Combined In Situ Hybridization and Immunohistochemistry on Archival Tissues Reveals Stromal microRNA-204 as Prognostic Biomarker for Oral Squamous Cell Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. In Situ Hybridization (ISH)
2.2. Immunohistochemistry (IHC)
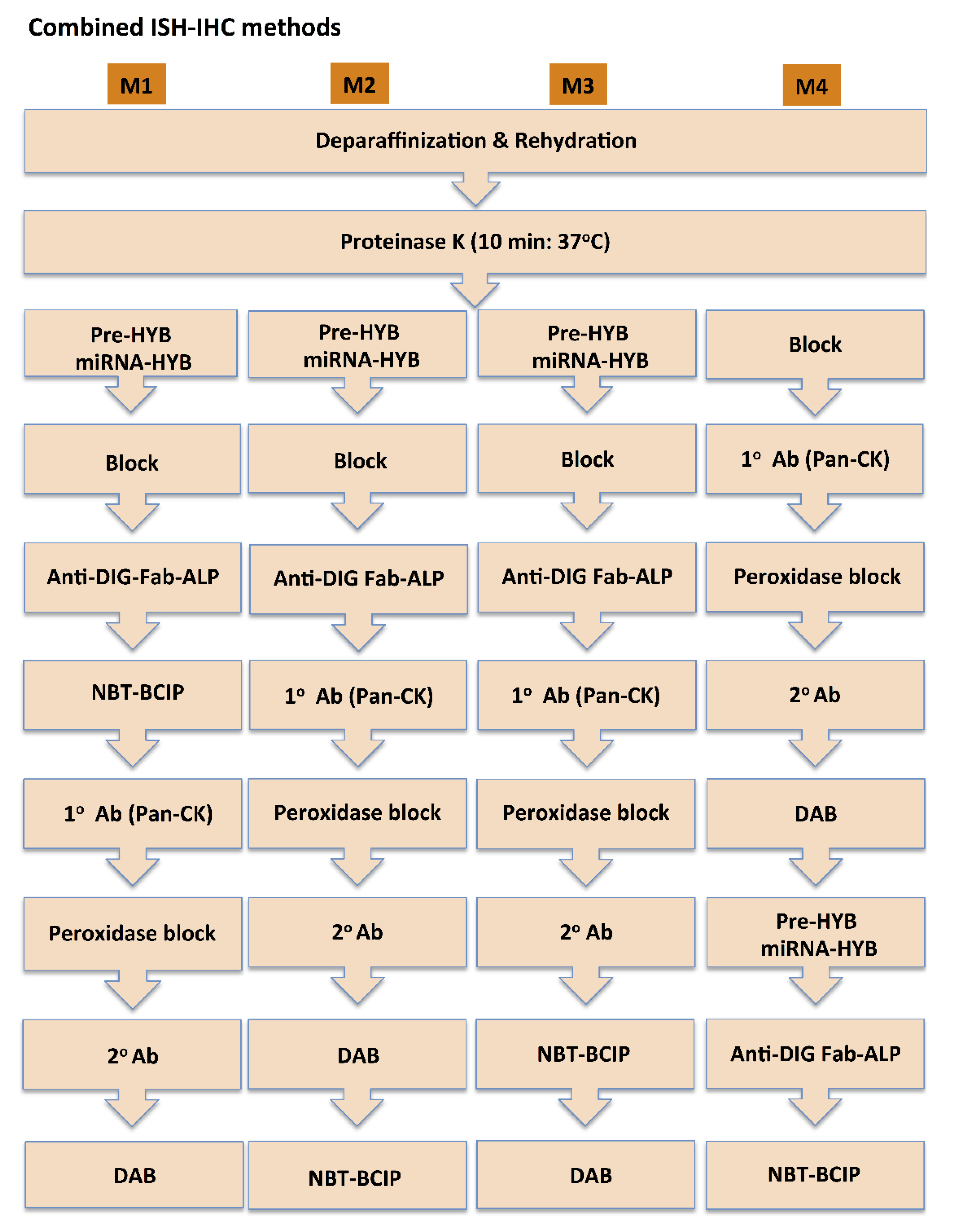
2.3. Combined miR ISH and IHC staining
2.4. Image Acquisition and Quantification
2.5. Study Cohort
2.6. Evaluation of Clinical and Pathological Parameters
2.7. RNA Extraction and Quantitative Reverse-Transcriptase Polymerase Chain Reaction (qRT-PCR)
2.8. Statistical Analysis
3. Results
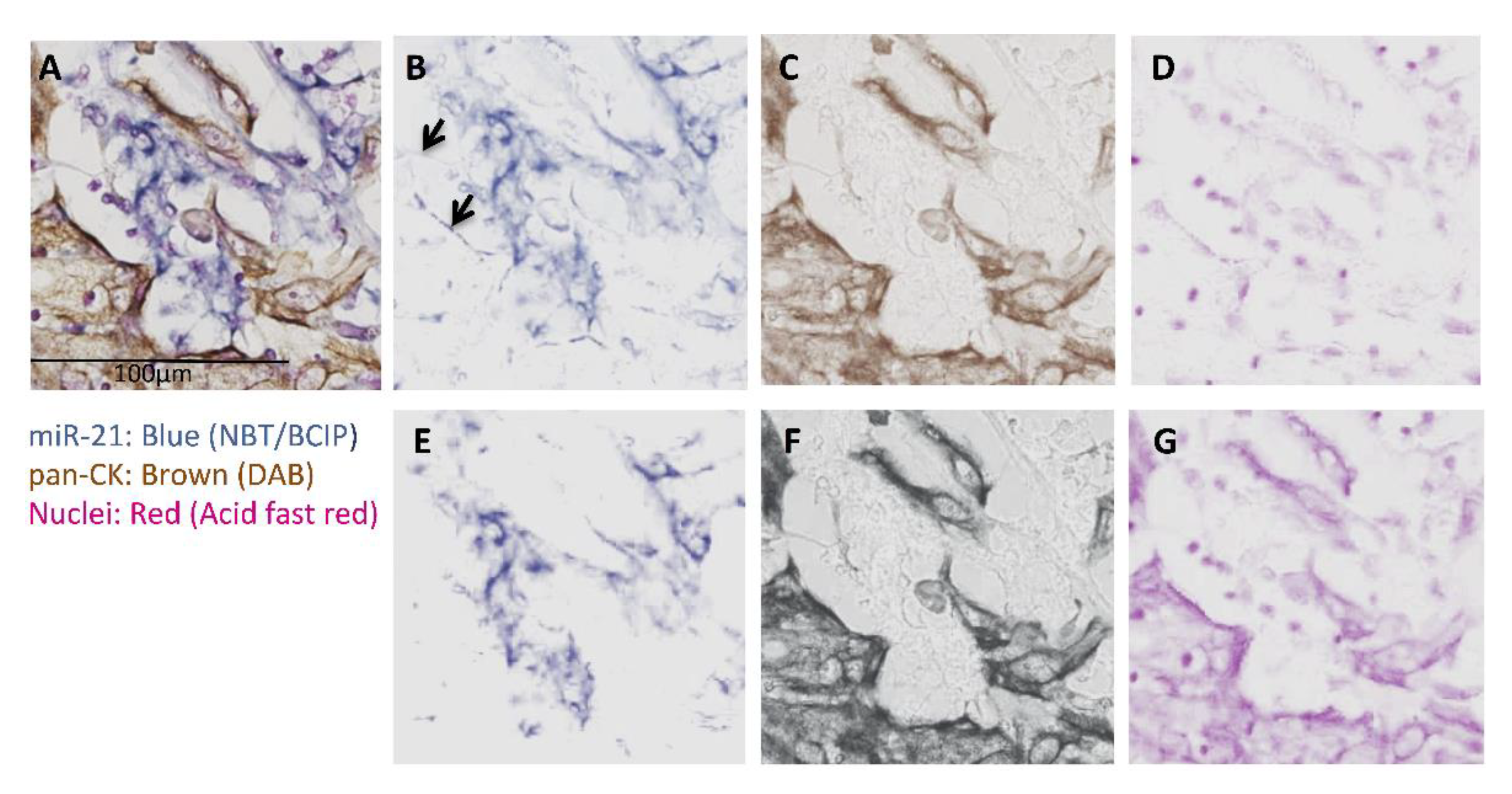
3.1. miRs Expression and Their Co-Localization with pan-CK
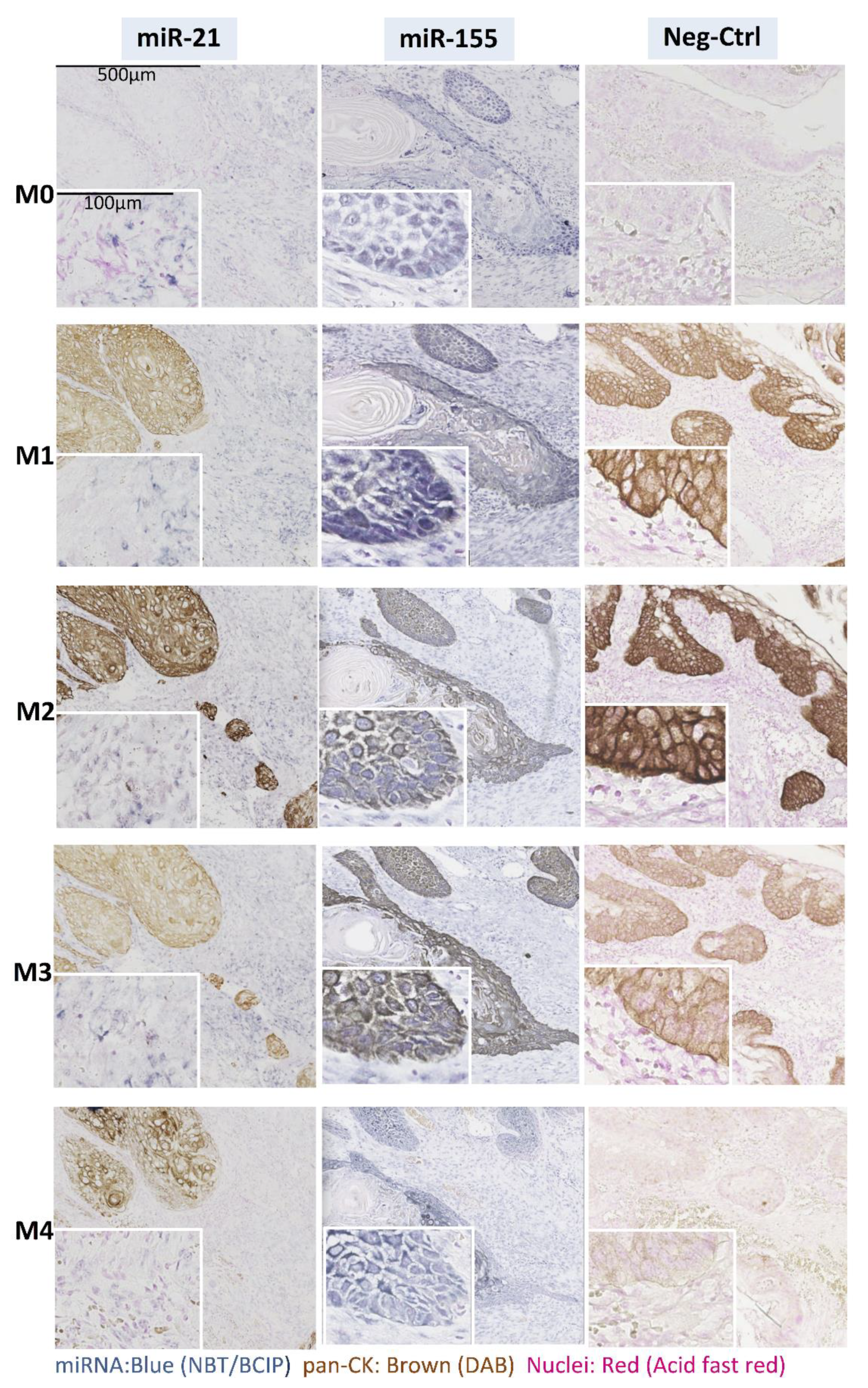
3.2. Effects of IHC on ISH and Vice Versa
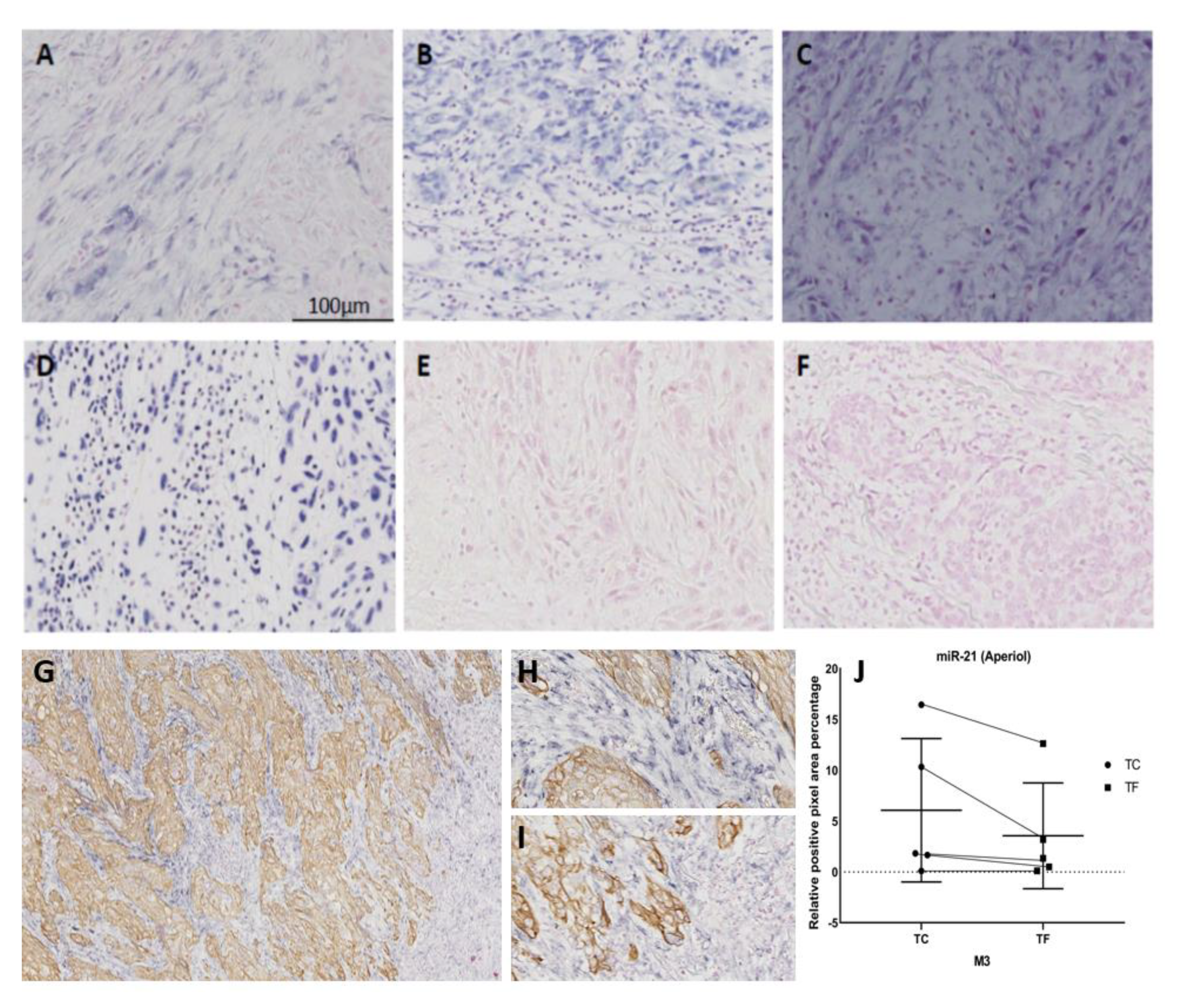
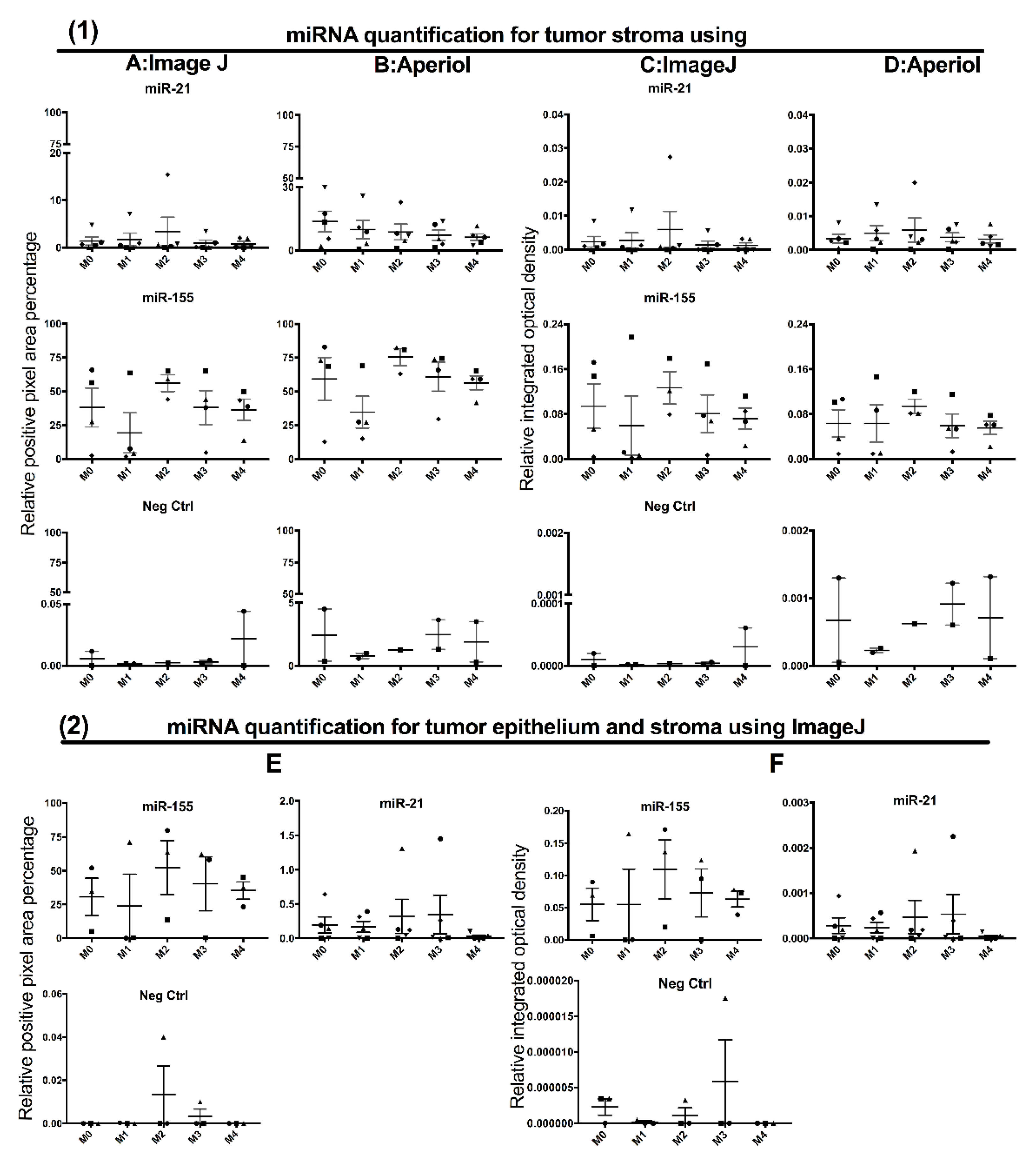
3.3. Contribution of Noise to miR Signal in the Double Staining Methods
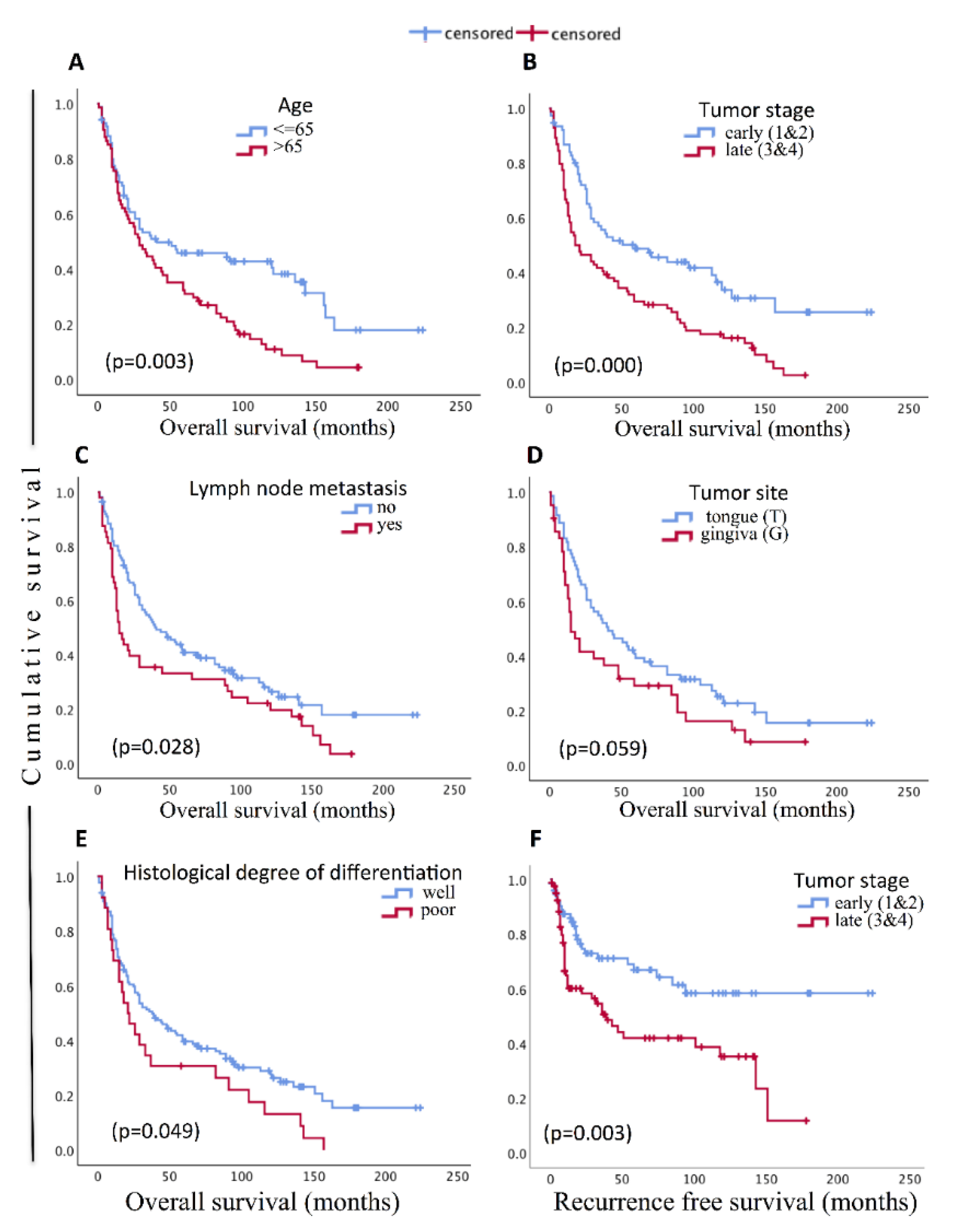
3.4. Cohort Description and Prognostic Significance of Clinico-Pathological Parameters
3.5. Expression of miR-204 in NHOM and OSCC
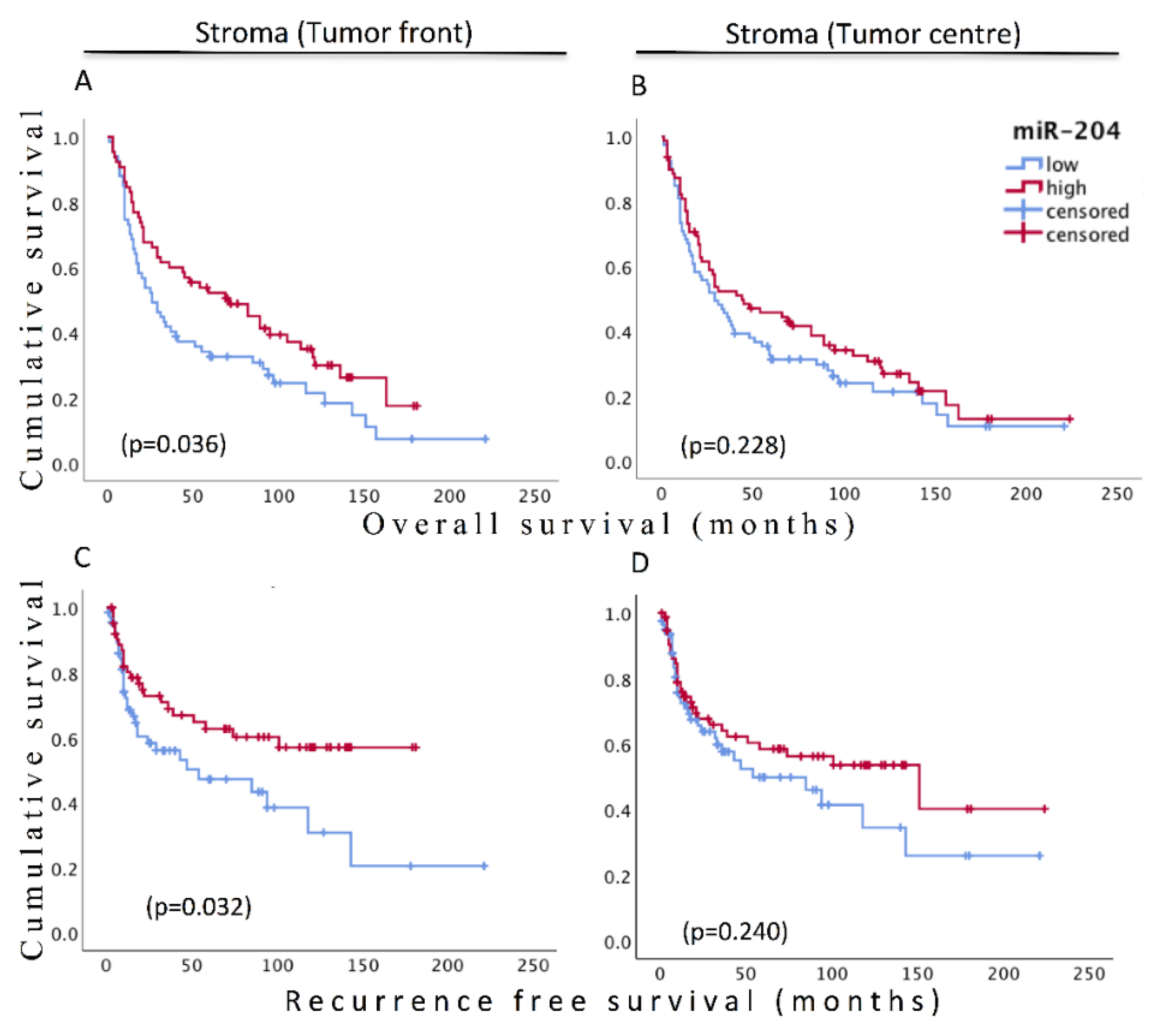
3.6. Prognostic Significance of Stromal miR-204
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kalluri, R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 2016, 16, 582–598. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Zeisberg, M. Fibroblasts in cancer. Nat. Rev. Cancer 2006, 6, 392–401. [Google Scholar] [CrossRef] [PubMed]
- RonnovJessen, L.; Petersen, O.W.; Bissell, M.J. Cellular changes involved in conversion of normal to malignant breast: Im-portance of the stromal reaction. Physiol. Rev. 1996, 76, 69–125. [Google Scholar] [CrossRef]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Shenouda, S.K.; Alahari, S.K. MicroRNA function in cancer: Oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009, 28, 369–378. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar] [CrossRef]
- Min, A.; Zhu, C.; Peng, S.; Rajthala, S.; Costea, D.E.; Sapkota, D. MicroRNAs as Important Players and Biomarkers in Oral Carcinogenesis. BioMed Res. Int. 2015, 2015, 1–10. [Google Scholar] [CrossRef]
