scholar] [CrossRef] [Green Version]
- An, Y.; Adams, J.R.; Hollern, D.P.; Zhao, A.; Chang, S.G.; Gams, M.S.; Chung, P.E.D.; He, X.; Jangra, R.; Shah, J.S.; et al. Cdh1 and Pik3ca Mutations Cooperate to Induce Immune-Related Invasive Lobular Carcinoma of the Breast. Cell Rep. 2018, 25, 702–714.e706. [Google Scholar] [CrossRef] [Green Version]
- Jacot, W.; Mollevi, C.; Fina, F.; Lopez-Crapez, E.; Martin, P.M.; Colombo, P.E.; Bibeau, F.; Romieu, G.; Lamy, P.J. High EGFR protein expression and exon 9 PIK3CA mutations are independent prognostic factors in triple negative breast cancers. BMC Cancer 2015, 15, 986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narang, P.; Chen, M.; Sharma, A.A.; Anderson, K.S.; Wilson, M.A. The neoepitope landscape of breast cancer: Implications for immunotherapy. BMC Cancer 2019, 19, 200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibrahim, E.M.; Al-Foheidi, M.E.; Al-Mansour, M.M.; Kazkaz, G.A. The prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancer: A meta-analysis. Breast Cancer Res. Treat. 2014, 148, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.; Gray, R.J.; Demaria, S.; Goldstein, L.; Perez, E.A.; Shulman, L.N.; Martino, S.; Wang, M.; Jones, V.E.; Saphner, T.J.; et al. Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J. Clin. Oncol. 2014, 32, 2959–2966. [Google Scholar] [CrossRef]
- Gao, G.; Wang, Z.; Qu, X.; Zhang, Z. Prognostic value of tumor-infiltrating lymphocytes in patients with triple-negative breast cancer: A systematic review and meta-analysis. BMC Cancer 2020, 20, 179. [Google Scholar] [CrossRef] [Green Version]
- Pruneri, G.; Vingiani, A.; Bagnardi, V.; Rotmensz, N.; De Rose, A.; Palazzo, A.; Colleoni, A.M.; Goldhirsch, A.; Viale, G. Clinical validity of tumor-infiltrating lymphocytes analysis in patients with triple-negative breast cancer. Ann. Oncol. 2016, 27, 249–256. [Google Scholar] [CrossRef]
- Salgado, R.; Denkert, C.; Demaria, S.; Sirtaine, N.; Klauschen, F.; Pruneri, G.; Wienert, S.; Van den Eynden, G.; Baehner, F.L.; Penault-Llorca, F.; et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: Recommendations by an International TILs Working Group 2014. Ann. Oncol. 2015, 26, 259–271. [Google Scholar] [CrossRef]
- Stovgaard, E.S.; Nielsen, D.; Hogdall, E.; Balslev, E. Triple negative breast cancer-prognostic role of immune-related factors: A systematic review. Acta Oncol. 2018, 57, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Harano, K.; Wang, Y.; Lim, B.; Seitz, R.S.; Morris, S.W.; Bailey, D.B.; Hout, D.R.; Skelton, R.L.; Ring, B.Z.; Masuda, H.; et al. Rates of immune cell infiltration in patients with triple-negative breast cancer by molecular subtype. PLoS ONE 2018, 13, e0204513. [Google Scholar] [CrossRef] [Green Version]
- Bottai, G.; Raschioni, C.; Losurdo, A.; Di Tommaso, L.; Tinterri, C.; Torrisi, R.; Reis-Filho, J.S.; Roncalli, M.; Sotiriou, C.; Santoro, A.; et al. An immune stratification reveals a subset of PD-1/LAG-3 double-positive triple-negative breast cancers. Breast Cancer Res. 2016, 18, 121. [Google Scholar] [CrossRef] [Green Version]
