NF2 and Canonical Hippo-YAP Pathway Define Distinct Tumor Subsets Characterized by Different Immune Deficiency and Treatment Implications in Human Pleural Mesothelioma
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
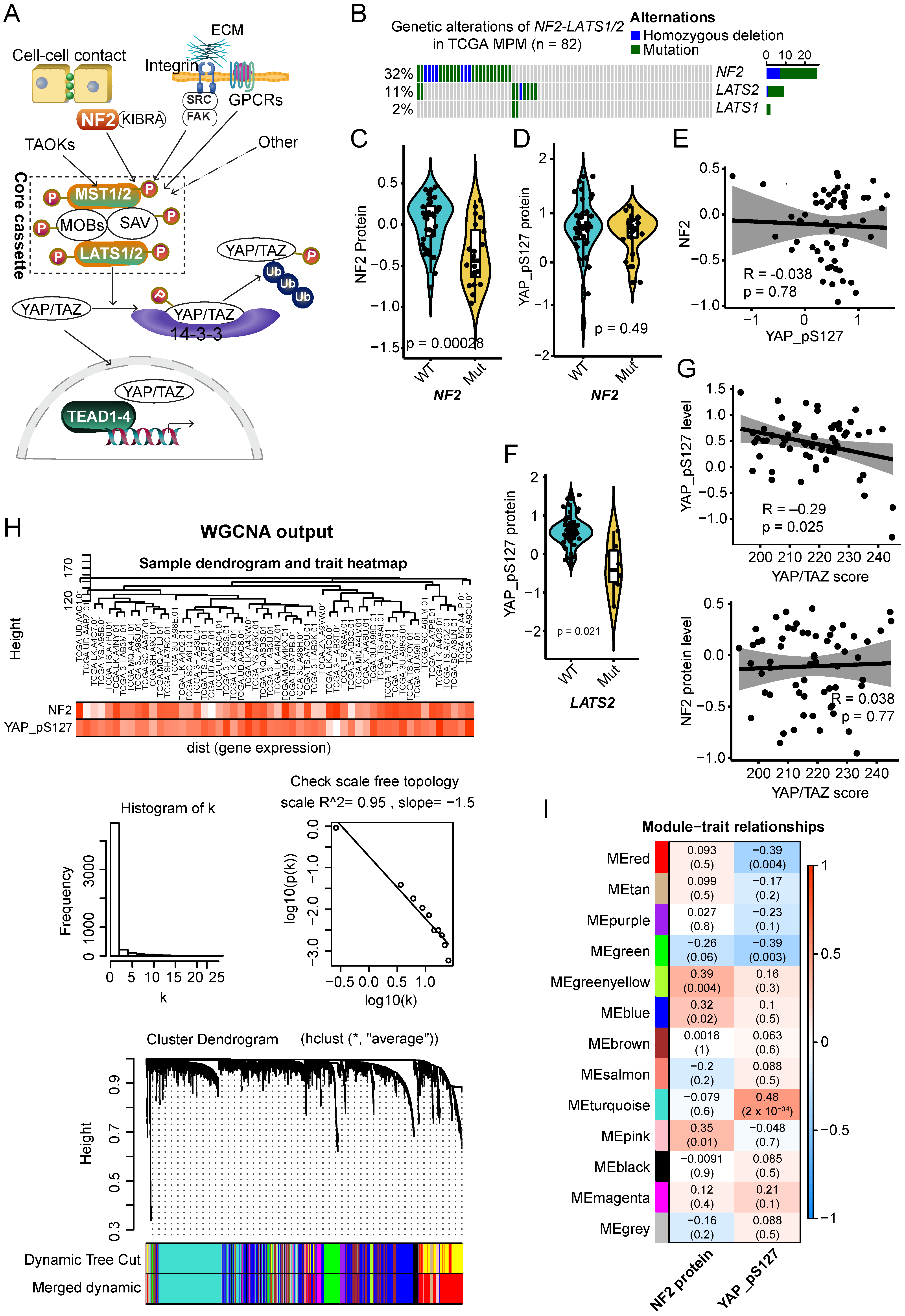
2.1. Molecular Clusters Correlated with NF2 and p-YAP Protein Levels in Clinical MPM Samples
2.2. NF2 Loss-of-Function Is Characterized by a Deficient B-Cell Receptor (BCR) Signaling Pathway
2.3. MPM Tumors with Lower p-YAP Level Display an Exhausted T-Cell-Mediated Immune Phenotype
2.4. Dysregulation of NF2 and Hippo-YAP Exhibits Different Infiltrative Immune Signatures in MPM
2.5. NF2 and Hippo-YAP Engage Different Protein Interactors and Are Differentially Associated with Patient Prognosis
3. Discussion
3.1. A Pleiotropic Role of NF2
3.2. Hippo Pathway Dysregulation
3.3. Compromised Tumor Immunity in MPM
3.4. Combined Targeted Therapy and Immunotherapy for an MPM Subset
3.5. Study Limitations
4. Materials and Methods
4.1. WGCNA and Function Enrichment Analyses
4.2. Cell Culture and Cell Viability Assay
4.3. Immunoblots
4.4. Public Databases
4.5. Gene Signature Scores
4.5.1. YAP-TAZ Target Gene Signature Score
4.5.2. Hub Genes-Based Signature Score
4.6. Survival Analysis
4.7. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, H.; Xu, D.; Schmid, R.A.; Peng, R.-W. Biomarker-guided targeted and immunotherapies in malignant pleural mesothelioma. Ther. Adv. Med Oncol. 2020, 12, 1758835920971421. [Google Scholar] [CrossRef]
- Xu, D.; Yang, H.; Schmid, R.A.; Peng, R.-W. Therapeutic Landscape of Malignant Pleural Mesothelioma: Collateral Vulnerabilities and Evolutionary Dependencies in the Spotlight. Front. Oncol. 2020, 10, 579464. [Google Scholar] [CrossRef] [PubMed]
- Bueno, R.; Stawiski, E.W.; Goldstein, L.D.; Durinck, S.; De Rienzo, A.; Modrusan, Z.; Gnad, F.; Nguyen, T.T.; Jaiswal, B.S.; Chirieac, L.R.; et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat. Genet. 2016, 48, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Hmeljak, J.; Sanchez-Vega, F.; Hoadley, K.A.; Shih, J.; Stewart, C.; Heiman, D.I.; Tarpey, P.; Danilova, L.; Drill, E.; Gibb, E.A.; et al. Integrative Molecular Characterization of Malignant Pleural Mesothelioma. Cancer Discov. 2018, 8, 1548–1565. [Google Scholar] [CrossRef]
- Guo, G.; Chmielecki, J.; Goparaju, C.; Heguy, A.; Dolgalev, I.; Carbone, M.; Seepo, S.; Meyerson, M.; Pass, H.I. Whole-Exome Sequencing Reveals Frequent Genetic Alterations in BAP1, NF2, CDKN2A, and CUL1 in Malignant Pleural Mesothelioma. Cancer Res. 2014, 75, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Harvey, K.F.; Zhang, X.; Thomas, D.M. The Hippo pathway and human cancer. Nat. Rev. Cancer 2013, 13, 246–257. [Google Scholar] [CrossRef] [PubMed]
- Totaro, A.; Panciera, T.; Piccolo, S. YAP/TAZ upstream signals and downstream responses. Nat. Cell Biol. 2018, 20, 888–899. [Google Scholar] [CrossRef]