- Chen, L.; Jin, H. MicroRNAs as novel biomarkers in the diagnosis of non-small cell lung cancer: A meta-analysis based on 20 studies. Tumor Biol. 2014, 35, 9119–9129. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Wang, Y.; Roth, J.; Wu, X. Abstract 298: Serum MicroRNAs as biomarkers in early stage non-small cell lung cancer. Epidemiology 2014, 74, 298. [Google Scholar] [CrossRef]
- Zahran, F.; Ghalwash, D.; Shaker, O.; Al-Johani, K.; Scully, C. Salivary microRNAs in oral cancer. Oral Dis. 2015, 21, 739–747. [Google Scholar] [CrossRef] [PubMed]
- Mlcochova, H.; Hezova, R.; Stanik, M.; Slaby, O. Urine microRNAs as potential noninvasive biomarkers in urologic cancers. Urol. Oncol. Semin. Orig. Investig. 2014, 32, 41.e1–41.e9. [Google Scholar] [CrossRef]
- Yang, J.-Y.; Sun, Y.-W.; Liu, D.-J.; Zhang, J.F.; Li, J.; Hua, R. MicroRNAs in stool samples as potential screening biomarkers for pancreatic ductal adenocarcinoma cancer. Am. J. Cancer Res. 2014, 4, 663–673. [Google Scholar]
- Hedbäck, N.; Jensen, D.H.; Specht, L.; Fiehn, A.-M.K.; Therkildsen, M.H.; Friis-Hansen, L.; Dabelsteen, E.; Von Buchwald, C. MiR-21 Expression in the Tumor Stroma of Oral Squamous Cell Carcinoma: An Independent Biomarker of Disease Free Survival. PLoS ONE 2014, 9, e95193. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Xia, Y.; Zhu, Y.; Ma, T.; Pan, C.; Wang, J.; He, Z.; Li, Z.; Qi, X.; Chen, Y. miR-204 functions as a tumor suppressor by regulating SIX1 in NSCLC. FEBS Lett. 2014, 588, 3703–3712. [Google Scholar] [CrossRef]
- Sacconi, A.; Biagioni, F.; Canu, V.; Mori, F.; Di Benedetto, A.; Lorenzon, L.; Ercolani, C.; Di Agostino, S.; Cambria, A.; Germoni, S.; et al. miR-204 targets Bcl-2 expression and enhances responsiveness of gastric cancer. Cell Death Dis. 2012, 3, e423. [Google Scholar] [CrossRef]
- Hong, B.S.; Ryu, H.S.; Kim, N.; Kim, J.; Lee, E.; Moon, H.; Kim, K.H.; Jin, M.-S.; Kwon, N.H.; Kim, S.; et al. Tumor Suppressor miRNA-204-5p Regulates Growth, Metastasis, and Immune Microenvironment Remod-eling in Breast Cancer. Cancer Res. 2019, 79, 1520–1534. [Google Scholar] [PubMed]
