AKT3 Is a Novel Regulator of Cancer-Associated Fibroblasts in Head and Neck Squamous Cell Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. The Cancer Genome Atlas (TCGA) Dataset
2.2. Isolation of Fibroblasts
2.3. RNA Sequencing (RNA-seq)
2.4. Transfection
2.5. RT-PCR
2.6. Preparation of Peripheral Blood Mononuclear Cells and Isolation of CD14+ Cells
2.7. Cell Proliferation Assay
2.8. Migration Assay
2.9. T-cell Proliferation Assay
2.10. Macrophage Induction Assay
2.11. Flow Cytometry Staining and Analysis
2.12. Preparation of Patient Tissue Samples for Immunohistochemistry
2.13. Immunohistochemistry
2.14. Evaluation of Immunohistochemistry
2.15. Statistical Analysis
3. Results
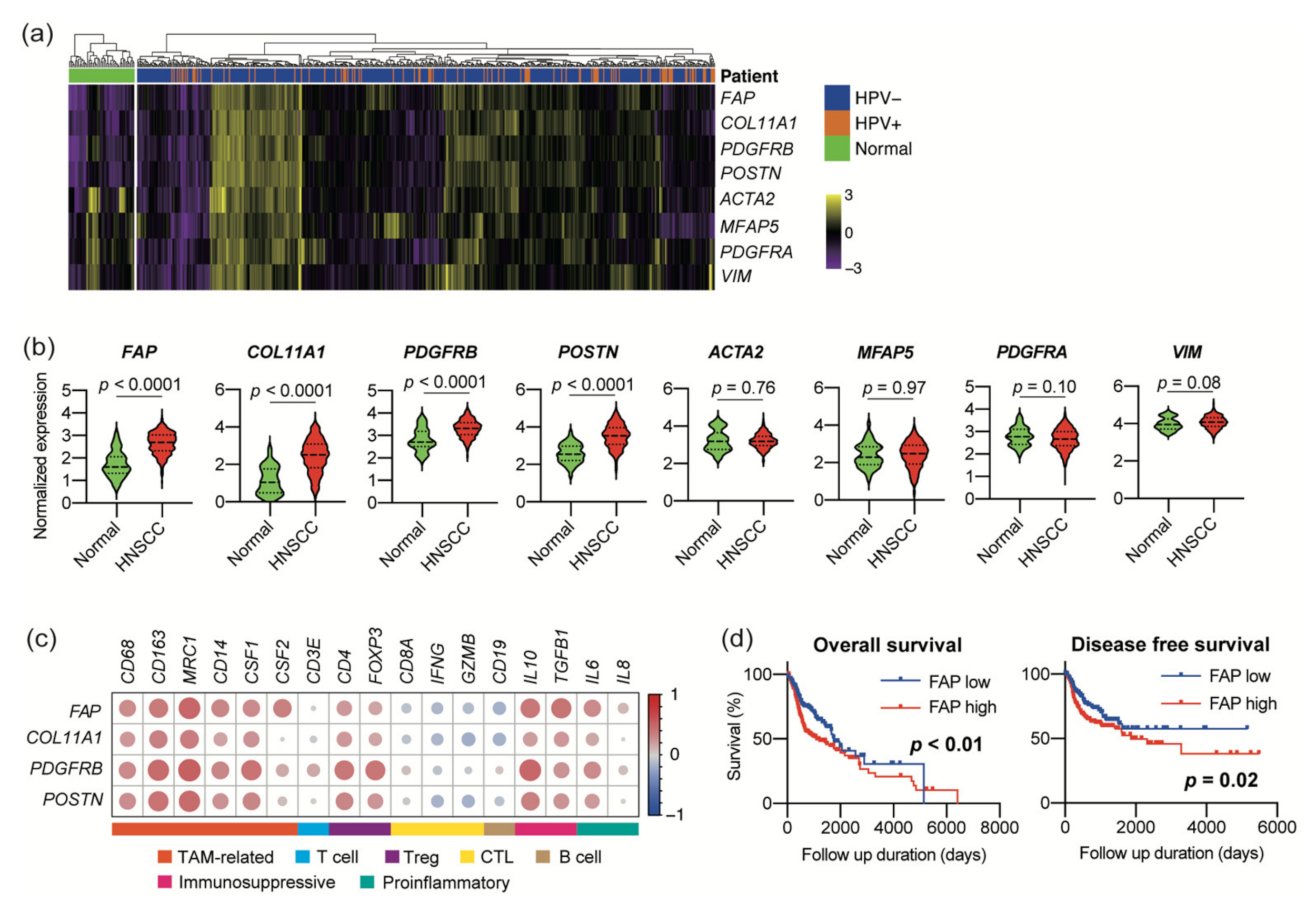
3.1. Expression of CAF Marker Mrnas Correlates with Immunosuppressive Microenvironment in Large HNSCC Cohort
3.2. Sequencing of CAF mRNA Revealed a Distinct Signature of Upregulated Genes
3.3. AKT3 Depletion Reprogrammed CAF Activity
3.4. AKT3 Depletion Impaired CAF Immunosuppressive Activity
3.5. AKT3 Expression in CAFs Correlated with Immune Cell Infiltration into The Tumor Microenvironment
3.6. AKT3 Expression in CAFs Correlated with a Poor Prognosis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Leemans, C.R.; Braakhuis, B.J.; Brakenhoff, R.H. The molecular biology of head and neck cancer. Nat. Rev. Cancer 2011, 11, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Ferris, R.L.; Blumenschein, G.; Fayette, J.; Guigay, J.; Colevas, A.D.; Licitra, L.; Harrington, K.; Kasper, S.; Vokes, E.E.; Even, C.; et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N. Engl. J. Med. 2016, 375, 1856–1867. [Google Scholar] [CrossRef] [PubMed]
- Burtness, B.; Harrington, K.J.; Greil, R.; Soulières, D.; Tahara, M.; de Castro, G.; Psyrri, A.; Basté, N.; Neupane, P.; Bratland, Å.; et al. Investigators, Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): A randomised, open-label, phase 3 study. Lancet 2019, 394, 1915–1928. [Google Scholar] [CrossRef]
- Chen, S.M.Y.; Krinsky, A.L.; Woolaver, R.A.; Wang, X.; Chen, Z.; Wang, J.H. Tumor immune microenvironment in head and neck cancers. Mol. Carcinog. 2020, 59, 766–774. [Google Scholar] [CrossRef]
- Puram, S.V.; Tirosh, I.; Parikh, A.S.; Patel, A.P.; Yizhak, K.; Gillespie, S.; Rodman, C.; Luo, C.L.; Mroz, E.A.; Emerick, K.S.; et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017, 171, 1611–1624. [Google Scholar] [CrossRef]
- Sewell-Loftin, M.K.; Bayer, S.V.H.; Crist, E.; Hughes, T.; Joison, S.M.; Longmore, G.D.; George, S.C. Cancer-associated fibroblasts support vascular growth through mechanical force. Sci. Rep. 2017, 7, 12574. [Google Scholar] [CrossRef] [PubMed]