- Yan, F.; Qian, M.; He, Q.; Zhu, H.; Yang, B. The posttranslational modifications of Hippo-YAP pathway in cancer. Biochim. Biophys. Acta (BBA) Gen. Subj. 2020, 1864, 129397. [Google Scholar] [CrossRef]
- Yang, H.; Xu, D.; Yang, Z.; Yao, F.; Zhao, H.; Schmid, R.A.; Peng, R.-W. Systematic Analysis of Aberrant Biochemical Networks and Potential Drug Vulnerabilities Induced by Tumor Suppressor Loss in Malignant Pleural Mesothelioma. Cancers 2020, 12, 2310. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 1–13. [Google Scholar] [CrossRef]
- Sato, T.; Sekido, Y. NF2/Merlin Inactivation and Potential Therapeutic Targets in Mesothelioma. Int. J. Mol. Sci. 2018, 19, 988. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Karas, P.J.; Hadley, C.C.; Bayley, V.J.C.; Khan, A.B.; Jalali, A.; Sweeney, A.D.; Klisch, T.J.; Patel, A.J. The Role of Merlin/NF2 Loss in Meningioma Biology. Cancers 2019, 11, 1633. [Google Scholar] [CrossRef]
- Petrilli, A.M.; Fernández-Valle, C. Role of Merlin/NF2 inactivation in tumor biology. Oncogene 2016, 35, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Thurneysen, C.; Opitz, I.; Kurtz, S.; Weder, W.; Stahel, R.A.; Felley-Bosco, E. Functional inactivation of NF2/merlin in human mesothelioma. Lung Cancer 2009, 64, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, X.; Maglic, D.; Dill, M.T.; Mojumdar, K.; Ng, P.K.-S.; Jeong, K.J.; Tsang, Y.H.; Moreno, D.; Bhavana, V.H.; et al. Comprehensive Molecular Characterization of the Hippo Signaling Pathway in Cancer. Cell Rep. 2018, 25, 1304–1317.e5. [Google Scholar] [CrossRef] [PubMed]
- Sekido, Y. Inactivation of Merlin in malignant mesothelioma cells and the Hippo signaling cascade dysregulation. Pathol. Int. 2011, 61, 331–344. [Google Scholar] [CrossRef]
- He, M.; Zhou, Z.; Shah, A.A.; Hong, Y.; Chen, Q.; Wan, Y. New insights into posttranslational modifications of Hippo pathway in carcinogenesis and therapeutics. Cell Div. 2016, 11, 1–10. [Google Scholar] [CrossRef]
- Gallant, S.; Gilkeson, G. ETS transcription factors and regulation of immunity. Arch. Immunol. Ther. Exp. 2006, 54, 149–163. [Google Scholar] [CrossRef]
- Zhao, C.; Inoue, J.; Imoto, I.; Otsuki, T.; Iida, S.; Ueda, R.; Inazawa, J. POU2AF1, an amplification target at 11q23, promotes growth of multiple myeloma cells by directly regulating expression of a B-cell maturation factor, TNFRSF17. Oncogene 2007, 27, 63–75. [Google Scholar] [CrossRef]
- Gregorc, V.; Gaafar, R.M.; Favaretto, A.; Grossi, F.; Jassem, J.; Polychronis, A.; Bidoli, P.; Tiseo, M.; Shah, R.; Taylor, P.; et al. NGR-hTNF in combination with best investigator choice in previously treated malignant pleural mesothelioma (NGR015): A randomised, double-blind, placebo-controlled phase 3 trial. Lancet Oncol. 2018, 19, 799–811. [Google Scholar] [CrossRef]
- Zalcman, G.; Mazieres, J.; Margery, J.; Greillier, L.; Audigier-Valette, C.; Moro-Sibilot, D.; Molinier, O.; Corre, R.; Monnet, I.; Gounant, V.; et al. Bevacizumab for newly diagnosed pleural mesothelioma in the Mesothelioma Avastin Cisplatin Pemetrexed Study (MAPS): A randomised, controlled, open-label, phase 3 trial. Lancet 2016, 387, 1405–1414. [Google Scholar] [CrossRef]