- Flores-Pérez, A.; Marchat, L.A.; Rodríguez-Cuevas, S.; Bautista-Piña, V.; Hidalgo-Miranda, A.; Ocampo, E.A.; Martínez, M.S.; Palma-Flores, C.; Fonseca-Sánchez, M.A.; La Vega, H.A.-D.; et al. Dual targeting of ANGPT1 and TGFBR2 genes by miR-204 controls angiogenesis in breast cancer. Sci. Rep. 2016, 6, 34504. [Google Scholar] [CrossRef] [PubMed]
- Bao, W.; Wang, H.-H.; Tian, F.-J.; He, X.-Y.; Qiu, M.-T.; Wang, J.-Y.; Zhang, H.-J.; Wang, L.H.; Wan, X.-P. A TrkB-STAT3-miR-204-5p regulatory circuitry controls proliferation and invasion of endometrial carcinoma cells. Mol. Cancer 2013, 12, 155. [Google Scholar] [CrossRef] [PubMed]
- Bharambe, H.S.; Paul, R.; Panwalkar, P.; Jalali, R.; Sridhar, E.; Gupta, T.; Moiyadi, A.; Shetty, P.; Kazi, S.; Deogharkar, A.; et al. Downregulation of miR-204 expression defines a highly aggressive subset of Group 3/Group 4 me-dulloblastomas. Acta Neuropathol. Commun. 2019, 7, 52. [Google Scholar] [CrossRef] [PubMed]
- Imam, J.S.; Plyler, J.R.; Bansal, H.; Prajapati, S.; Bansal, S.; Rebeles, J.; Chen, H.-I.H.; Chang, Y.F.; Panneerdoss, S.; Zoghi, B.; et al. Genomic Loss of Tumor Suppressor miRNA-204 Promotes Cancer Cell Migration and Invasion by Activat-ing AKT/mTOR/Rac1 Signaling and Actin Reorganization. PLoS ONE 2012, 7, e52397. [Google Scholar] [CrossRef]
- Butrym, A.; Rybka, J.; Baczyńska, D.; Tukiendorf, A.; Kuliczkowski, K.; Mazur, G. Low expression of microRNA-204 (miR-204) is associated with poor clinical outcome of acute myeloid leukemia (AML) patients. J. Exp. Clin. Cancer Res. 2015, 34, 1–5. [Google Scholar] [CrossRef]
- Ryan, J.; Tivnan, A.; Fay, J.; Bryan, K.; Meehan, M.; Creevey, L.; Lynch, J.; Bray, I.M.; O'Meara, A.; Tracey, L.; et al. MicroRNA-204 increases sensitivity of neuroblastoma cells to cisplatin and is associated with a favourable clinical outcome. Br. J. Cancer 2012, 107, 967–976. [Google Scholar] [CrossRef]
