miR-21 Plays a Dual Role in Tumor Formation and Cytotoxic Response in Breast Tumors
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Cell Culture
2.2. Transfection and Cell Viability Assays
2.3. Treatment for Autophagy Determination
2.4. Mouse Models
2.4.1. Breast Cancer Metastases Murine Model
2.4.2. MMTV-PyMT Orthotopic Mouse Model
2.4.3. MMTV-PyMT Genetically Engineered Mouse Model
2.5. Human Sera
2.6. Irradiation and Chemotherapeutic Treatments
2.7. Genotyping PCR, RNA Isolation and qRT-PCR
2.8. Hematoxylin Eosin Staining
2.8.1. cDNA Profiling
2.8.2. Tissue Lysis, Western Blotting and Statistical Analysis
3. Results
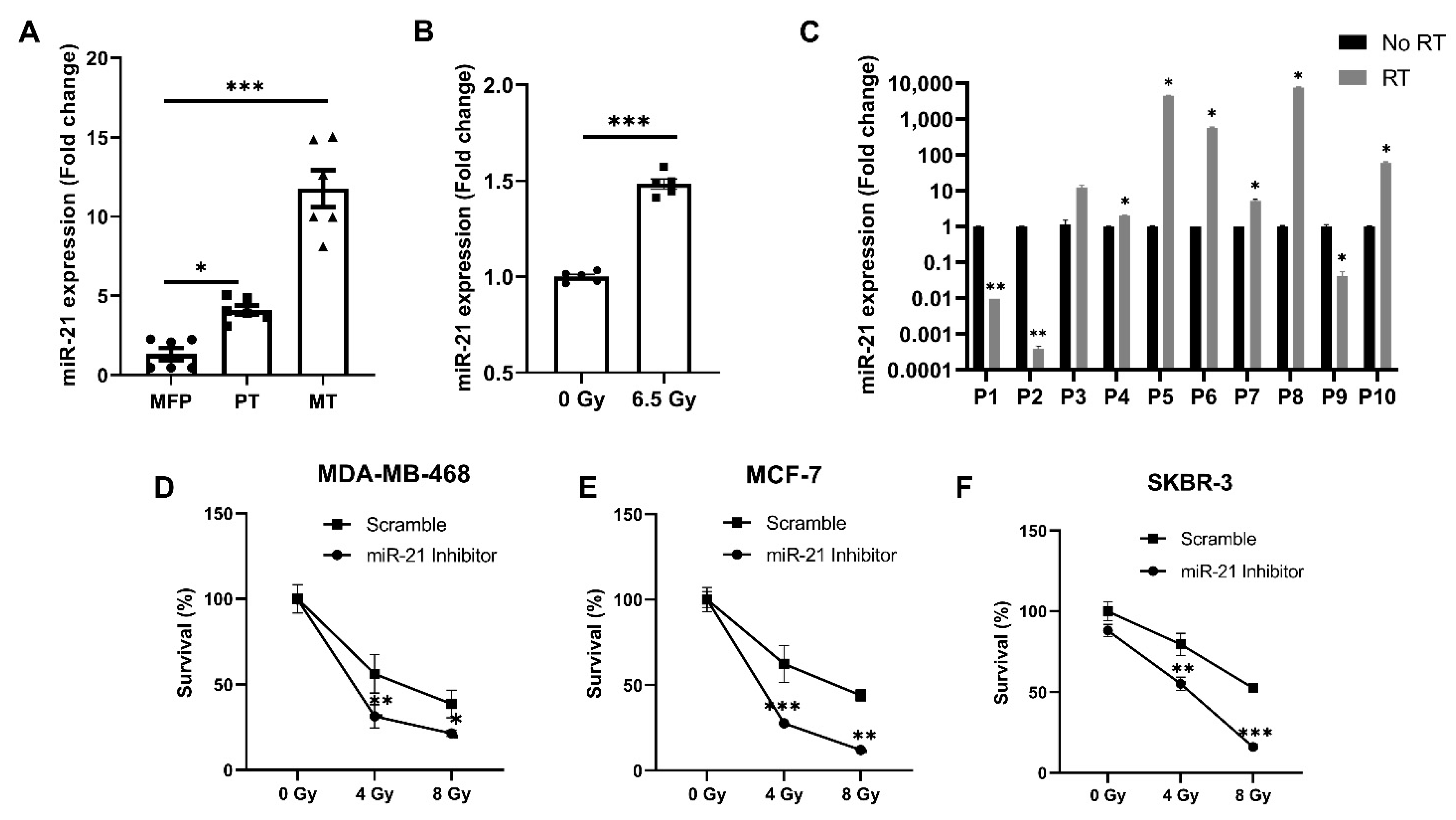
3.1. miR-21 Expression Plays a Critical Role in Breast Cancer Growth, Metastases and Treatment
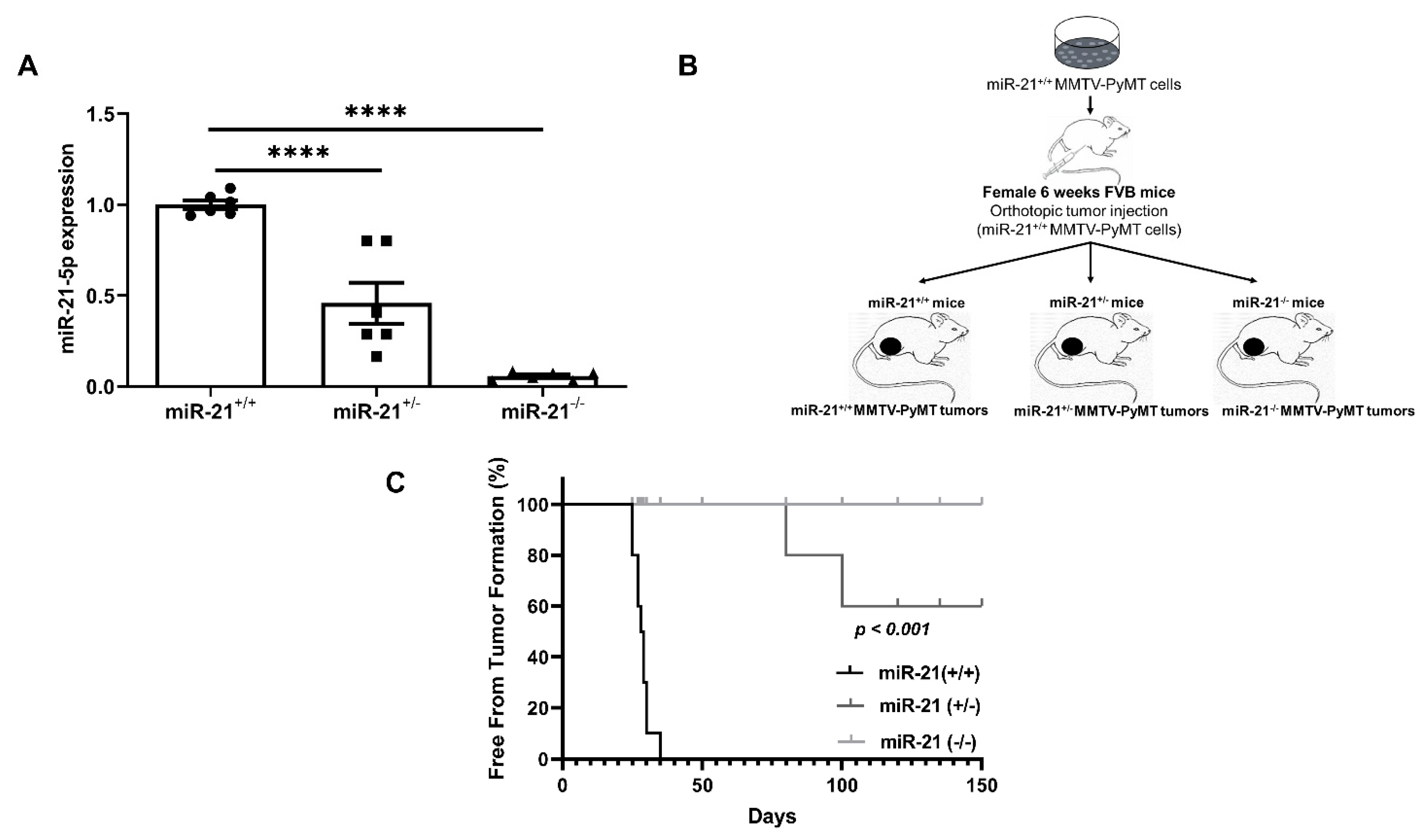
3.2. Loss of miR-21 Protects Mice from Breast Cancer Initiation: Orthotopic Model
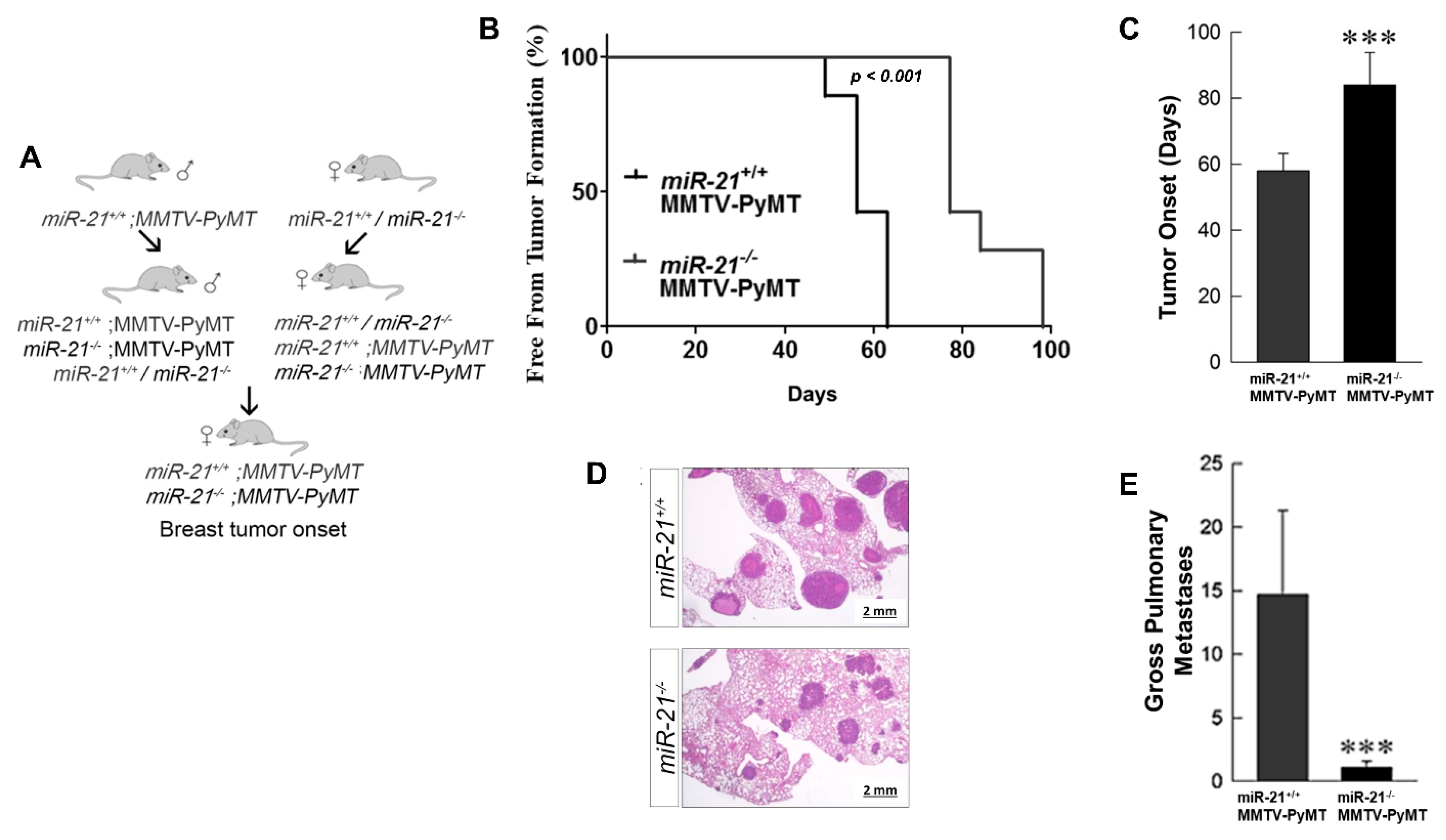
3.3. Loss of miR-21 Delays Tumor Growth and Reduces Metastases: Genetically Engineered Model
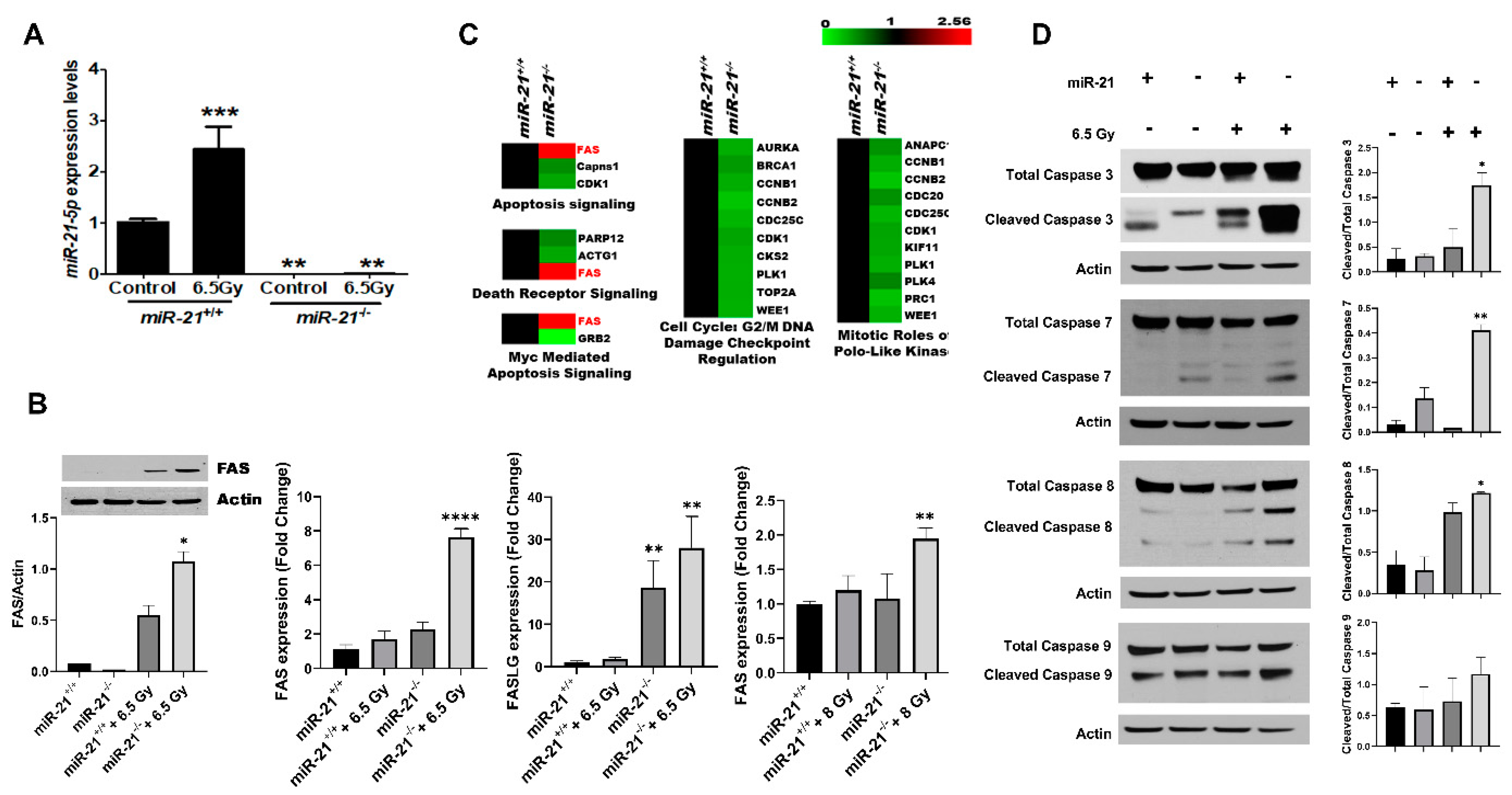
3.4. Radiation Increases Apoptosis in MMTV-PyMT Tumors Lacking miR-21
3.5. The Inhibition of miR-21 Sensitizes Breast Cancer Cells to Cytotoxic Therapies
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wein, L.; Loi, S. Mechanisms of resistance of chemotherapy in early-stage triple negative breast cancer (TNBC). Breast 2017, 34 (Suppl. 1), S27–S30. [Google Scholar] [CrossRef]
- Niu, J.; Shi, Y.; Tan, G.; Yang, C.H.; Fan, M.; Pfeffer, L.M.; Wu, Z.H. DNA damage induces NF-kappaB-dependent microRNA-21 up-regulation and promotes breast cancer cell invasion. J. Biol. Chem. 2012, 287, 21783–21795. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Meng, H.; Peng, Q.; Yang, X.; Gan, R.; Zhao, L.; Chen, Z.; Lu, J.; Meng, Q.H. Downregulation of microRNA-21 expression restrains non-small cell lung cancer cell proliferation and migration through upregulation of programmed cell death 4. Cancer Gene Ther. 2015, 22, 23–29. [Google Scholar] [CrossRef] [Green Version]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Di Leva, G.; Garofalo, M.; Croce, C.M. MicroRNAs in cancer. Annu. Rev. Pathol. Mech. Dis. 2014, 9, 287–314. