A Drastic Shift in Lipid Adducts in Colon Cancer Detected by MALDI-IMS Exposes Alterations in Specific K+ Channels
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Colon Biopsy Collection
2.2. Sample Preparation for MALDI-Imaging and Data Analysis
2.3. Cell Culture Experiments
2.3.1. Cells and Reagents
2.3.2. Pharmacological Inhibition of K+ Channels
2.3.3. Clonogenic Survival Assay
2.4. Interrogation of CRC Patient Gene Expression Data Sets
2.4.1. Interrogation of Patient-Derived CRC Biopsies
2.4.2. Interrogation of Macro- and Micro-Dissected Datasets
2.4.3. Interrogation of Fluorescent Activated Cell Sorting CRC Cells Datasets
2.4.4. Patient Survival Analysis and CMS Association
2.5. Statistics
3. Results
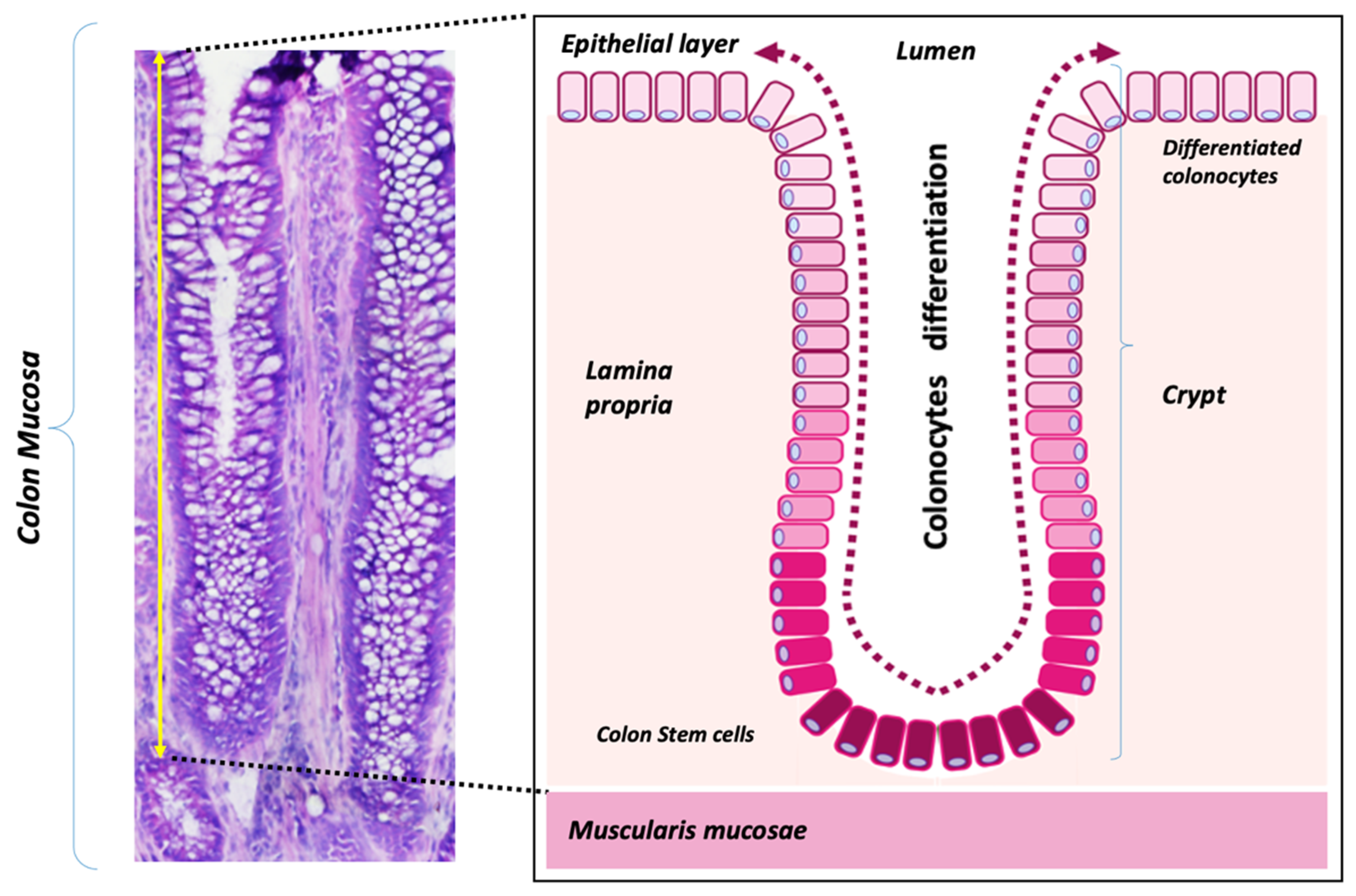
3.1. Colon Mucosa Malignization Induces a Drastic Shift in the [Na+]/[K+] Adduct Ratio
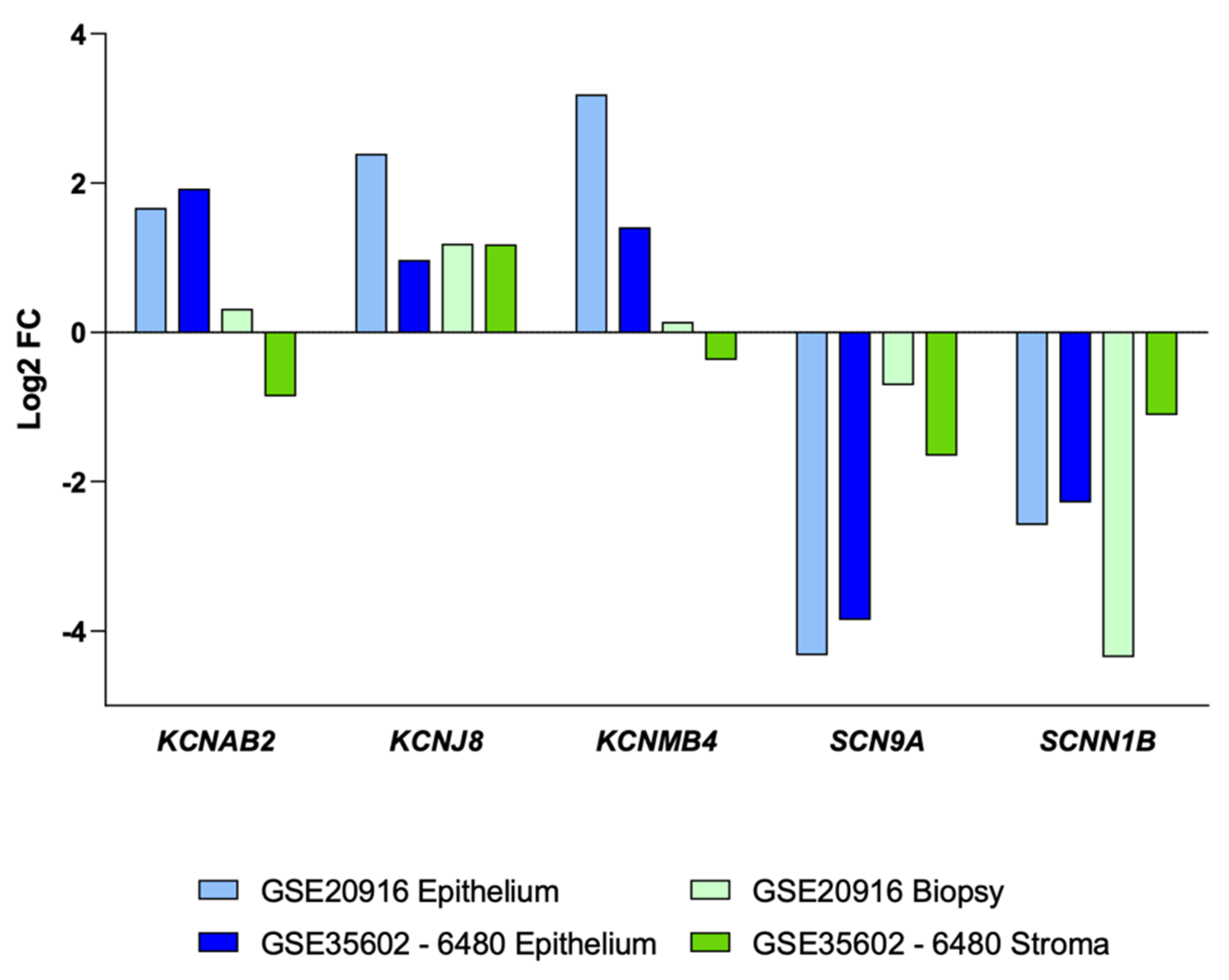
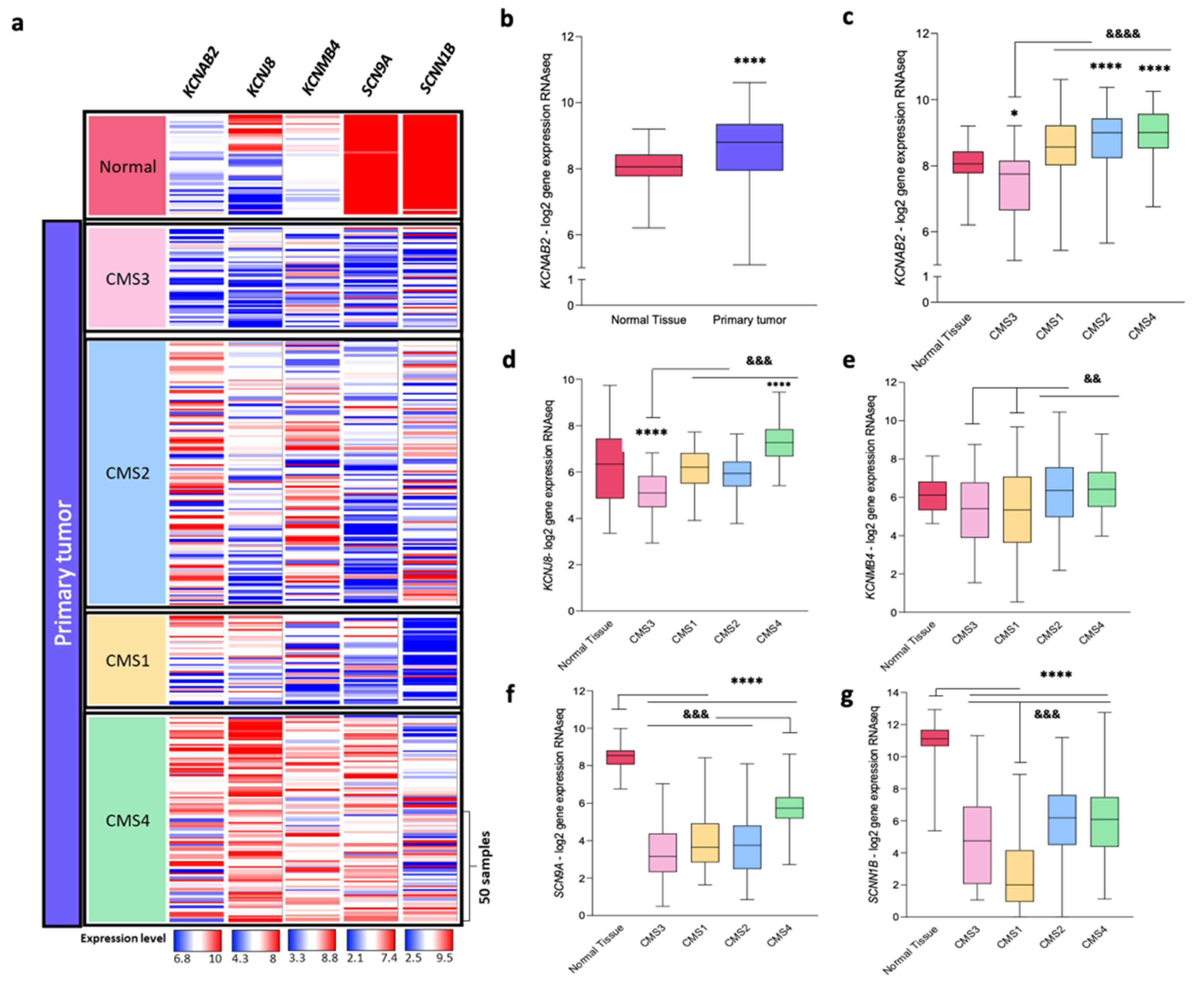
3.2. Impact of Colon Cancer on K+ and Na+ Channels at the Transcriptional Level
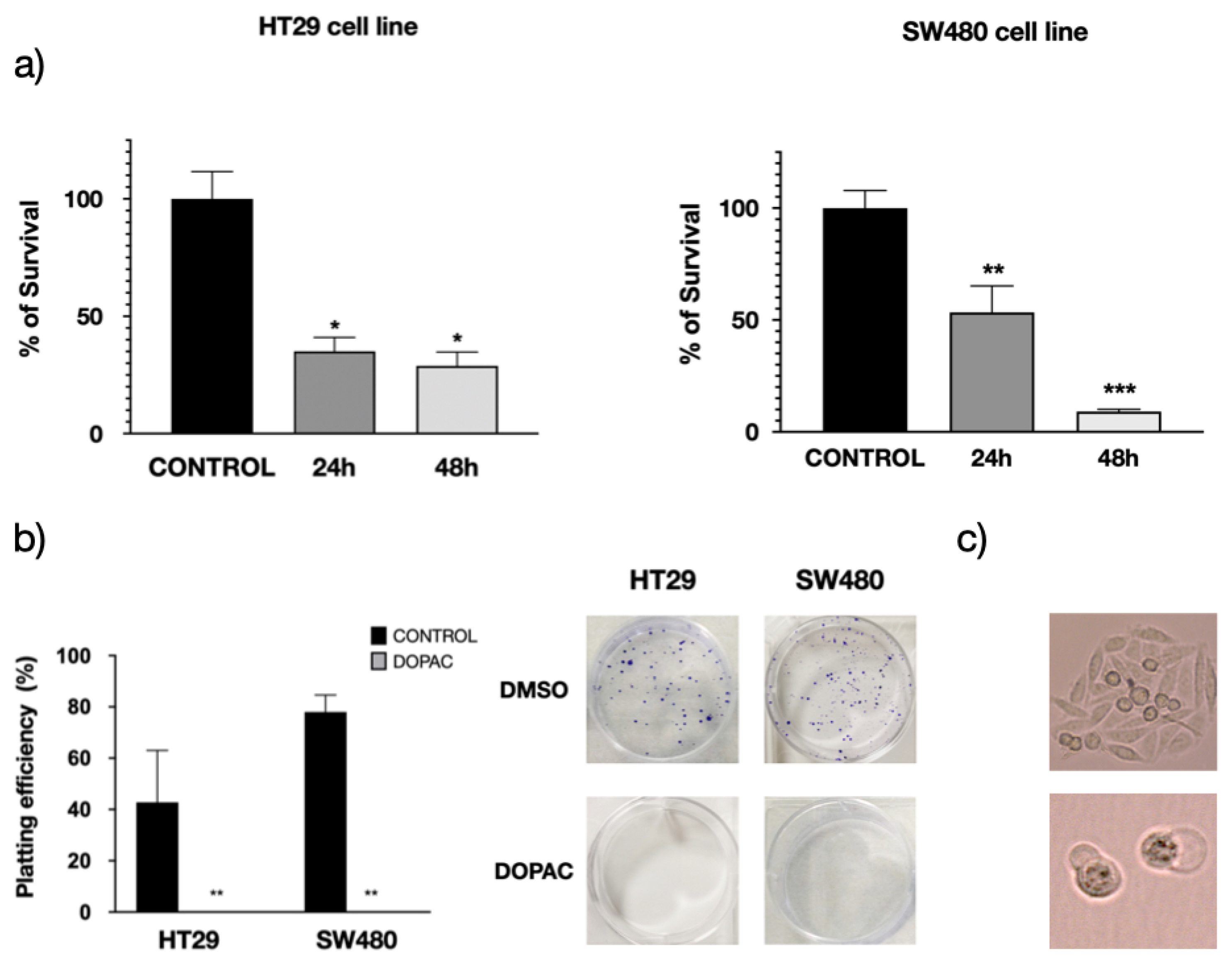
3.3. Pharmacological Inhibition of KCNAB2 Drastically Halts Colon Cancer Proliferation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Araghi, M.; Soerjomataram, I.; Bardot, A.; Ferlay, J.; Cabasag, C.J.; Morrison, D.S.; De, P.; Tervonen, H.; Walsh, P.M.; Bucher, O.; et al. Changes in colorectal cancer incidence in seven high-income countries: A population-based study. Lancet Gastroenterol. Hepatol. 2019, 4, 511–518. [Google Scholar] [CrossRef]
- Meyerson, M.; Gabriel, S.; Getz, G. Advances in understanding cancer genomes through second-generation sequencing. Nat. Rev. Genet. 2010, 11, 685–696. [Google Scholar] [CrossRef]
- Mouradov, D.; Domingo, E.; Gibbs, P.; Jorissen, R.N.; Li, S.; Soo, P.Y.; Lipton, L.; Desai, J.; Danielsen, H.E.; Oukrif, D.; et al. Survival in stage II/III colorectal cancer is independently predicted by chromosomal and microsatellite instability, but not by specific driver mutations. Am. J. Gastroenterol. 2013, 108, 1785–1793. [Google Scholar] [CrossRef]
- Carethers, J.M.; Jung, B.H. Genetics and Genetic Biomarkers in Sporadic Colorectal Cancer. Gastroenterology 2015, 149, 1177–1190.e3. [Google Scholar] [CrossRef] [PubMed]
- Samowitz, W.S.; Sweeney, C.; Herrick, J.; Albertsen, H.; Levin, T.R.; Murtaugh, M.A.; Wolff, R.K.; Slattery, M.L. Poor Survival Associated with the BRAF V600E Mutation in Microsatellite-Stable Colon Cancers. Cancer Res. 2005, 65, 6063–6069. [Google Scholar] [CrossRef]