- Tsai, S.-C.; Huang, S.-F.; Chiang, J.-H.; Chen, Y.-F.; Huang, C.-C.; Tsai, M.-H.; Tsai, F.-J.; Kao, M.-C.; Yang, J.-S. The differential regulation of microRNAs is associated with oral cancer. Oncol. Rep. 2017, 38, 1613–1620. [Google Scholar] [CrossRef]
- Chattopadhyay, E.; Singh, R.; Ray, A.; Roy, R.; De Sarkar, N.; Paul, R.R.; Pal, M.; Aich, R.; Roy, B. Expression deregulation of mir31 and CXCL12 in two types of oral precancers and cancer: Im-portance in progression of precancer and cancer. Sci. Rep. 2016, 6, 32735. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Yang, X.; Huang, Y.; Fan, H.; Zhang, Q.; Wu, Y.; Li, J.; Hasina, R.; Cheng, C.; Lingen, M.W.; et al. Network Modeling Identifies Molecular Functions Targeted by miR-204 to Suppress Head and Neck Tumor Metastasis. PLoS Comput. Biol. 2010, 6, e1000730. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.-C.; Chen, P.-N.; Peng, C.-Y.; Yu, C.-H.; Chou, M.Y. Suppression of miR-204 enables oral squamous cell carcinomas to promote cancer stemness, EMT traits, and lymph node metastasis. Oncotarget 2016, 7, 20180–20192. [Google Scholar] [CrossRef]
- Shi, L.-J.; Zhang, C.-Y.; Zhou, Z.-T.; Ma, J.-Y.; Liu, Y.; Bao, Z.-X.; Jiang, W.-W. MicroRNA-155 in oral squamous cell carcinoma: Overexpression, localization, and prognostic potential. Head Neck 2014, 37, 970–976. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.-H.; Huang, X.-F.; Wang, Z.-Y.; Han, W.; Deng, R.-Z.; Mou, Y.-B.; Ding, L.; Hou, Y.-Y.; Hu, Q.-G. Upregulation of a potential prognostic biomarker, miR-155, enhances cell proliferation in patients with oral squamous cell carcinoma. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2014, 117, 227–233. [Google Scholar] [CrossRef]
- Concato, J.; Peduzzi, P.; Holford, T.R.; Feinstein, A.R. The Importance of Events Per Independent Variable in Proportional Hazards and Other Multivariable Analyses. I. Background, goals, and general strategy. J. Clin. Epidemiol. 1995, 48, 1495–1501. [Google Scholar] [CrossRef]
- Altman, D.G.; McShane, L.M.; Sauerbrei, W.; Taube, S.E. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): Explanation and elabo-ration. PLoS Med. 2012, 9, e1001216. [Google Scholar] [CrossRef]