- Pavlasova, G.; Mraz, M. The regulation and function of CD20: An “enigma” of B-cell biology and targeted therapy. Haematology 2020, 105, 1494–1506. [Google Scholar] [CrossRef]
- Tedder, T.F.; Boyd, A.W.; Freedman, A.S.; Nadler, L.M.; Schlossman, S.F. The B cell surface molecule B1 is functionally linked with B cell activation and differentiation. J. Immunol. 1985, 135, 973–979. [Google Scholar]
- Helmink, B.A.; Reddy, S.M.; Gao, J.; Zhang, S.; Basar, R.; Thakur, R.; Yizhak, K.; Sade-Feldman, M.; Blando, J.; Han, G.; et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature 2020, 577, 549–555. [Google Scholar] [CrossRef]
- Petitprez, F.; De Reyniès, A.; Keung, E.Z.; Chen, T.W.-W.; Sun, C.-M.; Calderaro, J.; Jeng, Y.-M.; Hsiao, L.-P.; Lacroix, L.; Bougoüin, A.; et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature 2020, 577, 556–560. [Google Scholar] [CrossRef] [PubMed]
- Beltrami, S.; Kim, R.; Gordon, J. Neurofibromatosis type 2 protein, NF2: An uncoventional cell cycle regulator. Anticancer. Res. 2013, 33, 1–11. [Google Scholar] [PubMed]
- Curto, M.; Cole, B.K.; Lallemand, D.; Liu, C.-H.; McClatchey, A.I. Contact-dependent inhibition of EGFR signaling by Nf2/Merlin. J. Cell Biol. 2007, 177, 893–903. [Google Scholar] [CrossRef] [PubMed]
- Cole, B.K.; Curto, M.; Chan, A.W.; McClatchey, A.I. Localization to the Cortical Cytoskeleton Is Necessary for Nf2/Merlin-Dependent Epidermal Growth Factor Receptor Silencing. Mol. Cell. Biol. 2007, 28, 1274–1284. [Google Scholar] [CrossRef]
- Lallemand, D.; Manent, J.; Couvelard, A.; Watilliaux, A.; Siena, M.; Chareyre, F.; Lampin, A.; Niwa-Kawakita, M.; Kalamarides, M.; Giovannini, M. Merlin regulates transmembrane receptor accumulation and signaling at the plasma membrane in primary mouse Schwann cells and in human schwannomas. Oncogene 2008, 28, 854–865. [Google Scholar] [CrossRef]
- Hsu, P.-C.; Jablons, D.M.; Yang, C.-T.; You, L. Epidermal Growth Factor Receptor (EGFR) Pathway, Yes-Associated Protein (YAP) and the Regulation of Programmed Death-Ligand 1 (PD-L1) in Non-Small Cell Lung Cancer (NSCLC). Int. J. Mol. Sci. 2019, 20, 3821. [Google Scholar] [CrossRef]
- Nilsson, M.B.; Sun, H.; Robichaux, J.; Pfeifer, M.; McDermott, U.; Travers, J.; Diao, L.; Xi, Y.; Tong, P.; Shen, L.; et al. A YAP/FOXM1 axis mediates EMT-associated EGFR inhibitor resistance and increased expression of spindle assembly checkpoint components. Sci. Transl. Med. 2020, 12, eaaz4589. [Google Scholar] [CrossRef] [PubMed]
- Ehmer, U.; Sage, J. Control of Proliferation and Cancer Growth by the Hippo Signaling Pathway. Mol. Cancer Res. 2016, 14, 127–140. [Google Scholar] [CrossRef] [PubMed]
- Crawford, J.J.; Bronner, S.M.; Zbieg, J.R. Hippo pathway inhibition by blocking the YAP/TAZ–TEAD interface: A patent review. Expert Opin. Ther. Patents 2018, 28, 867–873. [Google Scholar] [CrossRef] [PubMed]