- Melo, F.D.S.E.; Wang, X.; Jansen, M.; Fessler, E.; Trinh, A.; De Rooij, L.P.M.H.; De Jong, J.H.; De Boer, O.J.; Van Leersum, R.; Bijlsma, M.F.; et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat. Med. 2013, 19, 614–618. [Google Scholar] [CrossRef] [PubMed]
- Marisa, L.; de Reyniès, A.; Duval, A.; Selves, J.; Gaub, M.P.; Vescovo, L.; Etienne-Grimaldi, M.-C.; Schiappa, R.; Guenot, D.; Ayadi, M.; et al. Gene Expression Classification of Colon Cancer into Molecular Subtypes: Characterization, Validation, and Prognostic Value. PLoS Med. 2013, 10, e1001453. [Google Scholar] [CrossRef] [PubMed]
- Fontana, E.; Eason, K.; Cervantes, A.; Salazar, R.; Sadanandam, A. Context matters—consensus molecular subtypes of colorectal cancer as biomarkers for clinical trials. Ann. Oncol. 2019, 30, 520–527. [Google Scholar] [CrossRef]
- Calon, A.; Lonardo, E.; Berenguer-Llergo, A.; Espinet, E.; Hernando-Momblona, X.; Iglesias, M.; Sevillano, M.; Palomo-Ponce, S.; Tauriello, D.V.F.; Byrom, D.; et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat. Genet. 2015, 47, 320–329. [Google Scholar] [CrossRef]
- Tauriello, D.V.F.; Palomo-Ponce, S.; Stork, D.; Berenguer-Llergo, A.; Badia-Ramentol, J.; Iglesias, M.; Sevillano, M.; Ibiza, S.; Cañellas, A.; Hernando-Momblona, X.; et al. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 2018, 554, 538–543. [Google Scholar] [CrossRef]
- Korte, A.R.; Yandeau-Nelson, M.D.; Nikolau, B.J.; Lee, Y.J.I. Subcellular-level resolution MALDI-MS imaging of maize leaf metabolites by MALDI-linear ion trap-Orbitrap mass spectrometer. Anal. Bioanal. Chem. 2015, 407, 2301–2309. [Google Scholar] [CrossRef]
- Boggio, K.J.; Obasuyi, E.; Sugino, K.; Nelson, S.B.; Agar, N.Y.; Agar, J.N. Recent advances in single-cell MALDI mass spectrometry imaging and potential clinical impact. Expert Rev. Proteom. 2011, 8, 591–604. [Google Scholar] [CrossRef]
- Lagarrigue, M.; Becker, M.; Lavigne, R.; Deininger, S.-O.; Walch, A.; Aubry, F.; Suckau, D.; Pineau, C. Revisiting Rat Spermatogenesis with MALDI Imaging at 20-μm Resolution. Mol. Cell. Proteom. 2011, 10, M110.005991. [Google Scholar] [CrossRef]
- Bestard-Escalas, J.; Maimó-Barceló, A.; Pérez-Romero, K.; Lopez, D.H.; Barceló-Coblijn, G. Ins and Outs of Interpreting Lipidomic Results. J. Mol. Biol. 2019, 431, 5039–5062. [Google Scholar] [CrossRef]