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.-L.; Wang, H.; Liu, J.; Wang, Z.-X. MicroRNA-21 (miR-21) expression promotes growth, metastasis, and chemo- or radioresistance in non-small cell lung cancer cells by targeting PTEN. Mol. Cell. Biochem. 2013, 372, 35–45. [Google Scholar] [CrossRef]
- Gong, C.; Yao, Y.; Wang, Y.; Liu, B.; Wu, W.; Chen, J.; Su, F.; Yao, H.; Song, E. Up-regulation of miR-21 mediates resistance to trastuzumab therapy for breast cancer. J. Biol. Chem. 2011, 286, 19127–19137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacKenzie, T.A.; Schwartz, G.N.; Calderone, H.M.; Graveel, C.R.; Winn, M.E.; Hostetter, G.; Wells, W.A.; Sempere, L.F. Stromal expression of miR-21 identifies high-risk group in triple-negative breast cancer. Am. J. Pathol. 2014, 184, 3217–3225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Y.; Zhou, X.; Ji, J.; Chen, L.; Cao, J.; Luo, J.; Zhang, S. High expression levels of miR-21 and miR-210 predict unfavorable survival in breast cancer: A systemic review and meta-analysis. Int. J. Biol. Markers 2015, 30, 347–358. [Google Scholar] [CrossRef]
- Wu, M.-F.; Yang, J.; Xiang, T.; Shi, Y.-Y.; Liu, L.-J. miR-21 targets Fas ligand-mediated apoptosis in breast cancer cell line MCF-7. Acta Acad. Med. Wuhan 2014, 34, 190–194. [Google Scholar] [CrossRef]
- Chao, T.-F.; Xiong, H.-H.; Liu, W.; Chen, Y.; Zhang, J.-X. MiR-21 mediates the radiation resistance of glioblastoma cells by regulating PDCD4 and hMSH2. Acta Acad. Med. Wuhan 2013, 33, 525–529. [Google Scholar] [CrossRef] [PubMed]
- Das, N.A.; Carpenter, A.J.; Belenchia, A.; Aroor, A.R.; Noda, M.; Siebenlist, U.; Chandrasekar, B.; DeMarco, V.G. Empagliflozin reduces high glucose-induced oxidative stress and miR-21-dependent TRAF3IP2 induction and RECK suppression, and inhibits human renal proximal tubular epithelial cell migration and epithelial-to-mesenchymal transition. Cell. Signal. 2020, 68, 109506. [Google Scholar] [CrossRef]