- Woolgar, J.A.; Scott, J. Prediction of cervical lymph node metastasis in squamous cell carcinoma of the tongue/floor of mouth. Head Neck 1995, 17, 463–472. [Google Scholar] [CrossRef]
- Almangush, A.; Bello, I.O.; Keski–Säntti, H.; Mäkinen, L.K.; Kauppila, J.H.; Pukkila, M.; Hagström, J.; Laranne, J.; Tommola, S.; Nieminen, O.; et al. Depth of invasion, tumor budding, and worst pattern of invasion: Prognostic indicators in early-stage oral tongue cancer. Head Neck 2013, 36, 811–818. [Google Scholar] [CrossRef]
- Brandwein-Gensler, A.; Teixeira, M.S.; Lewis, C.M.; Lee, B.; Rolnitzky, L.; Hille, J.J.; Genden, E.; Urken, M.L.; Wang, B.Y. Oral squamous cell carcinoma—Histologic risk assessment, but not margin status, is strongly predictive of local disease-free and overall survival. Am. J. Surg. Pathol. 2005, 29, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Sempere, L.F.; Preis, M.; Yezefski, T.; Ouyang, H.; Suriawinata, A.A.; Silahtaroglu, A.; Conejo-Garcia, J.R.; Kauppinen, S.; Wells, W.; Korc, M. Fluorescence-Based Codetection with Protein Markers Reveals Distinct Cellular Compartments for Al-tered MicroRNA Expression in Solid Tumors. Clin. Cancer Res. 2010, 16, 4246–4255. [Google Scholar] [CrossRef]
- Chaudhuri, A.D.; Yelamanchili, S.V.; Fox, H.S. Combined fluorescent in situ hybridization for detection of microRNAs and immunofluorescent labeling for cell-type markers. Front. Cell. Neurosci. 2013, 7, 160. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, B.S.; Holmstrøm, K. Combined MicroRNA In Situ Hybridization and Immunohistochemical Detection of Protein Markers. Methods Mol. Biol. 2013, 986, 353–365. [Google Scholar] [CrossRef]
- Nuovo, G.J. In situ detection of microRNAs in paraffin embedded, formalin fixed tissues and the co-localization of their putative targets. Methods 2010, 52, 307–315. [Google Scholar] [CrossRef]
- Ruifrok, A.C.; Johnston, D.A. Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Histol. 2001, 23, 291–299. [Google Scholar] [PubMed]