- De Jaeghere, E.A.; Denys, H.G.; de Wever, O. Fibroblasts Fuel Immune Escape in the Tumor Microenvironment. Trends Cancer 2019, 5, 704–723. [Google Scholar] [CrossRef]
- Takahashi, H.; Sakakura, K.; Kawabata-Iwakawa, R.; Rokudai, S.; Toyoda, M.; Nishiyama, M.; Chikamatsu, K. Immunosuppressive activity of cancer-associated fibroblasts in head and neck squamous cell carcinoma. Cancer Immunol. Immunother. 2015, 64, 1407–1417. [Google Scholar] [CrossRef]
- Takahashi, H.; Sakakura, K.; Kudo, T.; Toyoda, M.; Kaira, K.; Oyama, T.; Chikamatsu, K. Cancer-associated fibroblasts promote an immunosuppressive microenvironment through the induction and accumulation of protumoral macrophages. Oncotarget 2017, 8, 8633–8647. [Google Scholar] [CrossRef]
- Balsamo, M.; Scordamaglia, F.; Pietra, G.; Manzini, C.; Cantoni, C.; Boitano, M.; Queirolo, P.; Vermi, W.; Facchetti, F.; Moretta, A.; et al. Melanoma-associated fibroblasts modulate NK cell phenotype and antitumor cytotoxicity. Proc. Natl. Acad. Sci. USA. 2009, 106, 20847–20852. [Google Scholar] [CrossRef]
- Nazareth, M.R.; Broderick, L.; Simpson-Abelson, M.R.; Kelleher, R.J.; Yokota, S.J.; Bankert, R.B. Characterization of human lung tumor-associated fibroblasts and their ability to modulate the activation of tumor-associated T cells. J. Immunol. 2007, 178, 5552–5562. [Google Scholar] [CrossRef]
- Sun, Q.; Zhang, B.; Hu, Q.; Qin, Y.; Xu, W.; Liu, W.; Yu, X.; Xu, J. The impact of cancer-associated fibroblasts on major hallmarks of pancreatic cancer. Theranostics 2018, 8, 5072–5087. [Google Scholar] [CrossRef] [PubMed]
- Vara, J.A.F.; Casado, E.; de Castro, J.; Cejas, P.; Belda-Iniesta, C.; González-Barón, M. PI3K/Akt signalling pathway and cancer. Cancer Treat. Rev. 2004, 30, 193–204. [Google Scholar] [CrossRef]
- Yang, J.; Nie, J.; Ma, X.; Wei, Y.; Peng, Y.; Wei, X. Targeting PI3K in cancer: Mechanisms and advances in clinical trials. Mol. Cancer 2019, 18, 26. [Google Scholar] [CrossRef]
- Gonzalez, E.; McGraw, T.E. The Akt kinases: Isoform specificity in metabolism and cancer. Cell Cycle 2009, 8, 2502–2508. [Google Scholar] [CrossRef]
- Marquard, F.E.; Jücker, M. PI3K/AKT/mTOR signaling as a molecular target in head and neck cancer. Biochem. Pharmacol. 2020, 172, 113729. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Bode, A.M.; Dong, Z.; Lee, M.H. AKT as a Therapeutic Target for Cancer. Cancer Res. 2019, 79, 1019–1031. [Google Scholar] [CrossRef] [PubMed]