- Marsola, A.P.Z.C.; Simões, B.P.; Palma, L.C.; Berzoti-Coelho, M.G.; Burin, S.M.; De Castro, F.A. Expression of Hippo signaling pathway and Aurora kinase genes in chronic myeloid leukemia. Med. Oncol. 2018, 35, 26. [Google Scholar] [CrossRef]
- Wang, P.; Gong, Y.; Guo, T.; Li, M.; Fang, L.; Yin, S.; Kamran, M.; Liu, Y.; Xu, J.; Xu, L.; et al. Activation of Aurora A kinase increases YAP stability via blockage of autophagy. Cell Death Dis. 2019, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Harjunpää, H.; Guillerey, C. TIGIT as an emerging immune checkpoint. Clin. Exp. Immunol. 2020, 200, 108–119. [Google Scholar] [CrossRef]
- Samstein, R.M.; Lee, C.-H.; Shoushtari, A.N.; Hellmann, M.D.; Shen, R.; Janjigian, Y.Y.; Barron, D.A.; Zehir, A.; Jordan, E.J.; Omuro, A.; et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019, 51, 202–206. [Google Scholar] [CrossRef]
- Miyanaga, A.; Masuda, M.; Tsuta, K.; Kawasaki, K.; Nakamura, Y.; Sakuma, T.; Asamura, H.; Gemma, A.; Yamada, T. Hippo Pathway Gene Mutations in Malignant Mesothelioma: Revealed by RNA and Targeted Exon Sequencing. J. Thorac. Oncol. 2015, 10, 844–851. [Google Scholar] [CrossRef]
- Tu, M.M.; Lee, F.Y.F.; Jones, R.T.; Kimball, A.K.; Saravia, E.; Graziano, R.F.; Coleman, B.; Menard, K.; Yan, J.; Michaud, E.; et al. Targeting DDR2 enhances tumor response to anti–PD-1 immunotherapy. Sci. Adv. 2019, 5, eaav2437. [Google Scholar] [CrossRef]
- Krusch, M.; Salih, H.R. Effects of BCR-ABL inhibitors on anti-tumor immunity. Curr. Med. Chem. 2011, 18, 5174–5184. [Google Scholar] [CrossRef]
- Ke, Y.; Reddel, R.R.; Gerwin, B.I.; Reddel, H.K.; Somers, A.N.; McMenamin, M.G.; LaVeck, M.A.; Stahel, R.A.; Lechner, J.F.; Harris, C.C. Establishment of a human in vitro mesothelial cell model system for investigating mechanisms of asbestos-induced mesothelioma. Am. J. Pathol. 1989, 134, 979–991. [Google Scholar]
- Ray, F.; Meyne, J.; Kraemer, P.M. SV40 T antigen induced chromosomal changes reflect a process that is both clastogenic and aneuploidogenic and is ongoing throughout neoplastic progression of human fibroblasts. Mutat. Res. Mol. Mech. Mutagen. 1992, 284, 265–273. [Google Scholar] [CrossRef]
- Csibi, A.; Blenis, J. Hippo–YAP and mTOR pathways collaborate to regulate organ size. Nat. Cell Biol. 2012, 14, 1244–1245. [Google Scholar] [CrossRef]
- O’Donnell, J.S.; Massi, D.; Teng, M.W.; Mandala, M. PI3K-AKT-mTOR inhibition in cancer immunotherapy, redux. Semin. Cancer Biol. 2018, 48, 91–103. [Google Scholar] [CrossRef]
- Klebanoff, C.A.; Crompton, J.G.; Leonardi, A.J.; Yamamoto, T.N.; Chandran, S.S.; Eil, R.L.; Sukumar, M.; Vodnala, S.K.; Madhusudhanan, S.; Ji, Y.; et al. Inhibition of AKT signaling uncouples T cell differentiation from expansion for receptor-engineered adoptive immunotherapy. JCI Insight 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef] [PubMed]