- Najjary, S.; Mohammadzadeh, R.; Mokhtarzadeh, A.; Mohammadi, A.; Kojabad, A.B.; Baradaran, B. Role of miR-21 as an authentic oncogene in mediating drug resistance in breast cancer. Gene 2020, 738, 144453. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; Wu, Y.Q.; Zhang, S.P. MiR-21-5p enhances the progression and paclitaxel resistance in drug-resistant breast cancer cell lines by targeting PDCD4. Neoplasma 2019, 66, 746–755. [Google Scholar] [CrossRef] [PubMed]
- van Jaarsveld, M.T.; Wouters, M.D.; Boersma, A.W.; Smid, M.; van IJcken, W.F.; Mathijssen, R.H.; Hoeijmakers, J.H.; Martens, J.W.; van Laere, S.; Wiemer, E.A.; et al. DNA damage responsive microRNAs misexpressed in human cancer modulate therapy sensitivity. Mol. Oncol. 2014, 8, 458–468. [Google Scholar] [CrossRef] [PubMed]
- Guy, C.T.; Cardiff, R.D.; Muller, W.J. Induction of mammary tumors by expression of polyomavirus middle T oncogene: A transgenic mouse model for metastatic disease. Mol. Cell. Biol. 1992, 12, 954–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khoury, S.; Ajuyah, P.; Tran, N. Isolation of small noncoding RNAs from human serum. J. Vis. Exp. 2014, 51443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Mattos-Arruda, L.; Bottai, G.; Nuciforo, P.G.; Di Tommaso, L.; Giovannetti, E.; Peg, V.; Losurdo, A.; Pérez-Garcia, J.; Masci, G.; Corsi, F.; et al. MicroRNA-21 links epithelial-to-mesenchymal transition and inflammatory signals to confer resistance to neoadjuvant trastuzumab and chemotherapy in HER2-positive breast cancer patients. Oncotarget 2015, 6, 37269–37280. [Google Scholar] [CrossRef] [Green Version]