- Harayama, T.; Riezman, H. Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol. 2018, 19, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Barceló-Coblijn, G.; Fernández, J.A. Mass spectrometry coupled to imaging techniques: The better the view the greater the challenge. Front. Physiol. 2015, 6, 3. [Google Scholar] [CrossRef]
- Garate, J.; Fernández, R.; Lage, S.; Bestard-Escalas, J.; Lopez, D.H.; Reigada, R.; Khorrami, S.; Ginard, D.; Reyes, J.; Amengual, I.; et al. Imaging mass spectrometry increased resolution using 2-mercaptobenzothiazole and 2,5-diaminonaphtalene matrices: Application to lipid distribution in human colon. Anal. Bioanal. Chem. 2015, 407, 4697–4708. [Google Scholar] [CrossRef] [PubMed]
- Bestard-Escalas, J.; Garate, J.; Maimó-Barceló, A.; Fernández, R.; Lopez, D.H.; Lage, S.; Reigada, R.; Khorrami, S.; Ginard, D.; Reyes, J.; et al. Lipid fingerprint image accurately conveys human colon cell pathophysiologic state: A solid candidate as biomarker. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2016, 1861, 1942–1950. [Google Scholar] [CrossRef]
- Lopez, D.H.; Bestard-Escalas, J.; Garate, J.; Maimó-Barceló, A.; Fernández, R.; Reigada, R.; Khorrami, S.; Ginard, D.; Okazaki, T.; Fernández, J.A.; et al. Tissue-selective alteration of ethanolamine plasmalogen metabolism in dedifferentiated colon mucosa. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 928–938. [Google Scholar] [CrossRef] [PubMed]
- Garate, J.; Lage, S.; Martín-Saiz, L.; Perez-Valle, A.; Ochoa, B.; Boyano, M.D.; Fernández, R.; Fernández, J.A. Influence of Lipid Fragmentation in the Data Analysis of Imaging Mass Spectrometry Experiments. J. Am. Soc. Mass Spectrom. 2020, 31, 517–526. [Google Scholar] [CrossRef]
- Fernández, R.; González, P.; Lage, S.; Garate, J.; Maqueda, A.; Marcaida, I.; Maguregui, M.; Ochoa, B.; Rodríguez, F.J.; Fernández, J.A. Influence of the Cation Adducts in the Analysis of Matrix-Assisted Laser Desorption Ionization Imaging Mass Spectrometry Data from Injury Models of Rat Spinal Cord. Anal. Chem. 2017, 89, 8565–8573. [Google Scholar] [CrossRef]
- Fernández, R.; Garate, J.; Lage, S.; Terés, S.; Higuera, M.; Bestard-Escalas, J.; López, D.H.; Guardiola-Serrano, F.; Escribá, P.V.; Barceló-Coblijn, G.; et al. Identification of Biomarkers of Necrosis in Xenografts Using Imaging Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2016, 27, 244–254. [Google Scholar] [CrossRef]