- Li, J.; Huang, H.; Sun, L.; Yang, M.; Pan, C.; Chen, W.; Wu, D.; Lin, Z.; Zeng, C.; Yao, Y.; et al. MiR-21 Indicates Poor Prognosis in Tongue Squamous Cell Carcinomas as an Apoptosis Inhibitor. Clin. Cancer Res. 2009, 15, 3998–4008. [Google Scholar] [CrossRef] [PubMed]
- Irimie-Aghiorghiesei, A.I.; Pop-Bica, C.; Pintea, S.; Braicu, C.; Cojocneanu, R.; Zimța, A.-A.; Gulei, D.; Slabý, O.; Berindan-Neagoe, I. Prognostic Value of MiR-21: An Updated Meta-Analysis in Head and Neck Squamous Cell Carcinoma (HNSCC). J. Clin. Med. 2019, 8, 2041. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Yang, J.M.; Ahn, S.-H.; Jeong, W.-J.; Chung, J.-H.; Paik, J.H. Potential Oncogenic Role and Prognostic Implication of MicroRNA-155-5p in Oral Squamous Cell Carcinoma. Anticancer. Res. 2018, 38, 5193–5200. [Google Scholar] [CrossRef]
- Parkin, D.M.; Pisani, P.; Ferlay, J. Global cancer statistics. CA A Cancer J. Clin. 1999, 49, 33–64. [Google Scholar] [CrossRef]
- Lo, W.-L.; Kao, S.-Y.; Chi, L.-Y.; Wong, Y.-K.; Chang, R.C.-S. Outcomes of oral squamous cell carcinoma in Taiwan after surgical therapy: Factors affecting survival. J. Oral Maxillofac. Surg. 2003, 61, 751–758. [Google Scholar] [CrossRef]
- Ding, M.; Lin, B.; Li, T.; Liu, Y.; Li, Y.; Zhou, X.; Miao, M.; Gu, J.; Pan, H.; Yang, F.; et al. A dual yet opposite growth-regulating function of miR-204 and its target XRN1 in prostate adenocarcinoma cells and neuroendocrine-like prostate cancer cells. Oncotarget 2015, 6, 7686–7700. [Google Scholar] [CrossRef]

| Parameters | N (%) | Overall Survival | Recurrence Free Survival | ||
|---|---|---|---|---|---|
| p-Value | HR (95% CI) | p-Value | HR (95% CI) | ||
| miR-204_TF | 0.04 | 0.036 | |||
| Low | 67 (41.9) | 1 | 1 | ||
| High | 65 (40.8) | 0.657 (0.44–0.98) | 0.56 (0.33–0.96) | ||
| miR-204_TC | 0.234 | 0.245 | |||
| Low | 79 (49.4) | 1 | 1 | ||
| High | 79 (49.4) | 0.804 (0.56–1.15) | 0.75 (80.46–1.22) | ||
| Age (years) | 0.003 | 0.333 | |||
| ≤65 | 85 (53.1) | 1 | 1 | ||
| >65 | 74 (46.3) | 1.73 (1.20–2.48) | 0.27 (0.78–2.08) | ||