- Jackaman, C.; Cornwall, S.; Graham, P.T.; Nelson, D.J. CD40-activated B cells contribute to mesothelioma tumor regression. Immunol. Cell Biol. 2010, 89, 255–267. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.; Wolf, D.; Bortone, D.S.; Ouyang, T.-H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e14. [Google Scholar] [CrossRef]
- Bott, M.; Brevet, M.; Taylor, B.S.; Shimizu, S.; Ito, T.; Wang, L.; Creaney, J.; Lake, R.A.; Zakowski, M.F.; Reva, B.; et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat. Genet. 2011, 43, 668–672. [Google Scholar] [CrossRef]
- Shen, H.; Yang, E.S.-H.; Conry, M.; Fiveash, J.; Contreras, C.; Bonner, J.A.; Shi, L.Z. Predictive biomarkers for immune checkpoint blockade and opportunities for combination therapies. Genes Dis. 2019, 6, 232–246. [Google Scholar] [CrossRef] [PubMed]
- Yarchoan, M.; Albacker, L.A.; Hopkins, A.C.; Montesion, M.; Murugesan, K.; Vithayathil, T.T.; Zaidi, N.; Azad, N.S.; Laheru, D.A.; Frampton, G.M.; et al. PD-L1 expression and tumor mutational burden are independent biomarkers in most cancers. JCI Insight 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Lavado, A.; He, Y.; Paré, J.; Neale, G.; Olson, E.N.; Giovannini, M.; Cao, X. Tumor suppressor Nf2 limits expansion of the neural progenitor pool by inhibiting Yap/Taz transcriptional coactivators. Development 2013, 140, 3323–3334. [Google Scholar] [CrossRef]
- Matsuda, T.; Zhai, P.; Sciarretta, S.; Zhang, Y.; Jeong, J.I.; Ikeda, S.; Park, J.; Hsu, C.-P.; Tian, B.; Pan, D.; et al. NF2 Activates Hippo Signaling and Promotes Ischemia/Reperfusion Injury in the Heart. Circ. Res. 2016, 119, 596–606. [Google Scholar] [CrossRef]
- Reginensi, A.; Enderle, L.; Gregorieff, A.; Johnson, R.L.; Wrana, J.L.; McNeill, H. A critical role for NF2 and the Hippo pathway in branching morphogenesis. Nat. Commun. 2016, 7, 12309. [Google Scholar] [CrossRef]
- Hong, A.W.; Meng, Z.; Plouffe, S.W.; Lin, Z.; Zhang, M.; Guan, K.-L. Critical roles of phosphoinositides and NF2 in Hippo pathway regulation. Genes Dev. 2020, 34, 511–525. [Google Scholar] [CrossRef]
- Zanconato, F.; Cordenonsi, M.; Piccolo, S. YAP/TAZ at the Roots of Cancer. Cancer Cell 2016, 29, 783–803. [Google Scholar] [CrossRef]
- Garland, L.L.; Rankin, C.; Gandara, D.R.; Rivkin, S.E.; Scott, K.M.; Nagle, R.B.; Klein-Szanto, A.J.; Testa, J.R.; Altomare, D.A.; Borden, E.C. Phase II Study of Erlotinib in Patients With Malignant Pleural Mesothelioma: A Southwest Oncology Group Study. J. Clin. Oncol. 2007, 25, 2406–2413. [Google Scholar] [CrossRef]
- Govindan, R.; Kratzke, R.A.; Herndon, J.E.; Niehans, G.A.; Vollmer, R.; Watson, D.; Green, M.R.; Kindler, H.L. Gefitinib in Patients with Malignant Mesothelioma: A Phase II Study by the Cancer and Leukemia Group B. Clin. Cancer Res. 2005, 11, 2300–2304. [Google Scholar] [CrossRef]