- Fernández, R.; Lage, S.; Abad-García, B.; Barceló-Coblijn, G.; Terés, S.; López, D.H.; Guardiola-Serrano, F.; Martín, M.L.; Escribá, P.V.; Fernández, J.A. Analysis of the Lipidome of Xenografts Using MALDI-IMS and UHPLC-ESI-QTOF. J. Am. Soc. Mass Spectrom. 2014, 25, 1237–1246. [Google Scholar] [CrossRef]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Astigarraga, E.; Barreda-Gómez, G.; Lombardero, L.; Fresnedo, O.; Castaño, F.; Giralt, M.T.; Ochoa, B.; Rodríguez-Puertas, R.; Fernández, J.A. Profiling and Imaging of Lipids on Brain and Liver Tissue by Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Using 2-Mercaptobenzothiazole as a Matrix. Anal. Chem. 2008, 80, 9105–9114. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Xu, W.; Eberlin, L.S.; Wiseman, J.M.; Fang, X.; Jiang, Y.; Huang, Z.; Zhang, Y.; Cooks, R.G.; Ouyang, Z. Data Processing for 3D Mass Spectrometry Imaging. J. Am. Soc. Mass Spectrom. 2012, 23, 1147–1156. [Google Scholar] [CrossRef][Green Version]
- Fernández, R.; Carriel, V.; Lage, S.; Garate, J.; Díez-García, J.; Ochoa, B.; Castro, B.; Alaminos, M.; Fernández, J.A. Deciphering the Lipid Architecture of the Rat Sciatic Nerve Using Imaging Mass Spectrometry. ACS Chem. Neurosci. 2016, 7, 624–632. [Google Scholar] [CrossRef] [PubMed]
- Fernández, R.; Garate, J.; Lage, S.; Terés, S.; Higuera, M.; Bestard-Escalas, J.; Martin, M.L.; López, D.H.; Guardiola-Serrano, F.; Escribá, P.V.; et al. Optimized Protocol to Analyze Changes in the Lipidome of Xenografts after Treatment with 2-Hydroxyoleic Acid. Anal. Chem. 2016, 88, 1022–1029. [Google Scholar] [CrossRef]
- Barrett, T.; Edgar, R. Gene Expression Omnibus: Microarray Data Storage, Submission, Retrieval, and Analysis. Methods Enzymol. 2006, 411, 352–369. [Google Scholar] [PubMed]
- Skrzypczak, M.; Goryca, K.; Rubel, T.; Paziewska, A.; Mikula, M.; Jarosz, D.; Pachlewski, J.; Oledzki, J.; Ostrowsk, J. Modeling Oncogenic Signaling in Colon Tumors by Multidirectional Analyses of Microarray Data Directed for Maximization of Analytical Reliability. PLoS ONE 2010, 5, e13091. [Google Scholar] [CrossRef]
- Nishida, N.; Nagahara, M.; Sato, T.; Mimori, K.; Sudo, T.; Tanaka, F.; Shibata, K.; Ishii, H.; Sugihara, K.; Doki, Y.; et al. Microarray Analysis of Colorectal Cancer Stromal Tissue Reveals Upregulation of Two Oncogenic miRNA Clusters. Clin. Cancer Res. 2012, 18, 3054–3070. [Google Scholar] [CrossRef]
- Smyth, G.K. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3, 3. [Google Scholar] [CrossRef] [PubMed]