- Haghnavaz, N.; Asghari, F.; Komi, D.E.A.; Shanehbandi, D.; Baradaran, B.; Kazemi, T. HER2 positivity may confer resistance to therapy with paclitaxel in breast cancer cell lines. Artif. Cells Nanomed. Biotechnol. 2018, 46, 518–523. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Boopathi, E.; Addya, S.; Phillips, B.; Rigoutsos, I.; Penn, R.B.; Rattan, S. Aging-associated changes in microRNA expression profile of internal anal sphincter smooth muscle: Role of microRNA-133a. Am. J. Physiol. Liver Physiol. 2016, 311, G964–G973. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Lv, J.-H.; Liang, L.; Wang, J.-C.; Li, C.-R.; Sun, L.; Li, T. Downregulation of microRNA-21 inhibited radiation-resistance of esophageal squamous cell carcinoma. Cancer Cell Int. 2018, 18, 39. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zuo, Y.; Qian, X.-L.; Chen, Z.-P.; Wang, S.-K.; Song, L.; Peng, L.-P. Inhibition of MicroRNA-21-5p Promotes the Radiation Sensitivity of Non-Small Cell Lung Cancer Through HMSH2. Cell. Physiol. Biochem. 2017, 43, 1258–1272. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.; Nogales-Cadenas, R.; Zhang, Q.; Lin, J.R.; Zhang, W.; O’Brien, K.; Montagna, C.; Zhang, Z.D. Transcriptomic dynamics of breast cancer progression in the MMTV-PyMT mouse model. BMC Genom. 2017, 18, 185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puccetti, M.V.; Adams, C.M.; Dan, T.D.; Palagani, A.; Simone, B.A.; DeAngelis, T.; Eischen, C.M.; Simone, N.L. MicroRNA-21 is Required for Hematopoietic Cell Viability After Radiation Exposure. Int. J. Radiat. Oncol. Biol. Phys. 2019, 104, 1165–1174. [Google Scholar] [CrossRef]
- Li, J.; Chan, W.; Leung, W.; Wang, Y.; Xu, C. MicroRNA-21 promotes proliferation of rat hepatocyte BRL-3A by targeting FASLG. Genet. Mol. Res. 2015, 14, 4150–4160. [Google Scholar] [CrossRef] [PubMed]
- Sayed, D.; He, M.; Hong, C.; Gao, S.; Rane, S.; Yang, Z.; Abdellatif, M. MicroRNA-21 is a downstream effector of AKT that mediates its antiapoptotic effects via suppression of Fas ligand. J. Biol. Chem. 2010, 285, 20281–20290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Liu, Y.; Chen, X.; Zhang, M.; Xiao, Z. Impact of MiR-21 on the expression of FasL in the presence of TGF-beta1. Aesthetic Surg. J. 2013, 33, 