- Dieci, M.V.; Tsvetkova, V.; Griguolo, G.; Miglietta, F.; Tasca, G.; Giorgi, C.A.; Cumerlato, E.; Massa, D.; Lo Mele, M.; Orvieto, E.; et al. Integration of tumour infiltrating lymphocytes, programmed cell-death ligand-1, CD8 and FOXP3 in prognostic models for triple-negative breast cancer: Analysis of 244 stage I-III patients treated with standard therapy. Eur. J. Cancer 2020, 136, 7–15. [Google Scholar] [CrossRef]
- Wu, Y.; Kyle-Cezar, F.; Woolf, R.T.; Naceur-Lombardelli, C.; Owen, J.; Biswas, D.; Lorenc, A.; Vantourout, P.; Gazinska, P.; Grigoriadis, A.; et al. An innate-like Vdelta1(+) gammadelta T cell compartment in the human breast is associated with remission in triple-negative breast cancer. Sci. Transl. Med. 2019, 11, eaax9364. [Google Scholar] [CrossRef]
- Bouet-Toussaint, F.; Cabillic, F.; Toutirais, O.; Le Gallo, M.; de la Pintiere, C.T.; Daniel, P.; Genetet, N.; Meunier, B.; Dupont-Bierre, E.; Boudjema, K.; et al. Vgamma9Vdelta2 T cell-mediated recognition of human solid tumors. Potential for immunotherapy of hepatocellular and colorectal carcinomas. Cancer Immunol. Immunother. 2008, 57, 531–539. [Google Scholar] [CrossRef]
- Cordova, A.; Toia, F.; La Mendola, C.; Orlando, V.; Meraviglia, S.; Rinaldi, G.; Todaro, M.; Cicero, G.; Zichichi, L.; Donni, P.L.; et al. Characterization of human gammadelta T lymphocytes infiltrating primary malignant melanomas. PLoS ONE 2012, 7, e49878. [Google Scholar] [CrossRef] [Green Version]
- Corvaisier, M.; Moreau-Aubry, A.; Diez, E.; Bennouna, J.; Mosnier, J.F.; Scotet, E.; Bonneville, M.; Jotereau, F. V gamma 9V delta 2 T cell response to colon carcinoma cells. J. Immunol. 2005, 175, 5481–5488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meraviglia, S.; Eberl, M.; Vermijlen, D.; Todaro, M.; Buccheri, S.; Cicero, G.; La Mendola, C.; Guggino, G.; D’Asaro, M.; Orlando, V.; et al. In vivo manipulation of Vgamma9Vdelta2 T cells with zoledronate and low-dose interleukin-2 for immunotherapy of advanced breast cancer patients. Clin. Exp. Immunol. 2010, 161, 290–297. [Google Scholar]
- Raspollini, M.R.; Castiglione, F.; Degl’innocenti, D.R.; Amunni, G.; Villanucci, A.; Garbini, F.; Baroni, G.; Taddei, G.L. Tumour-infiltrating gamma/delta T-lymphocytes are correlated with a brief disease-free interval in advanced ovarian serous carcinoma. Ann. Oncol. 2005, 16, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Bonneville, M.; O’Brien, R.L.; Born, W.K. Gammadelta T cell effector functions: A blend of innate programming and acquired plasticity. Nat. Rev. Immunol. 2010, 10, 467–478. [Google Scholar] [CrossRef]
- Hidalgo, J.V.; Bronsert, P.; Orlowska-Volk, M.; Diaz, L.B.; Stickeler, E.; Werner, M.; Schmitt-Graeff, A.; Kayser, G.; Malkovsky, M.; Fisch, P. Histological Analysis of gammadelta T Lymphocytes Infiltrating Human Triple-Negative Breast Carcinomas. Front. Immunol. 2014, 5, 632. [Google Scholar] [CrossRef] [Green Version]