- Smyth, G.K. limma: Linear Models for Microarray Data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Springer: New York, NY, USA, 2005; pp. 397–420. [Google Scholar]
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Litan, A.; Langhans, S.A. Cancer as a channelopathy: Ion channels and pumps in tumor development and progression. Front. Cell. Neurosci. 2015, 9, 86. [Google Scholar] [CrossRef]
- Breuer, E.-K.; Fukushiro-Lopes, D.; Dalheim, A.; Burnette, M.; Zartman, J.; Kaja, S.; Wells, C.; Campo, L.; Curtis, K.J.; Romero-Moreno, R.; et al. Potassium channel activity controls breast cancer metastasis by affecting β-catenin signaling. Cell Death Dis. 2019, 10, 180. [Google Scholar] [CrossRef] [PubMed]
- Chao, C.; Carmical, J.R.; Ives, K.L.; Wood, T.G.; Aronson, J.F.; Gomez, G.A.; Djukom, C.D.; Hellmich, M.R. CD133+ colon cancer cells are more interactive with the tumor microenvironment than CD133− cells. Lab. Investig. 2012, 92, 420–436. [Google Scholar] [CrossRef]
- Alka, K.; Dolly, J.O.; Ryan, B.J.; Henehan, G.T.M. New inhibitors of the Kvβ2 subunit from mammalian Kv1 potassium channels. Int. J. Biochem. Cell Biol. 2014, 55, 35–39. [Google Scholar] [CrossRef]
- Eldrup, E.; Clausen, N.; Scherling, B.; Schmiegelow, K. Evaluation of plasma 3,4-dihydroxyphenylacetic acid (DOPAC) and plasma 3,4-dihydroxyphenylalanine (DOPA) as tumor markers in children with neuroblastoma. Scand. J. Clin. Lab. Investig. 2001, 61, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Westerink, B.H.C.; Korf, J. Regional rat brain levels of 3,4-dihydroxyphenylacetic acid and homovanillic acid: Concurrent fluorometric measurement and influence of drugs. Eur. J. Pharmacol. 1976, 38, 281–291. [Google Scholar] [CrossRef]
- Catalán, M.; Ferreira, J.; Carrasco-Pozo, C. The Microbiota-Derived Metabolite of Quercetin, 3,4-Dihydroxyphenylacetic Acid Prevents Malignant Transformation and Mitochondrial Dysfunction Induced by Hemin in Colon Cancer and Normal Colon Epithelia Cell Lines. Molecules 2020, 25, 4138. [Google Scholar] [CrossRef] [PubMed]
- Zabela, V.; Sampath, C.; Oufir, M.; Butterweck, V.; Hamburger, M. Single dose pharmacokinetics of intravenous 3,4-dihydroxyphenylacetic acid and 3-hydroxyphenylacetic acid in rats. Fitoterapia 2020, 142, 104526. [Google Scholar] [CrossRef]
- Comes, N.; Bielanska, J.; Vallejo-Gracia, A.; Serrano-Albarrás, A.; Marruecos, L.; Gómez, D.; Soler, C.; Condom, E.; Ramón y Cajal, S.; Hernández-Losa, J.; et al. The voltage-dependent K+ channels Kv1.3 and Kv1.5 in human cancer. Front. Physiol. 2013, 4, 282. [Google Scholar] [CrossRef]