- Mezzapelle, R.; Miglio, U.; Rena, O.; Paganotti, A.; Allegrini, S.; Antona, J.; Molinari, F.; Frattini, M.; Monga, G.; Alabiso, O.; et al. Mutation analysis of the EGFR gene and downstream signalling pathway in histologic samples of malignant pleural mesothelioma. Br. J. Cancer 2013, 108, 1743–1749. [Google Scholar] [CrossRef]
- Zhang, W.-Q.; Dai, Y.-Y.; Hsu, P.-C.; Wang, H.; Cheng, L.; Yang, Y.-L.; Wang, Y.-C.; Xu, Z.-D.; Liu, S.; Chan, G.; et al. Targeting YAP in malignant pleural mesothelioma. J. Cell. Mol. Med. 2017, 21, 2663–2676. [Google Scholar] [CrossRef]
- Murakami, H.; Mizuno, T.; Taniguchi, T.; Fujii, M.; Ishiguro, F.; Fukui, T.; Akatsuka, S.; Horio, Y.; Hida, T.; Kondo, Y.; et al. LATS2 Is a Tumor Suppressor Gene of Malignant Mesothelioma. Cancer Res. 2011, 71, 873–883. [Google Scholar] [CrossRef] [PubMed]
- Moroishi, T.; Hayashi, T.; Pan, W.-W.; Fujita, Y.; Holt, M.V.; Qin, J.; Carson, D.A.; Guan, K.-L. The Hippo Pathway Kinases LATS1/2 Suppress Cancer Immunity. Cell 2016, 167, 1525–1539.e17. [Google Scholar] [CrossRef]
- Taha, Z.; Van Rensburg, H.J.J.; Yang, X. The Hippo Pathway: Immunity and Cancer. Cancers 2018, 10, 94. [Google Scholar] [CrossRef]
- Hong, L.; Li, X.; Zhou, D.; Geng, J.; Chen, L. Role of Hippo signaling in regulating immunity. Cell. Mol. Immunol. 2018, 15, 1003–1009. [Google Scholar] [CrossRef] [PubMed]
- Van Rensburg, H.J.J.; Yang, X. The Hippo pathway and cancer immunity: Friend or foe? Oncoscience 2018, 5, 49–50. [Google Scholar] [CrossRef]
- Tranchant, R.; Quetel, L.; Tallet, A.; Meiller, C.; Renier, A.; De Koning, L.; De Reynies, A.; Le Pimpec-Barthes, F.; Zucman-Rossi, J.; Jaurand, M.-C.; et al. Co-occurring Mutations of Tumor Suppressor Genes, LATS2 and NF2, in Malignant Pleural Mesothelioma. Clin. Cancer Res. 2016, 23, 3191–3202. [Google Scholar] [CrossRef]
- Disselhorst, M.J.; Quispel-Janssen, J.; Lalezari, F.; Monkhorst, K.; de Vries, J.F.; van der Noort, V.; Harms, E.; Burgers, S.; Baas, P. Ipilimumab and nivolumab in the treatment of recurrent malignant pleural mesothelioma (INITIATE): Results of a prospective, single-arm, phase 2 trial. Lancet Respir. Med. 2019, 7, 260–270. [Google Scholar] [CrossRef]
- Hong, L.; Negrao, M.V.; Dibaj, S.S.; Chen, R.; Reuben, A.; Bohac, J.M.; Liu, X.; Skoulidis, F.; Gay, C.M.; Cascone, T.; et al. Programmed Death-Ligand 1 Heterogeneity and Its Impact on Benefit From Immune Checkpoint Inhibitors in NSCLC. J. Thorac. Oncol. 2020, 15, 1449–1459. [Google Scholar] [CrossRef]
- Maio, M.; Scherpereel, A.; Calabrò, L.; Aerts, J.; Perez, S.C.; Bearz, A.; Nackaerts, K.; Fennell, D.A.; Kowalski, D.; Tsao, A.S.; et al. Tremelimumab as second-line or third-line treatment in relapsed malignant mesothelioma (DETERMINE): A multicentre, international, randomised, double-blind, placebo-controlled phase 2b trial. Lancet Oncol. 2017, 18, 1261–1273. [Google Scholar] [CrossRef]
- Owonikoko, T.K.; Dwivedi, B.; Chen, Z.; Zhang, C.; Barwick, B.; Ernani, V.; Zhang, G.; Gilbert-Ross, M.; Carlisle, J.; Khuri, F.R.; et al. YAP1 Expression in Small Cell Lung Cancer Defines a Distinct Subtype with T-cell Inflamed Phenotype. J. Thorac. Oncol. 2021, 16, 464–476. [Google Scholar] [CrossRef]