- Amornphimoltham, P.; Sriuranpong, V.; Patel, V.; Benavides, F.; Conti, C.J.; Sauk, J.; Sausville, E.A.; Molinolo, A.A.; Gutkind, J.S. Persistent activation of the Akt pathway in head and neck squamous cell carcinoma: A potential target for UCN-01. Clin. Cancer Res. 2004, 10, 4029–4037. [Google Scholar] [CrossRef]
- Sun, J.; Nishiyama, T.; Shimizu, K.; Kadota, K. TCC: An R package for comparing tag count data with robust normalization strategies. BMC Bioinformatics 2013, 14, 219. [Google Scholar] [CrossRef] [PubMed]
- Fujii, N.; Shomori, K.; Shiomi, T.; Nakabayashi, M.; Takeda, C.; Ryoke, K.; Ito, H. Cancer-associated fibroblasts and CD163-positive macrophages in oral squamous cell carcinoma: Their clinicopathological and prognostic significance. J. Oral Pathol. Med. 2012, 41, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Kellermann, M.G.; Sobral, L.M.; da Silva, S.D.; Zecchin, K.G.; Graner, E.; Lopes, M.A.; Kowalski, L.P.; Coletta, R.D. Mutual paracrine effects of oral squamous cell carcinoma cells and normal oral fibroblasts: Induction of fibroblast to myofibroblast transdifferentiation and modulation of tumor cell proliferation. Oral Oncol. 2008, 44, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Rockey, D.C.; Weymouth, N.; Shi, Z. Smooth muscle α actin (Acta2) and myofibroblast function during hepatic wound healing. PLoS ONE 2013, 8, e77166. [Google Scholar] [CrossRef] [PubMed]
- Nurmik, M.; Ullmann, P.; Rodriguez, F.; Haan, S.; Letellier, E. In search of definitions: Cancer-associated fibroblasts and their markers. Int. J. Cancer 2020, 146, 895–905. [Google Scholar] [CrossRef]
- Noy, R.; Pollard, J.W. Tumor-associated macrophages: From mechanisms to therapy. Immunity 2014, 41, 49–61. [Google Scholar] [CrossRef]
- Sica, A.; Larghi, P.; Mancino, A.; Rubino, L.; Porta, C.; Totaro, M.G.; Rimoldi, M.; Biswas, S.K.; Allavena, P.; Mantovani, A. Macrophage polarization in tumour progression. Semin. Cancer Biol. 2008, 18, 349–355. [Google Scholar] [CrossRef]
- Yavuz, B.G.; Gunaydin, G.; Gedik, M.E.; Kosemehmetoglu, K.; Karakoc, D.; Ozgur, F.; Guc, D. Cancer associated fibroblasts sculpt tumour microenvironment by recruiting monocytes and inducing immunosuppressive PD-1. Sci. Rep. 2019, 9, 3172. [Google Scholar] [CrossRef]
- Cho, H.; Seo, Y.; Loke, K.M.; Kim, S.W.; Oh, S.M.; Kim, J.H.; Soh, J.; Kim, H.S.; Lee, H.; Kim, J.; et al. Cancer-Stimulated CAFs Enhance Monocyte Differentiation and Protumoral TAM Activation via IL6 and GM-CSF Secretion. Clin. Cancer Res. 2018, 24, 5407–5421. [Google Scholar] [CrossRef]