- Comes, N.; Serrano-Albarrás, A.; Capera, J.; Serrano-Novillo, C.; Condom, E.; Ramón y Cajal, S.; Ferreres, J.C.; Felipe, A. Involvement of potassium channels in the progression of cancer to a more malignant phenotype. Biochim. Biophys. Acta-Biomembr. 2015, 1848, 2477–2492. [Google Scholar] [CrossRef] [PubMed]
- Abdul, M.; Hoosein, N. Voltage-gated potassium ion channels in colon cancer. Oncol. Rep. 2002, 9, 961–964. [Google Scholar] [CrossRef] [PubMed]
- Maimó-Barceló, A.; Garate, J.; Bestard-Escalas, J.; Fernández, R.; Berthold, L.; Lopez, D.H.; Fernández, J.A.; Barceló-Coblijn, G. Confirmation of sub-cellular resolution using oversampling imaging mass spectrometry. Anal. Bioanal. Chem. 2019, 411, 7935–7941. [Google Scholar] [CrossRef] [PubMed]
- Dória, M.L.; McKenzie, J.S.; Mroz, A.; Phelps, D.L.; Speller, A.; Rosini, F.; Strittmatter, N.; Golf, O.; Veselkov, K.; Brown, R.; et al. Epithelial ovarian carcinoma diagnosis by desorption electrospray ionization mass spectrometry imaging. Sci. Rep. 2016, 6, 39219. [Google Scholar] [CrossRef]
- Guo, S.; Qiu, L.; Wang, Y.; Qin, X.; Liu, H.; He, M.; Zhang, Y.; Li, Z.; Chen, X. Tissue imaging and serum lipidomic profiling for screening potential biomarkers of thyroid tumors by matrix-assisted laser desorption/ionization- Fourier transform ion cyclotron resonance mass spectrometry. Anal. Bioanal. Chem. 2014, 406, 4357–4370. [Google Scholar] [CrossRef]
- Uchiyama, Y.; Hayasaka, T.; Masaki, N.; Watanabe, Y.; Masumoto, K.; Nagata, T.; Katou, F.; Setou, M. Imaging mass spectrometry distinguished the cancer and stromal regions of oral squamous cell carcinoma by visualizing phosphatidylcholine (16:0/16:1) and phosphatidylcholine (18:1/20:4). Anal. Bioanal. Chem. 2014, 406, 1307–1316. [Google Scholar] [CrossRef]
- Tata, A.; Woolman, M.; Ventura, M.; Bernards, N.; Ganguly, M.; Gribble, A.; Shrestha, B.; Bluemke, E.; Ginsberg, H.J.; Vitkin, A.; et al. Rapid Detection of Necrosis in Breast Cancer with Desorption Electrospray Ionization Mass Spectrometry. Sci. Rep. 2016, 6, 35374. [Google Scholar] [CrossRef] [PubMed]
- Eberlin, L.S.; Norton, I.; Dill, A.L.; Golby, A.J.; Ligon, K.L.; Santagata, S.; Cooks, R.G.; Agar, N.Y.R. Classifying Human Brain Tumors by Lipid Imaging with Mass Spectrometry. Cancer Res. 2012, 72, 645–654. [Google Scholar] [CrossRef]