1186–1198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cacan, E.; Spring, A.M.; Kumari, A.; Greer, S.F.; Garnett-Benson, C. Combination Treatment with Sublethal Ionizing Radiation and the Proteasome Inhibitor, Bortezomib, Enhances Death-Receptor Mediated Apoptosis and Anti-Tumor Immune Attack. Int. J. Mol. Sci. 2015, 16, 30405–30421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horton, J.K.; Siamakpour-Reihani, S.; Lee, C.-T.; Zhou, Y.; Chen, W.; Geradts, J.; Fels, D.R.; Hoang, P.; Ashcraft, K.A.; Groth, J.; et al. FAS Death Receptor: A Breast Cancer Subtype-Specific Radiation Response Biomarker and Potential Therapeutic Target. Radiat. Res. 2015, 184, 456–469. [Google Scholar] [CrossRef] [PubMed]
- Oshima, C.T.F.; Ribeiro, D.A.; Gomes, T.S.; Adios, P.C.; Egami, M.I.; Segreto, H.R.C. Amifostine Increases FAS and Caspase-3 Expression in Colonic Tissue of Irradiated Mice. Anticancer Res. 2015, 35, 2817–2822. [Google Scholar]
- Asangani, I.A.; Rasheed, S.A.K.; Nikolova, D.A.; Leupold, J.H.; Colburn, N.H.; Post, S.; Allgayer, H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 2007, 27, 2128–2136. [Google Scholar] [CrossRef] [Green Version]
- Frankel, L.B.; Christoffersen, N.R.; Jacobsen, A.; Lindow, M.; Krogh, A.; Lund, A.H. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J. Biol. Chem. 2008, 283, 1026–1033. [Google Scholar] [CrossRef] [Green Version]
- Seike, M.; Goto, A.; Okano, T.; Bowman, E.D.; Schetter, A.J.; Horikawa, I.; Mathe, E.A.; Jen, J.; Yang, P.; Sugimura, H.; et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc. Natl. Acad. Sci. USA 2009, 106, 12085–12090. [Google Scholar] [CrossRef] [Green Version]
- Han, M.; Liu, M.; Wang, Y.; Mo, Z.; Bi, X.; Liu, Z.; Fan, Y.; Chen, X.; Wu, C. Re-expression of miR-21 contributes to migration and invasion by inducing epithelial-mesenchymal transition consistent with cancer stem cell characteristics in MCF-7 cells. Mol. Cell. Biochem. 2012, 363, 427–436. [Google Scholar] [CrossRef]