- Siegers, G.M.; Dutta, I.; Kang, E.Y.; Huang, J.; Kobel, M.; Postovit, L.M. Aberrantly Expressed Embryonic Protein NODAL Alters Breast Cancer Cell Susceptibility to gammadelta T Cell Cytotoxicity. Front. Immunol. 2020, 11, 1287. [Google Scholar] [CrossRef] [PubMed]
- Janssen, A.; Hidalgo, J.V.; Beringer, D.X.; van Dooremalen, S.; Fernando, F.; van Diest, E.; Terrizi, A.R.; Bronsert, P.; Kock, S.; Schmitt-Graff, A.; et al. gammadelta T-cell Receptors Derived from Breast Cancer-Infiltrating T Lymphocytes Mediate Antitumor Reactivity. Cancer Immunol. Res. 2020, 8, 530–543. [Google Scholar] [CrossRef]
- Guiu, S.; Mollevi, C.; Charon-Barra, C.; Boissiere, F.; Crapez, E.; Chartron, E.; Lamy, P.J.; Gutowski, M.; Bourgier, C.; Romieu, G.; et al. Prognostic value of androgen receptor and FOXA1 co-expression in non-metastatic triple negative breast cancer and correlation with other biomarkers. Br. J. Cancer 2018, 119, 76–79. [Google Scholar] [CrossRef]
- Jacot, W.; Lopez-Crapez, E.; Mollevi, C.; Boissiere-Michot, F.; Simony-Lafontaine, J.; Ho-Pun-Cheung, A.; Chartron, E.; Theillet, C.; Lemoine, A.; Saffroy, R.; et al. BRCA1 Promoter Hypermethylation is Associated with Good Prognosis and Chemosensitivity in Triple-Negative Breast Cancer. Cancers 2020, 12, 828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jungbluth, A.A.; Frosina, D.; Fayad, M.; Pulitzer, M.P.; Dogan, A.; Busam, K.J.; Imai, N.; Gnjatic, S. Immunohistochemical Detection of gamma/delta T Lymphocytes in Formalin-fixed Paraffin-embedded Tissues. Appl. Immunohistochem. Mol. Morphol. 2019, 27, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.Y.; Chang, M.H.; Choi, Y.L.; Lee, J.E.; Nam, S.J.; Yang, J.H.; Park, Y.H.; Ahn, J.S.; Im, Y.H. Potential candidate biomarkers for heterogeneity in triple-negative breast cancer (TNBC). Cancer Chemother. Pharmacol. 2011, 68, 753–761. [Google Scholar] [CrossRef]
- Schmid, P.; Adams, S.; Rugo, H.S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Dieras, V.; Hegg, R.; Im, S.A.; Shaw Wright, G.; et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N. Engl. J. Med. 2018, 379, 2108–2121. [Google Scholar] [CrossRef] [PubMed]
- Schmid, P.; Cortes, J.; Pusztai, L.; McArthur, H.; Kummel, S.; Bergh, J.; Denkert, C.; Park, Y.H.; Hui, R.; Harbeck, N.; et al. Pembrolizumab for Early Triple-Negative Breast Cancer. N. Engl. J. Med. 2020, 382, 810–821. [Google Scholar] [CrossRef]
- Schmid, P.; Rugo, H.S.; Adams, S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Dieras, V.; Henschel, V.; Molinero, L.; Chui, S.Y.; et al. Atezolizumab plus nab-paclitaxel as first-line treatment for unresectable, locally advanced or metastatic triple-negative breast cancer (IMpassion130): Updated efficacy results from a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2020, 21, 44–59. [Google Scholar] [CrossRef]
- Yarchoan, M.; Hopkins, A.; Jaffee, E.M. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N. Engl. J. Med. 2017, 377, 2500–2501. [Google Scholar] [CrossRef] [PubMed]