- Patterson, N.H.; Alabdulkarim, B.; Lazaris, A.; Thomas, A.; Marcinkiewicz, M.M.; Gao, Z.H.; Vermeulen, P.B.; Chaurand, P.; Metrakos, P. Assessment of pathological response to therapy using lipid mass spectrometry imaging. Sci. Rep. 2016, 6, 26814. [Google Scholar] [CrossRef] [PubMed]
- Van Nuffel, S.; Elie, N.; Yang, E.; Nouet, J.; Touboul, D.; Chaurand, P.; Brunelle, A. Insights into the MALDI Process after Matrix Deposition by Sublimation Using 3D ToF-SIMS Imaging. Anal. Chem. 2018, 90, 1907–1914. [Google Scholar] [CrossRef]
- Rao, V.; Perez-Neut, M.; Kaja, S.; Gentile, S. Voltage-Gated Ion Channels in Cancer Cell Proliferation. Cancers 2015, 7, 849–875. [Google Scholar] [CrossRef] [PubMed]
- Schwab, A.; Stock, C. Ion channels and transporters in tumour cell migration and invasion. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130102. [Google Scholar] [CrossRef]
- Turner, K.L.; Sontheimer, H. Cl− and K+ channels and their role in primary brain tumour biology. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130095. [Google Scholar] [CrossRef]
- Pchelintseva, E.; Djamgoz, M.B.A. Mesenchymal stem cell differentiation: Control by calcium-activated potassium channels. J. Cell. Physiol. 2018, 233, 3755–3768. [Google Scholar] [CrossRef]
- Restrepo-Angulo, I.; Sánchez-Torres, C.; Camacho, J. Human EAG1 potassium channels in the epithelial-to-mesenchymal transition in lung cancer cells. Anticancer. Res. 2011, 31, 1265–1270. [Google Scholar] [PubMed]
- Hernández-Reséndiz, I.; Pacheu-Grau, D.; Sánchez, A.; Pardo, L.A. Inhibition of Kv10.1 Channels Sensitizes Mitochondria of Cancer Cells to Antimetabolic Agents. Cancers 2020, 12, 920. [Google Scholar] [CrossRef]
- Sontheimer, H. An Unexpected Role for Ion Channels in Brain Tumor Metastasis. Exp. Biol. Med. 2008, 233, 779–791. [Google Scholar] [CrossRef]
- Hatten, M.E.; Roussel, M.F. Development and cancer of the cerebellum. Trends Neurosci. 2011, 34, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Pardo, L.A.; Stühmer, W. The roles of K+ channels in cancer. Nat. Rev. Cancer 2014, 14, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Zhang, M.; Liu, W. Hypermethylated KCNQ1 acts as a tumor suppressor in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2018, 503, 3100–3107. [Google Scholar] [CrossRef] [PubMed]
- Goto, T.; Terada, N.; Inoue, T.; Kobayashi, T.; Nakayama, K.; Okada, Y.; Yoshikawa, T.; Miyazaki, Y.; Uegaki, M.; Utsunomiya, N.; et al. Decreased expression of lysophosphatidylcholine (16:0/OH) in high resolution imaging mass spectrometry independently predicts biochemical recurrence after surgical treatment for prostate cancer. Prostate 2015, 75, 1821–1830. [Google Scholar] [CrossRef] [PubMed]