- Ozgun, A.; Karagoz, B.; Bilgi, O.; Tuncel, T.; Baloğlu, H.; Kandemir, E.G. MicroRNA-21 as an indicator of aggressive phenotype in breast cancer. Onkologie 2013, 36, 115–118. [Google Scholar] [CrossRef]
- Si, M.-L.; Zhu, S.; Wu, H.; Lu, Z.; Wu, F.; Mo, Y.-Y. miR-21-mediated tumor growth. Oncogene 2007, 26, 2799–2803. [Google Scholar] [CrossRef] [Green Version]
- Yan, L.-X.; Huang, X.-F.; Shao, Q.; Huang, M.-Y.; Deng, L.; Wu, Q.-L.; Zeng, Y.-X.; Shao, J.-Y. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 2008, 14, 2348–2360. [Google Scholar] [CrossRef] [Green Version]
- Halimi, M.; Parsian, H.; Asghari, S.M.; Sariri, R.; Moslemi, D.; Yeganeh, F.; Zabihi, E. Clinical translation of human microRNA 21 as a potential biomarker for exposure to ionizing radiation. Transl. Res. 2014, 163, 578–584. [Google Scholar] [CrossRef] [PubMed]
- Rui, M.; Qu, Y.; Gao, T.; Ge, Y.; Feng, C.; Xu, X. Simultaneous delivery of anti-miR21 with doxorubicin prodrug by mimetic lipoprotein nanoparticles for synergistic effect against drug resistance in cancer cells. Int. J. Nanomed. 2017, 12, 217–237. [Google Scholar] [CrossRef] [Green Version]
- Simone, N.L.; Soule, B.P.; Ly, D.; Saleh, A.D.; Savage, J.E.; DeGraff, W.; Cook, J.; Harris, C.C.; Gius, D.; Mitchell, J.B. Ionizing radiation-induced oxidative stress alters miRNA expression. PLoS ONE 2009, 4, e6377. [Google Scholar] [CrossRef] [Green Version]
- Anastasov, N.; Höfig, I.; Vasconcellos, I.G.; Rappl, K.; Braselmann, H.; Ludyga, N.; Auer, G.; Aubele, M.; Atkinson, M.J. Radiation resistance due to high expression of miR-21 and G2/M checkpoint arrest in breast cancer cells. Radiat. Oncol. 2012, 7, 206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahraei, M.; Chaube, B.; Liu, Y.; Sun, J.; Kaplan, A.; Price, N.L.; Ding, W.; Oyaghire, S.; García-Milian, R.; Mehta, S.; et al. Suppressing miR-21 activity in tumor-associated macrophages promotes an antitumor immune response. J. Clin. Investig. 2019, 129, 5518–5536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, X.; Kumar, M.; Choudhury, S.N.; Buscaglia, L.E.B.; Barker, J.R.; Kanakamedala, K.; Liu, M.