- Filipovic, A.; Miller, G.; Bolen, J. Progress Toward Identifying Exact Proxies for Predicting Response to Immunotherapies. Front. Cell. Dev. Biol. 2020, 8, 155. [Google Scholar] [CrossRef]
- Gentles, A.J.; Newman, A.M.; Liu, C.L.; Bratman, S.V.; Feng, W.; Kim, D.; Nair, V.S.; Xu, Y.; Khuong, A.; Hoang, C.D.; et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015, 21, 938–945. [Google Scholar] [CrossRef]
- Bense, R.D.; Sotiriou, C.; Piccart-Gebhart, M.J.; Haanen, J.; van Vugt, M.; de Vries, E.G.E.; Schroder, C.P.; Fehrmann, R.S.N. Relevance of Tumor-Infiltrating Immune Cell Composition and Functionality for Disease Outcome in Breast Cancer. J. Natl. Cancer Inst. 2017, 109, djw192. [Google Scholar] [CrossRef] [Green Version]
- Tosolini, M.; Pont, F.; Poupot, M.; Vergez, F.; Nicolau-Travers, M.L.; Vermijlen, D.; Sarry, J.E.; Dieli, F.; Fournie, J.J. Assessment of tumor-infiltrating TCRVgamma9Vdelta2 gammadelta lymphocyte abundance by deconvolution of human cancers microarrays. Oncoimmunology 2017, 6, e1284723. [Google Scholar] [CrossRef] [PubMed]
- Chabab, G.; Boissiere-Michot, F.; Mollevi, C.; Ramos, J.; Lopez-Crapez, E.; Colombo, P.E.; Jacot, W.; Bonnefoy, N.; Lafont, V. Diversity of Tumor-Infiltrating, gammadelta T-Cell Abundance in Solid Cancers. Cells 2020, 9, 1537. [Google Scholar] [CrossRef]
- Lehmann-Che, J.; Hamy, A.S.; Porcher, R.; Barritault, M.; Bouhidel, F.; Habuellelah, H.; Leman-Detours, S.; de Roquancourt, A.; Cahen-Doidy, L.; Bourstyn, E.; et al. Molecular apocrine breast cancers are aggressive estrogen receptor negative tumors overexpressing either HER2 or GCDFP15. Breast Cancer Res. 2013, 15, R37. [Google Scholar] [CrossRef] [Green Version]
- Weisman, P.S.; Ng, C.K.; Brogi, E.; Eisenberg, R.E.; Won, H.H.; Piscuoglio, S.; De Filippo, M.R.; Ioris, R.; Akram, M.; Norton, L.; et al. Genetic alterations of triple negative breast cancer by targeted next-generation sequencing and correlation with tumor morphology. Mod. Pathol. 2016, 29, 476–488. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Zhang, Q.; Ye, J.; Wang, F.; Zhang, Y.; Wevers, E.; Schwartz, T.; Hunborg, P.; Varvares, M.A.; Hoft, D.F.; et al. Tumor-infiltrating gammadelta T lymphocytes predict clinical outcome in human breast cancer. J. Immunol. 2012, 189, 5029–5036. [Google Scholar] [CrossRef] [Green Version]
- Chabab, G.; Barjon, C.; Abdellaoui, N.; Salvador-Prince, L.; Dejou, C.; Michaud, H.A.; Boissiere-Michot, F.; Lopez-Crapez, E.; Jacot, W.; Pourquier, D.; et al. Identification of a regulatory Vdelta1 gamma delta T cell subpopulation expressing CD73 in human breast cancer. J. Leukoc. Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Peng, G.; Wang, H.Y.; Peng, W.; Kiniwa, Y.; Seo, K.H.; Wang, R.F. Tumor-infiltrating gammadelta T cells suppress T and dendritic cell function via mechanisms controlled by a unique toll-like receptor signaling pathway. Immunity 2007, 27, 334–348. [Google Scholar] [CrossRef] [Green Version]