- Hinsenkamp, I.; Schulz, S.; Roscher, M.; Suhr, A.M.; Meyer, B.; Munteanu, B.; Fuchser, J.; Schoenberg, S.O.; Ebert, M.P.A.; Wängler, B.; et al. Inhibition of Rho-Associated Kinase 1/2 Attenuates Tumor Growth in Murine Gastric Cancer. Neoplasia 2016, 18, 500–511. [Google Scholar] [CrossRef] [PubMed]
- Kurabe, N.; Hayasaka, T.; Ogawa, M.; Masaki, N.; Ide, Y.; Waki, M.; Nakamura, T.; Kurachi, K.; Kahyo, T.; Shinmura, K.; et al. Accumulated phosphatidylcholine (16:0/16:1) in human colorectal cancer; possible involvement of LPCAT4. Cancer Sci. 2013, 104, 1295–1302. [Google Scholar] [CrossRef]
- Tata, A.; Zheng, J.; Ginsberg, H.J.; Jaffray, D.A.; Ifa, D.R.; Zarrine-Afsar, A. Contrast Agent Mass Spectrometry Imaging Reveals Tumor Heterogeneity. Anal. Chem. 2015, 87, 7683–7689. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garate, J.; Maimó-Barceló, A.; Bestard-Escalas, J.; Fernández, R.; Pérez-Romero, K.; Martínez, M.A.; Payeras, M.A.; Lopez, D.H.; Fernández, J.A.; Barceló-Coblijn, G. A Drastic Shift in Lipid Adducts in Colon Cancer Detected by MALDI-IMS Exposes Alterations in Specific K+ Channels. Cancers 2021, 13, 1350. https://doi.org/10.3390/cancers13061350
Garate J, Maimó-Barceló A, Bestard-Escalas J, Fernández R, Pérez-Romero K, Martínez MA, Payeras MA, Lopez DH, Fernández JA, Barceló-Coblijn G. A Drastic Shift in Lipid Adducts in Colon Cancer Detected by MALDI-IMS Exposes Alterations in Specific K+ Channels. Cancers. 2021; 13(6):1350. https://doi.org/10.3390/cancers13061350
Chicago/Turabian StyleGarate, Jone, Albert Maimó-Barceló, Joan Bestard-Escalas, Roberto Fernández, Karim Pérez-Romero, Marco A. Martínez, Mª Antònia Payeras, Daniel H. Lopez, José Andrés Fernández, and Gwendolyn Barceló-Coblijn. 2021. "A Drastic Shift in Lipid Adducts in Colon Cancer Detected by MALDI-IMS Exposes Alterations in Specific K+ Channels" Cancers 13, no. 6: 1350. https://doi.org/10.3390/cancers13061350
APA StyleGarate, J., Maimó-Barceló, A., Bestard-Escalas, J., Fernández, R., Pérez-Romero, K., Martínez, M. A., Payeras, M. A., Lopez, D. H., Fernández, J. A., & Barceló-Coblijn, G. (2021). A Drastic Shift in Lipid Adducts in Colon Cancer Detected by MALDI-IMS Exposes Alterations in Specific K+ Channels. Cancers, 13(6), 1350. https://doi.org/10.3390/cancers13061350