- Pao, W.; Ooi, C.-H.; Birzele, F.; Ruefli-Brasse, A.; Cannarile, M.A.; Reis, B.; Scharf, S.H.; Schubert, D.A.; Hatje, K.; Pelletier, N.; et al. Tissue-Specific Immunoregulation: A Call for Better Understanding of the “Immunostat” in the Context of Cancer. Cancer Discov. 2018, 8, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Hall, S.R.R.; Sun, B.; Yao, F. Comment on “Heterogeneity in PD-L1 expression in malignant peritoneal mesothelioma with systemic or intraperitoneal chemotherapy”. Br. J. Cancer 2020, 1–2. [Google Scholar] [CrossRef]
- Yang, H.; Yao, F.; Davis, P.F.; Tan, S.T.; Hall, S.R.R. CD73, Tumor Plasticity and Immune Evasion in Solid Cancers. Cancers 2021, 13, 177. [Google Scholar] [CrossRef] [PubMed]
- Van Rensburg, H.J.J.; Azad, T.; Ling, M.; Hao, Y.; Snetsinger, B.; Khanal, P.; Minassian, L.M.; Graham, C.H.; Rauh, M.J.; Yang, X. The Hippo Pathway Component TAZ Promotes Immune Evasion in Human Cancer through PD-L1. Cancer Res. 2018, 78, 1457–1470. [Google Scholar] [CrossRef] [PubMed]
- DeFalco, J.; Harbell, M.; Manning-Bog, A.; Baia, G.; Scholz, A.; Millare, B.; Sumi, M.; Zhang, D.; Chu, F.; Dowd, C.; et al. Non-progressing cancer patients have persistent B cell responses expressing shared antibody paratopes that target public tumor antigens. Clin. Immunol. 2018, 187, 37–45. [Google Scholar] [CrossRef]
- Scherpereel, A.; Mazieres, J.; Greillier, L.; Lantuejoul, S.; Dô, P.; Bylicki, O.; Monnet, I.; Corre, R.; Audigier-Valette, C.; Locatelli-Sanchez, M.; et al. Nivolumab or nivolumab plus ipilimumab in patients with relapsed malignant pleural mesothelioma (IFCT-1501 MAPS2): A multicentre, open-label, randomised, non-comparative, phase 2 trial. Lancet Oncol. 2019, 20, 239–253. [Google Scholar] [CrossRef]
- Okada, M.; Kijima, T.; Aoe, K.; Kato, T.; Fujimoto, N.; Nakagawa, K.; Takeda, Y.; Hida, T.; Kanai, K.; Imamura, F.; et al. Clinical Efficacy and Safety of Nivolumab: Results of a Multicenter, Open-label, Single-arm, Japanese Phase II study in Malignant Pleural Mesothelioma (MERIT). Clin. Cancer Res. 2019, 25, 5485–5492. [Google Scholar] [CrossRef]
- Gotwals, P.; Cameron, S.; Cipolletta, D.; Cremasco, V.; Crystal, A.; Hewes, B.; Mueller, B.; Quaratino, S.; Sabatos-Peyton, C.; Petruzzelli, L.; et al. Prospects for combining targeted and conventional cancer therapy with immunotherapy. Nat. Rev. Cancer 2017, 17, 286–301. [Google Scholar] [CrossRef]
- Patel, S.A.; Minn, A.J. Combination Cancer Therapy with Immune Checkpoint Blockade: Mechanisms and Strategies. Immunity 2018, 48, 417–433. [Google Scholar] [CrossRef]
- Esteva, F.J.; Hubbard-Lucey, V.M.; Tang, J.; Pusztai, L. Immunotherapy and targeted therapy combinations in metastatic breast cancer. Lancet Oncol. 2019, 20, e175–e186. [Google Scholar] [CrossRef]