- Wu, P.; Wu, D.; Ni, C.; Ye, J.; Chen, W.; Hu, G.; Wang, Z.; Wang, C.; Zhang, Z.; Xia, W.; et al. gammadeltaT17 cells promote the accumulation and expansion of myeloid-derived suppressor cells in human colorectal cancer. Immunity 2014, 40, 785–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, S.; Cheng, Q.; Cai, Y.; Gong, H.; Wu, Y.; Yu, X.; Shi, L.; Wu, D.; Dong, C.; Liu, H. IL-17A produced by gammadelta T cells promotes tumor growth in hepatocellular carcinoma. Cancer Res. 2014, 74, 1969–1982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coffelt, S.B.; Kersten, K.; Doornebal, C.W.; Weiden, J.; Vrijland, K.; Hau, C.S.; Verstegen, N.J.M.; Ciampricotti, M.; Hawinkels, L.; Jonkers, J.; et al. IL-17-producing gammadelta T cells and neutrophils conspire to promote breast cancer metastasis. Nature 2015, 522, 345–348. [Google Scholar] [CrossRef]
- Rei, M.; Goncalves-Sousa, N.; Lanca, T.; Thompson, R.G.; Mensurado, S.; Balkwill, F.R.; Kulbe, H.; Pennington, D.J.; Silva-Santos, B. Murine CD27(-) Vgamma6(+) gammadelta T cells producing IL-17A promote ovarian cancer growth via mobilization of protumor small peritoneal macrophages. Proc. Natl. Acad. Sci. USA 2014, 111, E3562–E3570. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, Y.; Lim, S.O.; Yamaguchi, H. Oncogenic signaling pathways associated with immune evasion and resistance to immune checkpoint inhibitors in cancer. Semin. Cancer. Biol. 2020, 65, 51–64. [Google Scholar] [CrossRef]
- Crane, C.A.; Panner, A.; Murray, J.C.; Wilson, S.P.; Xu, H.; Chen, L.; Simko, J.P.; Waldman, F.M.; Pieper, R.O.; Parsa, A.T. PI(3) kinase is associated with a mechanism of immunoresistance in breast and prostate cancer. Oncogene 2009, 28, 306–312. [Google Scholar] [CrossRef] [Green Version]
- Borcoman, E.; De La Rochere, P.; Richer, W.; Vacher, S.; Chemlali, W.; Krucker, C.; Sirab, N.; Radvanyi, F.; Allory, Y.; Pignot, G.; et al. Inhibition of PI3K pathway increases immune infiltrate in muscle-invasive bladder cancer. Oncoimmunology 2019, 8, e1581556. [Google Scholar] [CrossRef]
- Bao, L.; Hao, C.; Wang, J.; Wang, D.; Zhao, Y.; Li, Y.; Yao, W. High-Dose Cyclophosphamide Administration Orchestrates Phenotypic and Functional Alterations of Immature Dendritic Cells and Regulates Th Cell Polarization. Front. Pharmacol. 2020, 11, 775. [Google Scholar] [CrossRef] [PubMed]
- Madondo, M.T.; Quinn, M.; Plebanski, M. Low dose cyclophosphamide: Mechanisms of T cell modulation. Cancer. Treat. Rev. 2016, 42, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Jacot, W.; Lopez-Crapez, E.; Thezenas, S.; Senal, R.; Fina, F.; Bibeau, F.; Romieu, G.; Lamy, P.J. Lack of EGFR-activating mutations in European patients with triple-negative breast cancer could emphasise geographic and ethnic variations in breast cancer mutation profiles. Breast Cancer Res. 2011, 13, R133. [Google Scholar] [CrossRef] [Green Version]