| Gender | 0.296 | 0.191 | |||
| Female | 58 (36.3) | 1 | 1 | ||
| Male | 102 (60.5) | 1.22 (0.84–0.18) | 1.42 (0.84–2.38) | ||
| Alcohol | 0.137 | 0.51 | |||
| Low-Normal | 51 (31.9) | 1 | 1 | ||
| Moderate-High | 35 (21.9) | 1.47 (0.89–2.43) | 1.23 (0.63–2.55) | ||
| Smoking | 0.44 | 0.287 | |||
| No | 49 (30.6) | 1 | 1 | ||
| Yes | 75 (46.9) | 1.18 (0.78–1.79) | 1.37 (0.77–2.46) | ||
| Tumor site | 0.063 | 0.591 | |||
| Tongue | 71 (44.4) | 1 | 1 | ||
| Gingiva | 42 (26.3) | 1.50 (0.98–2.30) | 1.10 (0.77–1.56) | ||
| Stage | 0.001 | 0.004 | |||
| Early (1&2) | 76 (47.5) | 1 | 1 | ||
| Late (3&4) | 84 (52.5) | 1.90 (1.32–2.75) | 2.11 (1.27–3.50) | ||
| T stage | 0.005 | 0.032 | |||
| T1 | 1 | 1 | |||
| T2 | 0.002 | 1.90 (1.13–3.21) | 0.107 | 1.82 (0.88–3.77) | |
| T3 | 0.104 | 1.63 (0.90–2.929 | 0.178 | 1.76 (0.77–4.02) | |
| T4 | 0 | 2.47 (1.50–4.07) | 0.003 | 2.86 (1.42–5.77) | |
| Lymph node | 0.031 | 0.108 | |||
| No metastasis | 112 870) | 1 | 1 | ||
| Metastasis | 48 (30) | 1.51 (1.04–2.2) | 1.52 (0.91–2.54) | ||
| Distant metastasis | 0.113 | 0.662 | |||
| No | 139 (86.9) | 1 | 1 | ||
| Yes | 21 (13.1) | 0.677 (0.42–0.10) | 1.19 (0.54–2.61) | ||
| Depth of invasion | 0.21 | 0.261 | |||
| Superficial (<4mm) | 41 (25.6) | 1 | 1 | ||
| Deep (≥4mm) | 46 (28.7) | 1.376 (0.84–0.27) | 1.51 (0.74–3.10) | ||
| Tumor budding score | 0.647 | 0.19 | |||
| Low (<5 buds) | 72 (45) | 1 | 1 | ||
| High (≥5 buds) | 58 (36) | 1.097 (0.74–1.63) | 1.43 (0.84–2.43) | ||
| Histological degree of differentiation | |||||
| Well diff | 72 (45) | 0.053 | 1 | 0.392 | 1 |
| Poor diff | 58 (36) | 1.55 (0.99–2.4) | 1.32 (0.70–2.47) | ||
| Worst pattern of invasion | |||||
| Type 1–3 | 19 (11.9) | 0.578 | 1 | 0.187 | 1 |
| Type 4 | 111 (69.4) | 1.47(0.89–2.43) | 1.99 (0.72–5.50) | ||
| Parameters | N (%) | a Overall Survival | b Recurrence Free Survival | ||
|---|---|---|---|---|---|
| p-Value | HR (95% CI) | p-Value | HR (95% CI) | ||
| miR-204_TF | 0.048 | 0.033 | |||
| Low | 67 (41.9) | 1 | 1 | ||
| High | 65 (40.8) | 0.668 (0.45–1.00) | 0.55(0.32–0.95) | ||