- Mace, T.A.; Ameen, Z.; Collins, A.; Wojcik, S.; Mair, M.; Young, G.S.; Fuchs, J.R.; Eubank, T.D.; Frankel, W.L.; Bekaii-Saab, T.; et al. Pancreatic cancer-associated stellate cells promote differentiation of myeloid-derived suppressor cells in a STAT3-dependent manner. Cancer Res. 2013, 73, 3007–3018. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Qi, F.; Zhao, F.; Li, G.; Shao, S.; Zhang, X.; Yuan, L.; Feng, Y. Cancer-associated fibroblasts enhance tumor-associated macrophages enrichment and suppress NK cells function in colorectal cancer. Cell Death Dis. 2019, 10, 273. [Google Scholar] [CrossRef]
- Liu, T.; Han, C.; Wang, S.; Fang, P.; Ma, Z.; Xu, L.; Yin, R. Cancer-associated fibroblasts: An emerging target of anti-cancer immunotherapy. J. Hematol. Oncol. 2019, 12, 86. [Google Scholar] [CrossRef]
- Togashi, Y.; Shitara, K.; Nishikawa, H. Regulatory T cells in cancer immunosuppression—Implications for anticancer therapy. Nat. Rev. Clin. Oncol. 2019, 16, 356–371. [Google Scholar] [CrossRef]
- Gaggioli, C.; Hooper, S.; Hidalgo-Carcedo, C.; Grosse, R.; Marshall, J.F.; Harrington, K.; Sahai, E. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell Biol. 2007, 9, 1392–1400. [Google Scholar] [CrossRef]
- Elyada, E.; Bolisetty, M.; Laise, P.; Flynn, W.F.; Courtois, E.T.; Burkhart, R.A.; Teinor, J.A.; Belleau, P.; Biffi, G.; Lucito, M.S.; et al. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discov. 2019, 9, 1102–1123. [Google Scholar] [CrossRef] [PubMed]
- Öhlund, D.; Handly-Santana, A.; Biffi, G.; Elyada, E.; Almeida, A.S.; Ponz-Sarvise, M.; Corbo, V.; Oni, T.E.; Hearn, S.A.; Lee, E.J.; et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 2017, 214, 579–596. [Google Scholar] [CrossRef] [PubMed]
- Pickard, A.; Cichon, A.C.; Barry, A.; Kieran, D.; Patel, D.; Hamilton, P.; Salto-Tellez, M.; James, J.; McCance, D.J. Inactivation of Rb in stromal fibroblasts promotes epithelial cell invasion. EMBO. J. 2012, 31, 3092–3103. [Google Scholar] [CrossRef] [PubMed]
- Cichon, A.C.; Pickard, A.; McDade, S.S.; Sharpe, D.J.; Moran, M.; James, J.A.; McCance, D.J. AKT in stromal fibroblasts controls invasion of epithelial cells. Oncotarget 2013, 4, 1103–1116. [Google Scholar] [CrossRef]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef]
- Chadli, L.; Sotthewes, B.; Li, K.; Andersen, S.N.; Cahir-McFarland, E.; Cheung, M.; Cullen, P.; Dorjée, A.; de Vries-Bouwstra, J.K.; Huizinga, T.W.J.; et al. Identification of regulators of the myofibroblast phenotype of primary dermal fibroblasts from early diffuse systemic sclerosis patients. Sci. Rep. 2019, 9, 4521. [Google Scholar] [CrossRef]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef]




| Variables | Overall Survival | Disease-Free Survival | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate | Univariate | Multivariate | ||||||