- Jacot, W.; Gutowski, M.; Azria, D.; Romieu, G. Adjuvant early breast cancer systemic therapies according to daily used technologies. Crit. Rev. Oncol. Hematol. 2012, 2, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Boissiere-Michot, F.; Jacot, W.; Fraisse, J.; Gourgou, S.; Timaxian, C.; Lazennec, G. Prognostic Value of CXCR2 in Breast Cancer. Cancers 2020, 12, 2076. [Google Scholar] [CrossRef] [PubMed]
- Lamy, P.J.; Fina, F.; Bascoul-Mollevi, C.; Laberenne, A.C.; Martin, P.M.; Ouafik, L.; Jacot, W. Quantification and clinical relevance of gene amplification at chromosome 17q12-q21 in human epidermal growth factor receptor 2-amplified breast cancers. Breast Cancer Res. 2011, 13, R15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacot, W.; Thezenas, S.; Senal, R.; Viglianti, C.; Laberenne, A.C.; Lopez-Crapez, E.; Bibeau, F.; Bleuse, J.P.; Romieu, G.; Lamy, P.J. BRCA1 promoter hypermethylation, 53BP1 protein expression and PARP-1 activity as biomarkers of DNA repair deficit in breast cancer. BMC Cancer 2013, 13, 523. [Google Scholar] [CrossRef] [Green Version]
- Esteller, M.; Silva, J.M.; Dominguez, G.; Bonilla, F.; Matias-Guiu, X.; Lerma, E.; Bussaglia, E.; Prat, J.; Harkes, I.C.; Repasky, E.A.; et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J. Natl. Cancer Inst. 2000, 92, 564–569. [Google Scholar] [CrossRef] [PubMed]





| Variables | N = 162 | % |
|---|---|---|
| Age (years), median [min–max] | 55.4 | [28.7–86.3] |
| <55 | 79 | 48.8 |
| ≥55 | 83 | 51.2 |
| Tumor size (pT) | ||
| T1 | 70 | 43.2 |
| T2 | 79 | 48.8 |
| T3/T4 | 13 | 8.0 |
| Nodal status (pN) | ||
| N− | 108 | 66.7 |
| N+ | 54 | 33.3 |
| Histological grade (SBR; 3 missing values) | ||
| 1–2 | 50 | 31.4 |
| 3 | 109 | 68.6 |
| Histology | ||
| Ductal | 134 | 82.7 |
| Lobular | 6 | 3.7 |
| Other | 22 | 13.6 |
| Adjuvant chemotherapy (1 missing value) | ||
| No | 45 | 28.0 |
| Yes | 116 | 72.0 |
| Basal-like phenotype (2 missing values) | ||
| No | 63 | 39.4 |
| Yes | 97 | 60.6 |
| AR/FOXA1 expression (1% cutoff; 6 missing values) | ||
| AR+/FOXA1− | 11 | 7.1 |
| AR+/FOXA1+ | 57 | 36.5 |
| AR− | 88 | 56.4 |
| BRCA1 promoter methylation (2 missing values) | ||
| No Yes | 124 | 77.5 |
| 36 | 22.5 | |
| PIK3CA mutations | ||
| None | 140 | 86.4 |
| Exon 9 | 10 | 6.2 |
| Exon 20 | 12 | 7.4 |
| TILs (%; 4 missing values) | ||
| ≤10 | 124 | 78.5 |
| >10 | 34 | 21.5 |
| CD3+ cell density (cells/mm2), median [min–max] (6 missing values) | 658.41 | [0.33–6821] |
| ≤658.41 | 78 | 50.0 |
| >658.41 | 78 | 50.0 |
| CD8+ cell density (cells/mm2), median [min–max] (5 missing values) | 73.81 | [0–1870] |
| ≤73.81 | 79 | 50.3 |
| >73.81 | 78 | 49.7 |
| PD-L1TC (8 missing values) | ||
| <1% | 84 | 54.6 |
| ≥1% | 70 | 45.4 |
| PD-L1SC (%; 8 missing values) | ||
| 0 | 29 | 18.8 |
| ]0–10] | 68 | 44.1 |
| ]10–50] | 34 | 22.1 |
| >50 | 23 | 15.0 |
| PD-1SC (%; 10 missing values) | ||
| 0 | 45 | 29.6 |
| ]0–10] | 50 | 32.9 |
| ]10–50] | 46 | 30.3 |
| >50 | 11 | 7.2 |
| γδ T cells density (cells/mm2), median [min–max] | 6.625 | [0–193.8] |