- Chen, R.; Lee, W.-C.; Fujimoto, J.; Li, J.; Hu, X.; Mehran, R.; Rice, D.; Swisher, S.G.; Sepesi, B.; Tran, H.T.; et al. Evolution of Genomic and T-cell Repertoire Heterogeneity of Malignant Pleural Mesothelioma Under Dasatinib Treatment. Clin. Cancer Res. 2020, 26, 5477–5486. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Yang, H.; Berezowska, S.; Gao, Y.; Liang, S.-Q.; Marti, T.M.; Hall, S.R.R.; Dorn, P.; Kocher, G.J.; Schmid, R.A.; et al. Endoplasmic Reticulum Stress Signaling as a Therapeutic Target in Malignant Pleural Mesothelioma. Cancers 2019, 11, 1502. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Liang, S.-Q.; Xu, D.; Yang, Z.; Marti, T.M.; Gao, Y.; Kocher, G.J.; Zhao, H.; Schmid, R.A.; Peng, R.-W. HSP90/AXL/eIF4E-regulated unfolded protein response as an acquired vulnerability in drug-resistant KRAS-mutant lung cancer. Oncogenesis 2019, 8, 1–13. [Google Scholar] [CrossRef]
- Li, J.; Lu, Y.; Akbani, R.; Ju, Z.; Roebuck, P.L.; Liu, W.; Yang, J.-Y.; Broom, B.M.; Verhaak, R.G.W.; Kane, D.W.; et al. TCPA: A resource for cancer functional proteomics data. Nat. Methods 2013, 10, 1046–1047. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Rees, M.G.; Seashore-Ludlow, B.; Cheah, J.H.; Adams, D.J.; Price, E.V.; Gill, S.; Javaid, S.; Coletti, M.E.; Jones, V.L.; Bodycombe, N.E.; et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat. Chem. Biol. 2016, 12, 109–116. [Google Scholar] [CrossRef]
- Yang, H.; Zhao, L.; Gao, Y.; Yao, F.; Marti, T.M.; Schmid, R.A.; Peng, R.-W. Pharmacotranscriptomic Analysis Reveals Novel Drugs and Gene Networks Regulating Ferroptosis in Cancer. Cancers 2020, 12, 3273. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, H.; Hall, S.R.R.; Sun, B.; Zhao, L.; Gao, Y.; Schmid, R.A.; Tan, S.T.; Peng, R.-W.; Yao, F. NF2 and Canonical Hippo-YAP Pathway Define Distinct Tumor Subsets Characterized by Different Immune Deficiency and Treatment Implications in Human Pleural Mesothelioma. Cancers 2021, 13, 1561. https://doi.org/10.3390/cancers13071561
Yang H, Hall SRR, Sun B, Zhao L, Gao Y, Schmid RA, Tan ST, Peng R-W, Yao F. NF2 and Canonical Hippo-YAP Pathway Define Distinct Tumor Subsets Characterized by Different Immune Deficiency and Treatment Implications in Human Pleural Mesothelioma. Cancers. 2021; 13(7):1561. https://doi.org/10.3390/cancers13071561
Chicago/Turabian StyleYang, Haitang, Sean R. R. Hall, Beibei Sun, Liang Zhao, Yanyun Gao, Ralph A. Schmid, Swee T. Tan, Ren-Wang Peng, and Feng Yao. 2021. "NF2 and Canonical Hippo-YAP Pathway Define Distinct Tumor Subsets Characterized by Different Immune Deficiency and Treatment Implications in Human Pleural Mesothelioma" Cancers 13, no. 7: 1561. https://doi.org/10.3390/cancers13071561
APA StyleYang, H., Hall, S. R. R., Sun, B., Zhao, L., Gao, Y., Schmid, R. A., Tan, S. T., Peng, R.-W., & Yao, F. (2021). NF2 and Canonical Hippo-YAP Pathway Define Distinct Tumor Subsets Characterized by Different Immune Deficiency and Treatment Implications in Human Pleural Mesothelioma. Cancers, 13(7), 1561. https://doi.org/10.3390/cancers13071561