| 5-y | p-Value | HR (95% CI) | p-Value | 5-y | p-Value | HR (95% CI) | p-Value | ||
| Rate (%) | Rate (%) | ||||||||
| Age (years) | |||||||||
| <71 | 70.9 | 0.045 | 1 | 0.035 | 66.3 | 0.816 | |||
| ≥71 | 47.1 | 2.769 (1.077–7.118) | 59.6 | ||||||
| Gender | |||||||||
| Male | 61 | 0.866 | 60.6 | 0.582 | |||||
| Female | 69.9 | 71.5 | |||||||
| Differentiation | |||||||||
| Well/moderate | 66.5 | 0.038 | 1 | 0.166 | 69.8 | 0.019 | 1 | 0.191 | |
| Poor | 40 | 2.097 (0.734–5.986) | 29.6 | 1.993 (0.709–5.604) | |||||
| Lymphatic invasion | |||||||||
| Negative | 75.8 | 0.006 | 1 | 0.099 | 77.4 | 0.007 | 1 | 0.463 | |
| Positive | 40.3 | 3.514 (0.790–15.630) | 40 | 1.634 (0.440–6.062) | |||||
| Vascular invasion | |||||||||
| Negative | 72.9 | 0.002 | 1 | 0.484 | 71.3 | 0.062 | 1 | 0.583 | |
| Positive | 35.9 | 1.466 (0.502–4.282) | 45.1 | 1.323 (0.487–3.598) | |||||
| T factor | |||||||||
| T1-2 | 66.6 | 0.195 | 66.3 | 0.657 | |||||
| T3-4 | 24.3 | N/A | |||||||
| N factor | |||||||||
| N0 | 67.1 | 0.062 | 1 | 0.932 | 74.3 | 0.001 | 1 | 0.063 | |
| N1-3 | 55.8 | 1.059 (0.282–3.987) | 42.6 | 3.771 (0.930–15.300) | |||||
| TNM stage | |||||||||
| I-II | 71.7 | 0.02 | 1 | 0.659 | 71 | 0.096 | 1 | 0.301 | |
| III-IV | 40.8 | 0.692 (0.135–3.548) | 45.7 | 0.514 (0.146–1.814) | |||||
| AKT3 in CAFs | |||||||||
| Negative | 76.7 | 0.037 | 1 | 0.035 | 72 | 0.158 | |||
| Positive | 46.2 | 2.860 (1.076–7.600) | 51.2 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takahashi, H.; Rokudai, S.; Kawabata-Iwakawa, R.; Sakakura, K.; Oyama, T.; Nishiyama, M.; Chikamatsu, K. AKT3 Is a Novel Regulator of Cancer-Associated Fibroblasts in Head and Neck Squamous Cell Carcinoma. Cancers 2021, 13, 1233. https://doi.org/10.3390/cancers13061233
Takahashi H, Rokudai S, Kawabata-Iwakawa R, Sakakura K, Oyama T, Nishiyama M, Chikamatsu K. AKT3 Is a Novel Regulator of Cancer-Associated Fibroblasts in Head and Neck Squamous Cell Carcinoma. Cancers. 2021; 13(6):1233. https://doi.org/10.3390/cancers13061233
Chicago/Turabian StyleTakahashi, Hideyuki, Susumu Rokudai, Reika Kawabata-Iwakawa, Koichi Sakakura, Tetsunari Oyama, Masahiko Nishiyama, and Kazuaki Chikamatsu. 2021. "AKT3 Is a Novel Regulator of Cancer-Associated Fibroblasts in Head and Neck Squamous Cell Carcinoma" Cancers 13, no. 6: 1233. https://doi.org/10.3390/cancers13061233
APA StyleTakahashi, H., Rokudai, S., Kawabata-Iwakawa, R., Sakakura, K., Oyama, T., Nishiyama, M., & Chikamatsu, K. (2021). AKT3 Is a Novel Regulator of Cancer-Associated Fibroblasts in Head and Neck Squamous Cell Carcinoma. Cancers, 13(6), 1233. https://doi.org/10.3390/cancers13061233