| ≤6.625 | 81 | 50.0 |
| >6.625 | 81 | 50.0 |
| Variables | γδ T Cell Infiltration (/mm2) | ||||
|---|---|---|---|---|---|
| ≤6.625 | >6.625 | p-Value | |||
| n = 81 | % | n = 81 | % | ||
| Age (years) | 0.008 | ||||
| <55 | 31 | 38.3 | 48 | 59.3 | |
| ≥55 | 50 | 61.7 | 33 | 40.7 | |
| Tumor size (pT) | 0.341 | ||||
| T1 | 32 | 39.5 | 38 | 46.9 | |
| T2/T3/T4 | 49 | 60.5 | 43 | 53.1 | |
| Nodal status (pN) | 0.096 | ||||
| N− | 49 | 60.5 | 59 | 72.8 | |
| N+ | 32 | 39.5 | 22 | 27.2 | |
| Histological grade (SBR) | 0.002 | ||||
| 1–2 | 34 | 43.0 | 16 | 20.0 | |
| 3 | 45 | 57.0 | 64 | 80.0 | |
| Histology | 0.325 | ||||
| Ductal | 65 | 80.2 | 69 | 85.2 | |
| Lobular | 5 | 6.2 | 1 | 1.2 | |
| Other | 11 | 13.6 | 11 | 13.6 | |
| Adjuvant chemotherapy | 0.010 | ||||
| No | 30 | 37.0 | 15 | 18.7 | |
| Yes | 51 | 63.0 | 65 | 81.3 | |
| Basal-like phenotype | 0.184 | ||||
| No | 36 | 44.4 | 27 | 34.2 | |
| Yes | 45 | 55.6 | 52 | 65.8 | |
| AR/FOXA1 (1% cutoff) | 0.072 | ||||
| AR+/FOXA1+ | 35 | 43.2 | 22 | 29.3 | |
| Other | 46 | 56.8 | 53 | 70.7 | |
| BRCA1 promoter methylation | 0.010 | ||||
| No | 68 | 86.1 | 56 | 69.1 | |
| Yes | 11 | 13.9 | 25 | 30.9 | |
| PIK3CA mutations | 0.107 | ||||
| None | 66 | 81.5 | 74 | 91.4 | |
| Exon 9/Exon 20 | 15 | 18.5 | 7 | 8.6 | |
| TILs | <0.001 | ||||
| ≤10% | 74 | 92.5 | 50 | 64.1 | |
| >10% | 6 | 7.5 | 28 | 35.9 | |
| CD3+ cell density (cells/mm2) | <0.001 | ||||
| ≤658.41 | 65 | 84.4 | 13 | 16.5 | |
| >658.41 | 12 | 15.6 | 66 | 83.5 | |
| CD8+ cell density (cells/mm2) | <0.001 | ||||
| ≤73.81 | 54 | 70.1 | 25 | 31.2 | |
| >73.81 | 23 | 29.9 | 55 | 68.8 | |
| PD-L1TC | <0.001 | ||||
| <1% | 58 | 74.4 | 26 | 34.2 | |
| ≥1% | 20 | 25.6 | 50 | 65.8 | |
| PD-L1SC | <0.001 | ||||
| ≤10% | 63 | 80.8 | 34 | 44.7 | |
| >10% | 15 | 19.2 | 42 | 55.3 | |
| PD-1SC | 0.040 | ||||
| ≤10% | 53 | 70.7 | 42 | 54.5 | |
| >10% | 22 | 29.3 | 35 | 45.5 | |
| Variables | OS | RFS | ||
|---|---|---|---|---|
| HR | 95% CI | HR | 95% CI | |
| Age (years) | p = 0.030 | p = 0.177 | ||
| <55 | 1 | 1 | ||
| ≥55 | 1.86 | 1.05–3.31 | 1.55 | 0.81–2.96 |
| Tumor size (pT) | p < 0.001 | p < 0.001 | ||
| T1 | 1 | 1 | ||
| T2/T3/T4 | 3.44 | 1.76–6.70 | 4.07 | 1.80–9.24 |
| Nodal status (pN) | p < 0.001 | p < 0.001 | ||
| N− | 1 | 1 | ||
| N+ | 2.66 | 1.54–4.60 | 5.10 | 2.62–9.93 |
| Histological grade (SBR) | p = 0.966 | p = 0.526 | ||
| 1–2 | 1 | 1 | ||
| 3 | 0.99 | 0.55–1.77 | 1.25 | 0.62–2.51 |
| Histology | p = 0.448 | p = 0.442 | ||
| Ductal | 1 | 1 | ||
| Other | 0.74 | 0.33–1.65 | 0.70 | 0.27–1.80 |
| Adjuvant chemotherapy | p < 0.001 | p = 0.036 | ||
| No | 1 | 1 | ||
| Yes | 0.32 | 0.19–0.56 | 0.49 | 0.26–0.94 |
| Basal-like phenotype | p = 0.329 | p = 0.964 | ||
| No (≤10%) | 1 | 1 | ||
| Yes (Basal) | 1.33 | 0.74–2.37 | 1.02 | 0.53–1.94 |
| AR/FOXA1 | p = 0.501 | p = 0.272 | ||
| AR+/FOXA1+ | 1 | 1 | ||
| Other | 0.83 | 0.47–1.44 | 0.70 | 0.37–1.32 |
| BRCA1 promoter methylation | p = 0.701 | p = 0.208 | ||