-F.; Li, Y. Loss of the miR-21 allele elevates the expression of its target genes and reduces tumorigenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 10144–10149. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Song, L.; Zhang, L.; Zeng, S.; Gao, F. miR-21 modulates resistance of HR-HPV positive cervical cancer cells to radiation through targeting LATS1. Biochem. Biophys. Res. Commun. 2015, 459, 679–685. [Google Scholar] [CrossRef]
- Campos-Parra, A.D.; Mitznahuatl, G.C.; Pedroza-Torres, A.; Romo, R.V.; Reyes, F.I.P.; López-Urrutia, E.; Pérez-Plasencia, C. Micro-RNAs as Potential Predictors of Response to Breast Cancer Systemic Therapy: Future Clinical Implications. Int. J. Mol. Sci. 2017, 18, 1182. [Google Scholar] [CrossRef]
- Radulovic, V.; Heider, T.; Richter, S.; Moertl, S.; Atkinson, M.J.; Anastasov, N. Differential response of normal and transformed mammary epithelial cells to combined treatment of anti-miR-21 and radiation. Int. J. Radiat. Biol. 2017, 93, 361–372. [Google Scholar] [CrossRef] [Green Version]
- Fang, H.; Xie, J.; Zhang, M.; Zhao, Z.; Wan, Y.; Yao, Y. miRNA-21 promotes proliferation and invasion of triple-negative breast cancer cells through targeting PTEN. Am. J. Transl. Res. 2017, 9, 953–961. [Google Scholar]
- Patutina, O.A.; Miroshnichenko, S.K.; Mironova, N.L.; Sen’Kova, A.V.; Bichenkova, E.V.; Clarke, D.J.; Vlassov, V.V.; Zenkova, M.A. Catalytic Knockdown of miR-21 by Artificial Ribonuclease: Biological Performance in Tumor Model. Front. Pharmacol. 2019, 10, 879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Tan, Z.; Hu, H.; Liu, H.; Wu, T.; Zheng, C.; Wang, X.; Luo, Z.; Wang, J.; Liu, S.; et al. microRNA-21 promotes breast cancer proliferation and metastasis by targeting LZTFL1. BMC Cancer 2019, 19, 738. [Google Scholar] [CrossRef] [Green Version]
- Abtin, M.; Alivand, M.R.; Khaniani, M.S.; Bastami, M.; Zaeifizadeh, M.; Derakhshan, S.M. Simultaneous downregulation of miR-21 and miR-155 through oleuropein for breast cancer prevention and therapy. J. Cell. Biochem. 2018, 119, 7151–7165. [Google Scholar] [CrossRef]