| miR-204_TC | 0.26 | 0.193 | |||
| Low | 79 (49.4) | 1 | 1 | ||
| High | 79 (49.4) | 0.812 (0.56–1.17) | 0.72 (0.44–1.22) | ||
| Age (years) | 0.004 | ||||
| ≤65 | 85 (53.1) | 1 | |||
| >65 | 74 (46.3) | 1.80 (1.20–2.70) | |||
| Stage | 0.005 | 0.004 | |||
| Early (1&2) | 76 (47.5) | 1 | 1 | ||
| Late (3&4) | 84 (52.5) | 1.78 (1.12–2.67) | 2.11 (1.27–3.50) | ||
| Lymph node | 0.108 | ||||
| No metastasis | 112 870) | 1 | |||
| Metastasis | 48 (30) | 1.057 (1.04–2.2) | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajthala, S.; Dongre, H.; Parajuli, H.; Min, A.; Nginamau, E.S.; Kvalheim, A.; Lybak, S.; Sapkota, D.; Johannessen, A.C.; Costea, D.E. Combined In Situ Hybridization and Immunohistochemistry on Archival Tissues Reveals Stromal microRNA-204 as Prognostic Biomarker for Oral Squamous Cell Carcinoma. Cancers 2021, 13, 1307. https://doi.org/10.3390/cancers13061307
Rajthala S, Dongre H, Parajuli H, Min A, Nginamau ES, Kvalheim A, Lybak S, Sapkota D, Johannessen AC, Costea DE. Combined In Situ Hybridization and Immunohistochemistry on Archival Tissues Reveals Stromal microRNA-204 as Prognostic Biomarker for Oral Squamous Cell Carcinoma. Cancers. 2021; 13(6):1307. https://doi.org/10.3390/cancers13061307
Chicago/Turabian StyleRajthala, Saroj, Harsh Dongre, Himalaya Parajuli, Anjie Min, Elisabeth Sivy Nginamau, Arild Kvalheim, Stein Lybak, Dipak Sapkota, Anne Christine Johannessen, and Daniela Elena Costea. 2021. "Combined In Situ Hybridization and Immunohistochemistry on Archival Tissues Reveals Stromal microRNA-204 as Prognostic Biomarker for Oral Squamous Cell Carcinoma" Cancers 13, no. 6: 1307. https://doi.org/10.3390/cancers13061307
APA StyleRajthala, S., Dongre, H., Parajuli, H., Min, A., Nginamau, E. S., Kvalheim, A., Lybak, S., Sapkota, D., Johannessen, A. C., & Costea, D. E. (2021). Combined In Situ Hybridization and Immunohistochemistry on Archival Tissues Reveals Stromal microRNA-204 as Prognostic Biomarker for Oral Squamous Cell Carcinoma. Cancers, 13(6), 1307. https://doi.org/10.3390/cancers13061307