| No | 1 | 1 | ||
| Yes | 0.87 | 0.44–1.75 | 0.59 | 0.25–1.41 |
| PIK3CA mutations | p = 0.061 | p = 0.202 | ||
| None | 1 | 1 | ||
| Exon 9/Exon 20 | 1.98 | 1.01–3.86 | 1.71 | 0.78–3.71 |
| TILs | p = 0.005 | p = 0.001 | ||
| ≤10% | 1 | 1 | ||
| >10% | 0.29 | 0.10–0.80 | 0.17 | 0.04–0.69 |
| CD3+ cell density (cells/mm2) | p = 0.010 | p < 0.001 | ||
| ≤658.41 | 1 | 1 | ||
| >658.41 | 0.48 | 0.27–0.85 | 0.26 | 0.12–0.55 |
| CD8+ cell density (cells/mm2) | p = 0.312 | p = 0.045 | ||
| ≤73.81 | 1 | 1 | ||
| >73.81 | 0.75 | 0.42–1.32 | 0.51 | 0.27–0.99 |
| PD-L1TC | p = 0.125 | p = 0.150 | ||
| <1% | 1 | 1 | ||
| ≥1% | 0.65 | 0.37–1.14 | 0.62 | 0.32–1.20 |
| PD-L1SC | p = 0.004 | p = 0.002 | ||
| ≤10% | 1 | 1 | ||
| >10% | 0.41 | 0.21–0.78 | 0.30 | 1.14–0.69 |
| PD-1SC | p = 0.403 | p = 0.808 | ||
| ≤10% | 1 | 1 | ||
| >10% | 1.27 | 0.72–2.22 | 1.08 | 0.57–2.07 |
| γδ T cell density (cells/mm2) | p = 0.001 | p < 0.001 | ||
| ≤6.625 | 1 | 1 | ||
| >6.625 | 0.39 | 0.22–0.70 | 0.28 | 0.14–0.58 |
| Variables | OS N = 161 | RFS N = 157 | ||
|---|---|---|---|---|
| HR | 95% CI | HR | 95% CI | |
| Tumor size | p = 0.003 | p = 0.002 | ||
| T1 | 1 | 1 | ||
| T2/T3/T4 | 2.73 | 1.34–5.56 | 3.37 | 1.45–7.83 |
| Nodal status | p = 0.002 | p < 0.001 | ||
| N− | 1 | 1 | ||
| N+ | 2.64 | 1.42–4.92 | 4.39 | 2.16–8.91 |
| Adjuvant chemotherapy | p < 0.001 | |||
| No | 1 | |||
| Yes | 0.27 | 0.14–0.49 | ||
| PIK3CA mutations | p = 0.032 | |||
| None | 1 | |||
| Exon 9/Exon 20 | 2.25 | 1.12–4.49 | ||
| CD8+ cell density (cells/mm2) | p = 0.017 | |||
| ≤73.81 | 1 | |||
| >73.81 | 0.43 | 0.21–0.87 | ||
| γδ T cell density (cells/mm2) | p = 0.031 | p = 0.011 | ||
| ≤6.625 | 1 | 1 | ||
| >6.625 | 0.51 | 0.28–0.96 | 0.39 | 0.18–0.84 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boissière-Michot, F.; Chabab, G.; Mollevi, C.; Guiu, S.; Lopez-Crapez, E.; Ramos, J.; Bonnefoy, N.; Lafont, V.; Jacot, W. Clinicopathological Correlates of γδ T Cell Infiltration in Triple-Negative Breast Cancer. Cancers 2021, 13, 765. https://doi.org/10.3390/cancers13040765
Boissière-Michot F, Chabab G, Mollevi C, Guiu S, Lopez-Crapez E, Ramos J, Bonnefoy N, Lafont V, Jacot W. Clinicopathological Correlates of γδ T Cell Infiltration in Triple-Negative Breast Cancer. Cancers. 2021; 13(4):765. https://doi.org/10.3390/cancers13040765
Chicago/Turabian StyleBoissière-Michot, Florence, Ghita Chabab, Caroline Mollevi, Séverine Guiu, Evelyne Lopez-Crapez, Jeanne Ramos, Nathalie Bonnefoy, Virginie Lafont, and William Jacot. 2021. "Clinicopathological Correlates of γδ T Cell Infiltration in Triple-Negative Breast Cancer" Cancers 13, no. 4: 765. https://doi.org/10.3390/cancers13040765
APA StyleBoissière-Michot, F., Chabab, G., Mollevi, C., Guiu, S., Lopez-Crapez, E., Ramos, J., Bonnefoy, N., Lafont, V., & Jacot, W. (2021). Clinicopathological Correlates of γδ T Cell Infiltration in Triple-Negative Breast Cancer. Cancers, 13(4), 765. https://doi.org/10.3390/cancers13040765