- Griveau, A.; Bejaud, J.; Anthiya, S.; Avril, S.; Autret, D.; Garcion, E. Silencing of miR-21 by locked nucleic acid–lipid nanocapsule complexes sensitize human glioblastoma cells to radiation-induced cell death. Int. J. Pharm. 2013, 454, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Haghpanah, V.; Fallah, P.; Tavakoli, R.; Naderi, M.; Samimi, H.; Soleimani, M.; Larijani, B. Antisense-miR-21 enhances differentiation/apoptosis and reduces cancer stemness state on anaplastic thyroid cancer. Tumour. Biol. 2016, 37, 1299–1308. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Huang, Y.; Chen, D.; He, J.; Zhu, W.; Zhang, Y.; Liu, X. Downregulation of miR-21 increases cisplatin sensitivity of non–small-cell lung cancer. Cancer Genet. 2014, 207, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Zou, W.; Hu, C.; Li, G.; Zhou, S.; He, Y.; Ma, F.; Deng, C.; Sun, L. Modulation of CASC2/miR-21/PTEN pathway sensitizes cervical cancer to cisplatin. Arch Biochem. Biophys. 2017, 623–624, 20–30. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dan, T.; Shastri, A.A.; Palagani, A.; Buraschi, S.; Neill, T.; Savage, J.E.; Kapoor, A.; DeAngelis, T.; Addya, S.; Camphausen, K.; et al. miR-21 Plays a Dual Role in Tumor Formation and Cytotoxic Response in Breast Tumors. Cancers 2021, 13, 888. https://doi.org/10.3390/cancers13040888
Dan T, Shastri AA, Palagani A, Buraschi S, Neill T, Savage JE, Kapoor A, DeAngelis T, Addya S, Camphausen K, et al. miR-21 Plays a Dual Role in Tumor Formation and Cytotoxic Response in Breast Tumors. Cancers. 2021; 13(4):888. https://doi.org/10.3390/cancers13040888
Chicago/Turabian StyleDan, Tu, Anuradha A. Shastri, Ajay Palagani, Simone Buraschi, Thomas Neill, Jason E. Savage, Aastha Kapoor, Tiziana DeAngelis, Sankar Addya, Kevin Camphausen, and et al. 2021. "miR-21 Plays a Dual Role in Tumor Formation and Cytotoxic Response in Breast Tumors" Cancers 13, no. 4: 888. https://doi.org/10.3390/cancers13040888






